Abstract
Background
Familial hypercholesterolaemia (FH) is the most common inherited metabolic disease with an autosomal dominant mode of inheritance. It is characterised by raised serum levels of total cholesterol (TC) and low-density lipoprotein cholesterol (LDL-c), leading to premature coronary artery disease. Children with FH are subjected to early and enhanced atherosclerosis, leading to greater risk of coronary events, including premature coronary artery disease. To the best of our knowledge, this is the first report of a pair of monochorionic diamniotic identical twins with a diagnosis of heterozygous FH, resulting from mutations in both LDLR and ABCG8 genes.
Case presentation
This is a rare case of a pair of 8-year-old monochorionic diamniotic identical twin, who on family cascade screening were diagnosed as definite FH, according to the Dutch Lipid Clinic Criteria (DLCC) with a score of 10. There were no lipid stigmata noted. Baseline lipid profiles revealed severe hypercholesterolaemia, (TC = 10.5 mmol/L, 10.6 mmol/L; LDL-c = 8.8 mmol/L, 8.6 mmol/L respectively). Their father is the index case who initially presented with premature CAD, and subsequently diagnosed as FH. Family cascade screening identified clinical FH in other family members including their paternal grandfather who also had premature CAD, and another elder brother, aged 10 years. Genetic analysis by targeted next-generation sequencing using MiSeq platform (Illumina) was performed to detect mutations in LDLR, APOB100, PCSK9, ABCG5, ABCG8, APOE and LDLRAP1 genes. Results revealed that the twin, their elder brother, father and grandfather are heterozygous for a missense mutation (c.530C > T) in LDLR that was previously reported as a pathogenic mutation. In addition, the twin has heterozygous ABCG8 gene mutation (c.55G > C). Their eldest brother aged 12 years and their mother both had normal lipid profiles with absence of LDLR gene mutation.
Conclusion
A rare case of Asian monochorionic diamniotic identical twin, with clinically diagnosed and molecularly confirmed heterozygous FH, due to LDLR and ABCG8 gene mutations have been reported. Childhood FH may not present with the classical physical manifestations including the pathognomonic lipid stigmata as in adults. Therefore, childhood FH can be diagnosed early using a combination of clinical criteria and molecular analyses.
Keywords: Familial hypercholesterolaemia, LDLR, ABCG8, Premature atherosclerosis, Coronary artery disease
Background
Familial hypercholesterolaemia (FH) is the most common inherited metabolic disease with an autosomal dominant mode of inheritance. It is characterised by raised serum levels of total cholesterol (TC) and low-density lipoprotein cholesterol (LDL-c), leading to premature coronary artery disease (CAD). Homozygous FH (HoFH) patients manifest a more severe clinical phenotype compared to heterozygous FH (HeFH). Globally in the community, the HeFH prevalence is reported to be 1:200–500, but more recent reports showed higher prevalence of 1:100–250 [1–5]. On the other hand, the prevalence of HoFH is much lower [6], although a greater prevalence is reported among certain populations, apparently due to founder effects [7].
Familial Hypercholesterolaemia is mainly caused by mutations in the low-density lipoprotein receptor (LDLR) gene that results in reduced uptake and clearance of LDL-c [8]. Less common causes of FH are mutations in the genes encoding apolipoprotein B-100 (APO B-100) and proprotein convertase subtilisin/kexin 9 (PCSK9)9. Rarely the autosomal recessive hypercholesterolemia (ARH) LDL receptor adaptor protein gene (LDLRAP1) can also result in a similar HoFH phenotype [9]. Over 1200 different types of mutations were described in the LDLR gene [10]. Few variants in APOB-100 gene, affecting the LDL receptor-binding domain of apolipoprotein B were reported leading to a disorder that is called familial defective Apo B (FDB) [5, 11] and almost all appear within a relatively small coding region near p.3527 [12]. More than 30 PCSK9 gain-of-function (GOF) mutations have been reported to be the cause of FH since the discovery of PCSK9 gene in 2003 [11, 13].
Sitosterolemia is an extremely rare autosomal recessive disorder of sterol metabolism caused by mutations in one of two genes ATP-binding cassette (ABC) sub-family G members 5 and 8 (ABCG5 and ABCG8) and it is characterised by increased absorption and decreased biliary excretion of plant sterols and cholesterol, resulting in elevated serum levels of plant sterols like sitosterol and campesterol [14, 15]. Subjects suffering from sitosterolemia also primarily develop xanthomas and even premature coronary atherosclerosis [16].
Genetic testing is helpful for confirmation of FH diagnosis. However, the causative mutation may remain undetected in certain patients, even after thorough screening for mutation using the currently available advance techniques [17]. Many mutations were ruled out as variants with uncertain significance probably due to lack of functional evidence, family co-segregation data or benign outcome from in silico analysis. However, there are evidences that accumulation of multiple small-effect common FH variants, including some autosomal recessive genes such as ABCG5/8, may still cause significant elevation of LDL-c, dubbed as polygenic FH [18, 19]. Lack of genetic finding among the suspected FH patients may also be caused by elevated LDL-c resulted from lifestyle factors and secondary causes (e.g. hypothyroidism or nephrotic syndrome), or a combination of these factors that might explain the clinical phenotype in terms of hypercholesterolemia or CAD with the absence of gene mutations [20].
The rapid advances in next generation sequencing (NGS)-based techniques have made them more accessible for diagnostic purposes. Most of the sequencing methods that are established to diagnose hereditary disorders usually concentrate on the exome, which constitutes about 1% of the whole human genome. Additionally, the computer-based prediction tools that are used to detect the pathogenicity of the gene sequence variants are now established and used to estimate the deleterious effect of the various amino acid changes [21].
There are potentially as many as 4.5 million individuals in Europe with HeFH and possibly 35 million around the world, of whom 20–25% of these are children and adolescents [5, 22]. Given the high prevalence of FH, it is estimated that one baby is born with FH every minute [22]. However, FH is very much under diagnosed and undertreated globally [5]. Children with FH have higher risk of early coronary events and death from myocardial infarction due to premature atherosclerosis. To the best of our knowledge, this is the first report of an Asian pair of monochorionic diamniotic identical twin with heterozygous FH caused by mutations in both LDLR and ABCG8 FH candidate genes. Furthermore, to the best of our knowledge, this is the first study which utilised targeted next-generation sequencing (TNGS) technique to identify the causative gene mutations for FH among the Malaysian population.
Case presentation
This is a rare case of a pair of 8-year-old monochorionic diamniotic identical twin, of Malaysian Indian descent, who on family cascade screening were diagnosed as definite FH, according to the Dutch Lipid Clinic Criteria (DLCC) with a score of 10. No lipid stigmata were identified. Baseline lipid profiles revealed severe hypercholesterolaemia, (TC = 10.5 mmol/L, 10.6 mmol/L; LDL-c = 8.8 mmol/L, 8.6 mmol/L respectively) (Table 1). Their father is the index case who initially presented with premature CAD, and subsequently diagnosed as FH. Family cascade screening identified clinical FH in other family members including their paternal grandfather who also had premature CAD, and another elder brother, aged 10 years.
Table 1.
Summary of the biochemical and molecular findings among the family members
| Family member | Clinical FH diagnosis | TC mmol/lL | TG mmol/L | HDL-C mmol/L | LDL-C mmol/L | dbSNP ID | Predicted effect |
|---|---|---|---|---|---|---|---|
| Proband | Definite FH | 10.0 | 1 | 1.5 | 8.6 | LDLR: rs121908026 | Probably Damaging |
| Older brother (Hypercholesterolemic) | Probable FH | 9.6 | 0.6 | 0.3 | 7 | LDLR: rs121908026 | Probably Damaging |
| Grandfather | Definite FH | 9.3 | 1.4 | 1 | 7.7 | LDLR: rs121908026 | Probably Damaging |
| Twin, youngest brother | Definite FH | 10.6 | 0.7 | 1.8 | 8.6 | LDLR: rs121908026 | Probably Damaging |
| ABCG8: rs11887534 | Possibly Damaging | ||||||
| Twin, youngest brother | Definite FH | 10.8 | 0.6 | 1.7 | 8.8 | LDLR: rs121908026 | Probably Damaging |
| ABCG8: rs11887534 | Possibly Damaging | ||||||
| Eldest brother (Normolipaemic) | Possible FH | 5.2 | 0.4 | 2.6 | 2.4 | LDLR: rs2738442 | Benign |
| ABCG8: rs11887534 | Possibly Damaging | ||||||
| Mother | Possible FH | 5.3 | 0..9 | 2 | 2.8 | LDLR: rs2738442 | Benign |
| ABCG8: rs11887534 | Possibly Damaging |
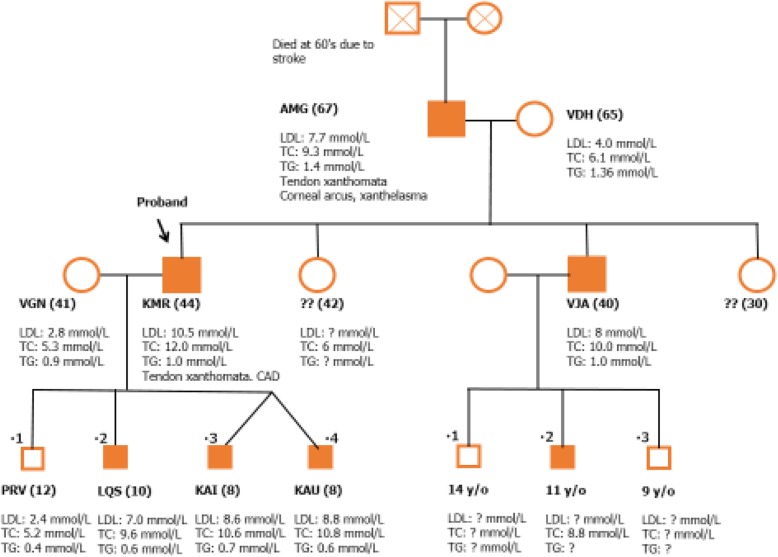
Their liver function test, renal profile and thyroid function test were normal. They have no history of any cardiovascular complications. Screening of the other family members revealed that, their older brother (10-year-old) was also diagnosed as probable FH according to the DLCC criteria with TC of 10.8 mmol/L and LDL-C of 7.0 mmol/L. Both their 43-year-old father (index case) and 63-year-old paternal grandfather were also diagnosed with FH and they had premature CAD with presence of tendon xanthomata and corneal arcus. Their mother and the eldest brother, who is 12 years old were healthy with normal lipid profile. The twin have no history of any cardiovascular events. Both of them were clinically healthy. Advices were given for a low cholesterol diet, appropriate caloric intake and regular physical activity. Pharmacological treatment has not been initiated for the twins and their affected 10-year-old brother due to parental refusal in view of their young age. However, they are monitored closely under our care in a Specialist Lipid and Coronary Risk Prevention Clinic of a teaching hospital. The family tree is illustrated in Fig. 1.
Fig. 1.
Family tree
In view of the strong family history of hypercholesterolemia and premature CAD, family cascade screening and genetic analyses were perfomed for the index case, affected and unaffected family members to detect for the pathogenic FH mutations following informed consent that were collected.
Blood samples were acquired from the proband and the other six family members including, the mother, the twins, two older brothers and the paternal grandfather. DNA was extracted from whole blood by using QIAamp Blood Mini Kit (Qiagen) according to standard manufacturer’s protocol. Extracted DNA was checked using agarose gel electrophoresis, followed by quantitation using Qubit fluorometer with dsDNA HS (High Sensitivity) Assay Kit (Thermo Fisher Scientific).
Targeted next-generation sequencing (TNGS) was implemented for comprehensive genetic analysis for the known and novel mutations in hot spots within exons and exon–intron boundaries of LDLR (39 amplicons), APOB100 (106 amplicons), PCSK9 (32 amplicons), ABCG5 (22 amplicons), ABCG8 (20 amplicons), APOE (8 amplicons) and LDLRAP1 (23 amplicons). Average amplicon length is 242 bp.
Fifty nanogram of genomic DNA per individual were used for library preparation (TruSeq Custom Amplicon, Illumina). Next-generation sequencing kit (MiSeq v3, Illumina) was used according to manufacturers’ protocols to prepare the library and the samples were sequenced on a MiSeq sequencer (MiSeq sequencer, Illumina). Amplification was considered passed the QC if 95% of the nucleotides has ≥20X coverage.
Genomic sequences were mapped to GRCh37 hg19 human reference assembly using TruSeq Amplicon V3.00 proprietary cloud-based software. Variant calling was performed using Somatic Variant Caller (Illumina) and generated into VCF format. BaseSpace Variant Interpreter (Illumina) was used to annotate the variants. Variants with minor allele frequency of ≤0.02 (ExAC browser and 1000 Genome) were considered as mutants. All mutations were checked for pathogenicity by searching in ClinVar. In silico prediction of pathogenicity in exonic mutations were interpreted using Mutation Taster and Polypophen2, while intronic mutations (≤10 bp away from exon) were checked for their effect on splicing by using Human Splicing Finder and NetGene 2.
Pedigree of the twins showed a vertical transmission of the hypercholesterolemia from the father to the twin suggesting an autosomal dominant mode of inheritance. The proband (the father who was clinically diagnosed as definite FH by the DLCC) was identified to have one heterozygous missense mutation in LDLR gene (c.530C > T, p.Ser177Leu) that is located at exon 4 of this gene. This heterozygous mutation was also found in the older son (clinically diagnosed as probable FH by the DLCC) and grandfather (clinically diagnosed as definite FH by the DLCC) (Table 1). The clinically diagnosed younger twin pair siblings (both are clinically diagnosed as definite FH by the DLCC) has the same heterozygous LDLR mutation that is reported in the proband (530C > T, p.Ser177Leu). This missense mutation that was identified in this family was observed to co-segregate with the hypercholesterolaemia and has already been reported as probably damaging by Polyphen2 and deleterious by SIFT. Another missense variant was also reported in this twin pair, c.55G > C (p.Asp19His) which is located in exon 1 of the ABCG8 gene and it was reported to be possibly damaging by Polyphen2 and SIFT. The mother and the other older brother who are normolipidemic were found to have heterozygous intronic variant (c.1060 + 7 T > C) that is located in intron 7 of the LDLR gene and was found to be a benign variant. Additionally, both of them were also found to be carriers for c.55G > C(p.Asp19His) variant in the ABCG8 gene.
There was no nonsense or frameshift mutations that were observed in the LDLR gene sequence. The analysis of the samples showed no other pathogenic variants in the known FH genes (APO B-100 and PCSK9 genes), which could contribute to the phenotype of this family. Table 2 represents a summary of the gene variants characteristics that were reported in this family.
Table 2.
Summary of genes and their variants which are identified by TNGS
| Gene | dbSNP ID | Chromosomal location | Exon (intron) | DNA Sequence | Variant name |
|---|---|---|---|---|---|
| LDLR | rs121908026 | Chr19:11216112 | Exon 4 | Reference: AGATGGCTCGGATGAGT Variant: AGATGGCTTGGATGAGT |
Missense variant NM_000527.4:c.530C > T p.Ser177Leu |
| LDLR | rs2738442 | Chr 19:11221454 | Intron 7 | Reference: GTGATTTCCGGGTGGGAC Variant: GTGATTCCCGGGTGGGAC |
Intronic variant NM_000527.4: c.1060 + 7 T > C |
| ABCG8 | rs11887534 | Chr 2:44066247 | Exon 1 | Reference: CTCCCCAGCATACCTCG Variant: CTCCCCAGGATACCTCG |
missense variant NM_022437.2:c.55G > C p.Asp19His |
Discussion and conclusion
This report highlights the importance of screening to identify FH in young children especially within family members diagnosed with FH. The 2016 ESC/EAS Guidelines for the Management of Dyslipidaemias outlines that family cascade screening is recommended when an index case of FH is being identified [23]. In children, the guideline also recommends testing from 5 years of age and even earlier if homozygous FH is highly suspicious [23].
Awareness on this disorder is somehow lacking even among the clinicians [5]. Early detection will allow immediate lipid-lowering medications to be commenced to reduce the risk of progression to CAD. Children diagnosed with FH require commencement of statin treatment as early as 8 to 10 years old [23]. Treatment with statin should be started with low doses and then increased to achieve the treatment goals. The goal in children above 10 years of age is LCL-c < 3.5 mmol/L whereas a level of at least a 50% reduction in the LDL-c level for younger children [23]. The concept of cumulative LDL-c burden highlights the importance of early commencement of treatment. For an individual with heterozygous FH, this LDL-c burden is reached by the age of 35 years if untreated, however this can be delayed to the age of 48 years if the treatment is started by the age of 18 years old and can be more delayed up to 53 years old with earlier treatment by the age of 10 years [5]. Therefore, early initiation of treatment is vital to reduce the risk of progression of atherosclerosis in later life.
Managing FH in young children is indeed challenging as evident in our case. Despite multiple counseling explaining the benefits of statin therapy which outweigh the risks, parents are still very reluctant to start treatment. Dietary and lifestyle modification alone are not sufficient and statins are still the cornerstone of FH management [22]. The thought of being dependent on medication lifelong possibly is the main reason behind their refusal. Furthermore, management challenges also include mental and socioeconomic factors particularly in the economically under-privileged countries in Asia. Treatment of psychosocial risk factors can counteract psychosocial stress, depression and anxiety faced by patients and their family members, thus facilitating behavioral change and improving quality of life and future prognosis [24]. Disease burden is not only faced by the affected family members but also the unaffected members caring for the patients.
The clinical diagnosis of FH is recommended to be confirmed by DNA analysis [23]. In view of the heterogeneous nature of the Malaysian population, it is very essential to screen the suspected FH patients for LDLR gene via testing for the unique allelic variants that were previously described plus the novel variants in this gene comprising the promoter and coding regions, splice sites and splice branch points in order to identify the causative mutations among the suspected FH patients [24]. Our research strategy is performed by confirming the genetic diagnosis of FH among the index case and the family members using the recently developed TNGS method to identify FH related gene mutations. To the best of our knowledge, that this is the first study which apply TNGS technique to identify the causative gene mutations for FH among Malaysian population. Two heterozygous mutations were identified in the LDLR gene: p.Ser177Leu (c.530C > T) and c.1060 + 7 T > C mutations. For p.Ser177Leu that is located in exon 4, at the region that is encoding the fourth repeat in the ligand binding domain of this gene [25], cytosine at nucleotide 530 was replaced by thymidine, with a subsequent changing of serine to leucine amino acid at codon 177. This amino acid is part of the highly conserved SerAspGlu sequence that is identified in the ligand binding domain of this gene [26]. This substitution slows the transport of the protein to the cell surface and the defective receptors will not be able to bind to the LDL-c in the proper normal way [27].
To date, this is the first study that reports this heterozygous disease-causing mutation (p.Ser177Leu) among Malaysian FH patients. However, it was previously reported among Portuguese [28], Polish [29], Spanish [30] and Czech FH patients [31].
It is challenging to predict the biological effects of missense mutations satisfactorily [32], principally in genes such as the LDLR where the rate of neutral alleles relative to disruptive-missense alleles is high [33]. Thormaehlen et al. established the biological effect of missense variants in the LDLR by an in vitro study. His group demonstrated that p.Ser177Leu mutation, identified among Italian subjects with acute myocardial infarction was described as a “disruptive-missense” variant as it strongly inhibited the LDLR protein function, by decreasing LDL-uptake to about 6–31% of the wild type LDLR [34]. This p.Ser177Leu mutation was found in ClinVar database as a pathogenic mutation for FH. For this family it is shown to co-segregate with hypercholesterolaemia and for this reason it is considered as a disease-causing mutation.
A second variant, c.1060 + 7 T > C in the LDLR gene was also reported in this study. It is located in the 3′ splice region of intron 7 of the LDLR gene and this variant was previously reported among the Malaysian population by Al-khateeb et al [35] Such alteration in non-coding regions close to the intron exon junction may potentially affect the splicing however this variant was reported to be a benign by the Polyphen2 and SIFT. Such evidence recommends that this variant may not have a damaging effect on LDLR function.
Another missense variant p.Asp19His, that was reported in the twin pair (hypercholesterolemic), mother and older brother (normolipidemic), but not in the other family members. It is located in exon 1 of ABCG8 gene and was reported to be as a susceptibility factor for gallstone disease among German [36] and Chinese population [37]. This variant was also found to be associated with the disease mentioned above among African American and Hispanic American ancestry additionally among Indian population [38–40]. Those reports are suggesting that such variant might be associated with a more successful transportation of cholesterol in the bile. It was reported as propably damaging in Clin Var database.
Mutations that cause sitosterolemia are very rare. Common sequence variants in ABCG8 may alter sterol metabolism and may lead to inter-individual variation in the plasma concentrations of plant sterols [41]. This p.Asp19His variant was reported among Hispanic subjects and was significantly associated with lower concentrations of plasma sterol concentration, suggesting that it may alter the function of ABCG8 by increasing its transporter function [42]. An association of this variant with plasma cholesterol concentration could not be observed [43] and this may explain the normal cholesterol level in the mother and eldest brother who are carriers of this variant. Our study showed that the twins who had both mutations (in LDLR and ABCG8 genes) had a more severe clinical phenotype with a higher TC and LDL-c compared to their father, grandfather and hypercholesterolemic brother who only had LDLR mutation. We postulate that ABCG8 gene variant on the top of LDLR mutation may enhance the severity of the clinical phenotype in terms of the hypercholesterolemia in the twin patients. This finding also suggests that if untreated, the twins may have worse lipid profile and clinical phenotype in their later life. A population based study with an appropriate sample size or a protein based study may be indicated to reveal the effect of this variant on the LDLR function.
Patients suffering from sitosterolemia have also been described to primarily develop xanthomata and even premature coronary atherosclerosis [16]. However, our twins did not manifest any sign of xanthomas. The absence of xanthomata was probably due to weak physical manifestation of the ABCG8 where it is only present in heterozygous form in the twins. However, there was evidence that the presence of rare ABCG5/G8 in excess, even without the presence of LDLR, can cause significance increase in LDL-c level and cholesterol hyperabsorption [44] which explains why the LDL-c in the twins were higher than in their father and grandfather who were absent of ABCG8 variant.
Diagnosis of sitosterolemia remains very challenging in children. Pediatric patients may manifest planar, tuberous, or tuberoeruptive xanthomata. Affected children may also present with profound hypercholesterolemia that is responsive to dietary management [45]. Sitosterolemia can be easily misdiagnosed as FH because of the similarity in clinical diagnosis [46, 47]. However, differentiation of sitosterolemia from FH is very important, since conventional high-vegetable (plant) oil cholesterol-lowering diet is contraindicated among those sitosterolemic patients as dietary therapy is aimed at reducing the phytosterol accumulation [45]. Both ABCG5 and ABCG8 were demonstrated to inhibit the absorption of cholesterol and plant sterols from the diet by mediating the efflux of these sterols from enterocytes back into gut lumen, and by promoting efficient secretion of cholesterol and plant sterols from hepatocytes into the bile [48]. In view of the essential role of ABCG5/ABCG8 in cholesterol removal from the body, it seems that overexpression of ABCG5 and ABCG8 would prevent atherosclerosis while a defect should promote atherosclerosis [49]. A large cohort study of patients with heterozygous FH showed that the ABCG8 rs11887534 variant have an association with higher risk of coronary heart disease [50].
This is the first in the literature to report a case of heterozygous FH in monochorionic diamniotic twin with mutations in both LDLR and ABCG8 gene. Miyagi et al. reported a dichorionic diamniotic twin with compound heterozygous LDLR mutation causing FH42. Rabacchi et al. on the other hand reported identical twins with compound heterozygous FH with two LDLR gene mutations [43].
In summary, a rare case of Asian monochorionic diamniotic identical twin, of Indian descent, with clinically diagnosed and molecularly confirmed heterozygous FH, due to LDLR and ABCG8 gene mutations have been reported. This is the first study that identified the disease-causing mutation p.Ser177Leu in the LDLR gene among Malaysian FH patients, suggesting that this mutation does not only exist among Caucasian population. Another variant in the ABCG8 gene, p.Asp19His, was also identified for the first time among Malaysian FH population.
Childhood FH may not present with the classical physical manifestations including the pathognomonic lipid stigmata as in adults. Therefore, childhood FH can possibly be better diagnosed early using a combination of both clinical criteria and molecular analyses. Furthermore, identification of index cases in childhood, may provide opportunities for reverse family cascade screening, allowing greater advantage of early detection and prevention of CAD in other family members. In addition, it is important to obtain an early diagnosis of childhood FH to prevent premature atherosclerosis and CAD.
Acknowledgements
We would like to thank our patients and their family who have kindly given the permission for us to report this case.
Funding
HMN was a recipient of a grant under the Long Term Research Grant Scheme (LRGS), grant code: 600-RMI/LRGS 5/3 (2/2011), from the Ministry of Higher Education, Malaysia and the MITRA Grant, grant code: 600-IRMI/MYRA 5/3MITRA (003/2017)-1, from Universiti Teknologi MARA (UiTM).
Availability of data and materials
The datasets generated during and/or analysed during the current study are not publicly available due to the aim to protect the confidentiality of the family but are available from the corresponding author upon reasonable request.
Declaration
This case has been presented as a poster presentation at the 86th European Atherosclerosis Congress from 5 to 8 May 2018 at Lisbon, Portugal and only the abstract has been published in the Atherosclerosis Journal, August 2018, Volume 275, Pages e102-e103.
Abbreviations
- APO B-100
Apolipoprotein B-100 gene
- CAD
Coronary artery disease
- FH
Familial hypercholesterolaemia
- HeFH
Heterozygous familial hypercholesterolaemia
- HoFH
Homozygous familial hypercholesterolaemia
- LDL-c
Low-density lipoprotein cholesterol
- LDLR
Low-density lipoprotein receptor
- PCSK9
Proprotein convertase subtilisin/kexin 9
- TC
Total cholesterol
Authors’ contributions
NSMN, HMN and NAMK treated the patients. Y-AC and AMA-K performed and analysed the molecular data. All authors drafted the article, critically revised the manuscript and approved the final manuscript.
Ethics approval and consent to participate
Not applicable
Consent for publication
Written informed consent was obtained from the parent for publication of this case report and any accompanying images. A copy of the written consent is available for review by the Editor-in-Chief of this journal.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Noor Shafina Mohd Nor, Email: shafinamohdnor@yahoo.com.
Alyaa Mahmood Al-Khateeb, Email: alyaa_1974@yahoo.com.
Yung-An Chua, Email: yungan.chua@gmail.com.
Noor Alicezah Mohd Kasim, Email: alicezah_80@yahoo.com.
Hapizah Mohd Nawawi, Phone: +603-61267464, Phone: +6012-3838075, Email: hapizah.nawawi@gmail.com.
References
- 1.Vallejo-Vaz AJ, De Marco M, Stevens CAT, Akram A, Freiberger T, Hovingh GK, et al. Overview of the current status of familial hypercholesterolaemia care in over 60 countries - the EAS familial hypercholesterolaemia studies collaboration (FHSC) Atherosclerosis. 2018;277:234–255. doi: 10.1016/j.atherosclerosis.2018.08.051. [DOI] [PubMed] [Google Scholar]
- 2.Benn M, Watts GF, Tybjaerg-Hansen A, Nordestgaard BG. Familial hypercholesterolemia in the Danish general population: prevalence, coronary artery disease, and cholesterol-lowering medication. J Clin Endocrinol Metab. 2012;97(11):3956–3964. doi: 10.1210/jc.2012-1563. [DOI] [PubMed] [Google Scholar]
- 3.Do R, Stitziel NO, Won HH, Jorgensen AB, Duga S, Angelica MP. Exome sequencing identifies rare LDLR and APOA5 alleles conferring risk for myocardial infarction. Nature. 2015;518(7537):102–106. doi: 10.1038/nature13917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Consortium UK. Walter K, Min JL, Huang J, Crooks L, Memari Y. The UK10K project identifies rare variants in health and disease. Nature. 2015;526(7571):82–90. doi: 10.1038/nature14962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nordestgaard B, Chapman M, Humphries S, et al. Familial hypercholesterolaemia is underdiagnosed and undertreated in the general population: guidance for clinicians to prevent coronary heart disease: consensus statement of the European atherosclerosis society. Eur Heart J. 2013;34:3478–3490. doi: 10.1093/eurheartj/eht273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldstein JL, Hobbs HH, Brown MS, et al. Familial hypercholesterolemia. In: Scriver CR, Ellenson LH, Ellis NA, et al., editors. The metabolic and molecular bases of inherited disease. New York: McGraw-Hill, Medical Publishing Division; 2001. p. 2863e913. [Google Scholar]
- 7.Fahed AC, Safa RM, Haddad FF, et al. Homozygous familial hypercholesterolemia in Lebanon: a genotype/phenotype correlation. Mol Genet Metab. 2011;102:181e8. doi: 10.1016/j.ymgme.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 8.Goldstein JL, Brown MS. Molecular medicine. The cholesterol quartet. Science. 2001;292(5520):1310–1312. doi: 10.1126/science.1061815. [DOI] [PubMed] [Google Scholar]
- 9.Marais AD. Familial hypercholesterolaemia. Clin Biochem Rev. 2004;25:49e68. [PMC free article] [PubMed] [Google Scholar]
- 10.Usifo E, Leigh SE, Whittall RA, Lench N, Taylor A, Yeats C, Orengo CA, Martin AC, Celli J, Humphries SE. Low-density lipoprotein receptor gene familial hypercholesterolemia variant database: update and pathological assessment. Ann Hum Genet. 2012;76(5):387–401. doi: 10.1111/j.1469-1809.2012.00724.x. [DOI] [PubMed] [Google Scholar]
- 11.Landrum MJ, Lee JM, Benson M, et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44:D862–D868. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muller PY, Miserez AR. Large heterogeneity of mutations in the gene encoding the low-density lipoprotein receptor in subjects with familial hypercholesterolaemia. Atheroscler Suppl. 2004;5:1–5. doi: 10.1016/j.atherosclerosissup.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 13.Varret M, Abifadel M, Rabès JP, Boileau C. Genetic heterogeneity of autosomal dominant hypercholesterolemia. Clin Genet. 2008;73:1–13. doi: 10.1111/j.1399-0004.2007.00915.x. [DOI] [PubMed] [Google Scholar]
- 14.Shulman RS, Bhattacharyya AK, Connor WE, Fredrickson DS. Beta-sitosterolemia and xanthomatosis. N Engl J Med. 1976;294:482–483. doi: 10.1056/NEJM197602262940907. [DOI] [PubMed] [Google Scholar]
- 15.Tada H, Kawashiri MA, Takata M, Matsunami K, Imamura A, Matsuyama M, Sawada H, Nunoi H, Konno T, Hayashi K, Nohara A, Inazu A, Kobayashi J, Mabuchi H, Yamagishi M. Infantile cases of sitosterolaemia with novel mutations in the ABCG5 gene: extreme hypercholesterolaemia is exacerbated by breastfeeding. JIMD Rep. 2015;21:115. doi: 10.1007/8904_2014_404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salen G, Patel S, Batta AK. Sitosterolemia. Cardiovasc Drug Rev. 2002;20:255–270. doi: 10.1111/j.1527-3466.2002.tb00096.x. [DOI] [PubMed] [Google Scholar]
- 17.Goldberg AC, Hopkins PN, Toth PP, Ballantyne CM, Rader DJ, Robinson JG, et al. Familial hypercholesterolemia: screening, diagnosis and management of pediatric and adult patients: clinical guidance from the National Lipid Association Expert Panel on Familial Hypercholesterolemia. J Clin Lipidol. 2011;5(3 Suppl):S1–S8. doi: 10.1016/j.jacl.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 18.Futema M, Shah S, Cooper JA, Li K, Whittal RA, Sharifi M, et al. Refinement of variant selection for the LDL cholesterol genetic risk score in the diagnosis of the polygenic form of clinical familial hypercholesterolemia and replication in samples from 6 countries. Clin Chem. 2015;61(1):231–238. doi: 10.1373/clinchem.2014.231365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang J, Dron JS, Ban MR, Robinson JF, McIntyre AD, Alazzam M, et al. Polygenic versus monogenic causes of hypercholesterolemia ascertained clinically. Arterioscler Thromb Vasc Biol. 2016;36(12):2439–2445. doi: 10.1161/ATVBAHA.116.308027. [DOI] [PubMed] [Google Scholar]
- 20.Khera AV, Won HH, Peloso GM, Lawson KS, Bartz TM, Deng X, et al. Diagnostic yield and clinical utility of sequencing familial hypercholesterolemia genes in patients with severe hypercholesterolemia. J Am Coll Cardiol. 2016;7(67):2578–2589. doi: 10.1016/j.jacc.2016.03.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Futema M, Plagnol V, Whittall RA, Neil HAW, on behalf of the Simon Broome Register Group, Steve Eric Humphries,1 UK10K4 Use of targeted exome sequencing as a diagnostic tool for Familial Hypercholesterolaemia. J Med Genet. 2012;49:644–649. doi: 10.1136/jmedgenet-2012-101189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wiegman A, Gidding SS, Watts GF, Chapman MJ, Ginsberg HN, Cuchel M. Familial hypercholesterolaemia in children and adolescents: gaining decades of life by optimizing detection and treatment. Eur Heart J. 2015;36:2425–2437. doi: 10.1093/eurheartj/ehv157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Catapano AL, Graham I, De Backer G, Wiklund O, Chapman MJ, Drexel H, Hoes AW, Jennings CS, Landmesser U, Pedersen TR, Reiner Ž, Riccardi G, Taskinen MR, Tokgozoglu L, Verschuren WM, Vlachopoulos C, Wood DA, Zamorano JL. 2016 ESC/EAS Guidelines for the Management of Dyslipidaemias: The Task Force for the Management of Dyslipidaemias of the European Society of Cardiology (ESC) and European Atherosclerosis Society (EAS) Developed with the special contribution of the European Assocciation for Cardiovascular Prevention & Rehabilitation (EACPR) Atherosclerosis. 2016;253:281–344. doi: 10.1016/j.atherosclerosis.2016.08.018. [DOI] [PubMed] [Google Scholar]
- 24.Piepoli MF, Hoes AW, Agewall S, Albus C, Brotons C, Catapano AL, et al. 2016 European guidelines on cardiovascular disease prevention in clinical practice. The Sixth Joint Task Force of the European Society of Cardiology and Other Societies on Cardiovascular Disease Prevention in Clinical Practice (constituted by representatives of 10 societies and by invited experts). Developed with the special contribution of the European Association for Cardiovascular Prevention and Rehabilitation. G Ital Cardiol (Rome) 2017;18(7):547–612. doi: 10.1714/2729.27821. [DOI] [PubMed] [Google Scholar]
- 25.Hopkins PN, Toth PP, Ballantyne CM, et al. Familial hypercholesterolemias: prevalence, genetics, diagnosis and screening recommendations from the National Lipid Association Expert Panel on Familial Hypercholesterolemia. J Clin Lipidol. 2011;5:S9e17. doi: 10.1016/j.jacl.2011.03.452. [DOI] [PubMed] [Google Scholar]
- 26.Sudhof TC, Goldstein JL, Brown MS, Russell DW. The LDL receptor gene: a mosaic of exons shared with different proteins. Science (Wash DC) 1985;228:815–822. doi: 10.1126/science.2988123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hobbs HH, Leitersdorf E, Leffert CC, Cryer DR, Brown MS, Goldstein JL. Evidence for a dominant gene that suppresses hypercholesterolaemia in a family with defective low density lipoprotein receptors. J Clin Invest. 1989;84:656–664. doi: 10.1172/JCI114212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bourbon M, Alves AC, Medeiros AM, Silva S, Soutar AK. Familial hypercholesterolaemia in Portugal. Atherosclerosis. 2008;196:633–642. doi: 10.1016/j.atherosclerosis.2007.07.019. [DOI] [PubMed] [Google Scholar]
- 29.Górski B, Kubalska J, Naruszewicz M, Lubiński J. LDL-R and Apo-B-100 gene mutations in Polish familial hypercholesterolaemias. Hum Genet. 1998;102(5):562–565. doi: 10.1007/s004390050740. [DOI] [PubMed] [Google Scholar]
- 30.Mozas P, Cenarro A, Civeira F, Castillo S, Ros E, Pocovi M. Mutation analysis in 36 unrelated Spanish subjects with familial hypercholesterolaemia: identification of 3 novel mutations in the LDL receptor gene. Hum Mutat. 2000;15:483. doi: 10.1002/(SICI)1098-1004(200005)15:5<483::AID-HUMU19>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 31.Dušková L, Kopečková L, Jansová E, Tichý L, Freiberger T, Zapletalová P, Soška V, Ravčuková B, Fajkusová L. An APEX-based genotyping microarray for the screening of 168 mutations associated with familial hypercholesterolaemia. Atherosclerosis. 2011;216(1):139–145. doi: 10.1016/j.atherosclerosis.2011.01.023. [DOI] [PubMed] [Google Scholar]
- 32.Richards CS, Bale S, Bellissimo DB, Das S, Grody WW, et al. ACMG recommendations for standards for interpretation and reporting of sequence variations: revisions 2007. Genet Med. 2008;10:294–300. doi: 10.1097/GIM.0b013e31816b5cae. [DOI] [PubMed] [Google Scholar]
- 33.Zuk O, Schaffner SF, Samocha K, Do R, Hechter E, et al. Searching for missing heritability: designing rare variant association studies. Proc Natl Acad Sci U S A. 2014;111:E455–E464. doi: 10.1073/pnas.1322563111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thormaehlen Aenne S., Schuberth Christian, Won Hong-Hee, Blattmann Peter, Joggerst-Thomalla Brigitte, Theiss Susanne, Asselta Rosanna, Duga Stefano, Merlini Pier Angelica, Ardissino Diego, Lander Eric S., Gabriel Stacey, Rader Daniel J., Peloso Gina M., Pepperkok Rainer, Kathiresan Sekar, Runz Heiko. Systematic Cell-Based Phenotyping of Missense Alleles Empowers Rare Variant Association Studies: A Case for LDLR and Myocardial Infarction. PLOS Genetics. 2015;11(2):e1004855. doi: 10.1371/journal.pgen.1004855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Al-Khateeb A, Zahri MK, Mohamed MS, Sasongko TH, Ibrahim S, Yusof Z, et al. Analysis of sequence variations in low-density lipoprotein receptor gene among Malaysian patients with familial hypercholesterolaemia. BMC Med Genet. 2011;12(1):1. doi: 10.1186/1471-2350-12-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buch S, Schafmayer C, Völzke H, Becker C, Franke A, von Eller-Eberstein H, Kluck C, Bässmann I, Brosch M, Lammert F, Miquel JF, Nervi F, Wittig M, Rosskopf D, Timm B, Höll C, Seeger M, ElSharawy A, Lu T, Egberts J, Fändrich F, Fölsch UR, Krawczak M, Schreiber S, Nürnberg P, Tepel J, Hampe J. A genome-wide association scan identifies the hepatic cholesterol transporter ABCG8 as a susceptibility factor for human gallstone disease. Nat Genet. 2007;39(8):995–999. doi: 10.1038/ng2101. [DOI] [PubMed] [Google Scholar]
- 37.Zhan L, Pan Y-Z, Chen L, Zhang H, Zhang H, Song J, Tzeng C-M, Sun C-Y. Prevalence of ABCB4 polymorphisms in gallstone disease in han-Chinese population. Am J Transl Res. 2016;8(2):1218–1227. [PMC free article] [PubMed] [Google Scholar]
- 38.Joshi, et al. Four susceptibility loci for gallstone disease identified in a meta-analysis of genome-wide association studies. Gastroenterology. 2016;151(2):351–363.e28. doi: 10.1053/j.gastro.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siddapuram SP, Mahurkar S, Duvvuru NR, et al. Hepatic cholesterol transporter ABCG8 polymorphisms in gallstone disease in an Indian population. J Gastroenterol Hepatol. 2010;25:1093–1098. doi: 10.1111/j.1440-1746.2010.06309.x. [DOI] [PubMed] [Google Scholar]
- 40.Srivastava A, Srivastava A, Srivastava K, Choudhuri G, Mittal B. Role of ABCG8 D19H (rs11887534) variant in gallstone susceptibility in northern India. J Gastroenterol Hepatol. 2010;25(11):1758–1762. doi: 10.1111/j.1440-1746.2010.06349.x. [DOI] [PubMed] [Google Scholar]
- 41.Berge KE, von Bergmann K, Lutjohann D, Guerra R, Grundy SM, Hobbs HH, Cohen JC. Heritability of plasma noncholesterol sterols and relationship to DNA sequence polymorphism in ABCG5 and ABCG8. J Lipid Res. 2002;43:486. [PubMed] [Google Scholar]
- 42.Miyagi Yoshitsune, Harada-Shiba Mariko, Ohta Takao. Effect of Statin Therapy in 4-Year-Old Dichorionic Diamniotic Twins with Familial Hypercholesterolemia Showing Multiple Xanthomas. Journal of Atherosclerosis and Thrombosis. 2016;23(1):112–117. doi: 10.5551/jat.31666. [DOI] [PubMed] [Google Scholar]
- 43.Rabacchi C, Bigazzi F, Puntoni M, Sbrana F, Sampietro T, Tarugi P, Bertolini S, Calandra S. Phenotypic variability in 4 homozygous familial hypercholesterolemia siblings compound heterozygous for LDLR mutations. J Clin Lipidol. 2016;10(4):944–952. doi: 10.1016/j.jacl.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 44.Lamiquiz-Moneo I, Baila-Rueda L, Bea A, Mateo-Galleo R, Perez-Callahorra S, Marco-Bennedi V, et al. ABCG5/G8 gene is associated with hypercholesterolemias without mutation in candidate genes and noncholesterol sterols. J Clin Lipidol. 2017;11:1432–1440. doi: 10.1016/j.jacl.2017.09.005. [DOI] [PubMed] [Google Scholar]
- 45.Salen G, Shefer S, Nguyen L, Nes GC, Tint GS, Shore V. Sitosterolemia. J Lipid Res. 1992;33(7):945. [PubMed] [Google Scholar]
- 46.Reiner Z, Guardamagna O, Nair D, et al. Lysosomal acid lipase deficiency – an under recognized cause of dyslipidaemia and liver dysfunction. Atherosclerosis. 2014;235:21–30. doi: 10.1016/j.atherosclerosis.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 47.Taher Z, Ibrahim B, Saifaldeen R, Albar R. Advanced case of sitosterolemia with a novel homozygous mutation. Curr Pediatr Res. 2018;22(1):4–8. [Google Scholar]
- 48.Brown JM, Yu L. Opposing gatekeepers of apical sterol transport: Nieman-Pick C1-like 1 (NPC1L1) and ATP-binding cassette transporters G5 and G8 (ABCG5/ABCG8) Immunol Endocr Metab Agents Med Chem. 2009;9:18–29. doi: 10.2174/187152209788009797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu XH, Qian K, Jiang N, Zheng XL, Cayabyab, Tang CK. ABCG5/ABCG8 in cholesterol excretion and atherosclerosis. Clin Chem Acta. 2014;428:82–88. doi: 10.1016/j.cca.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 50.Koeijvoets KC, van der Net JB, Dallinga-Thie GM, Steyerberg EW, Mensink RP, Kastelein JJ, et al. ABCG* gene polymorphism, plasma cholesterol concentration, and risk of cardiovascular disease in familial hypercholesterolemia. Atherosclerosis. 2009;204:453–458. doi: 10.1016/j.atherosclerosis.2008.09.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during and/or analysed during the current study are not publicly available due to the aim to protect the confidentiality of the family but are available from the corresponding author upon reasonable request.