Abstract
Periodontitis is characterized by a chronic inflammation produced in response to a disease-associated multispecies bacterial community in the subgingival region. Although the inflammatory processes occur locally in the oral cavity, several studies have determined that inflammatory mediators produced during periodontitis, as well as subgingival species and bacterial components, can disseminate from the oral cavity, contributing therefore, to various extraoral diseases like cancer. Interestingly, carcinogenesis associated with periodontal species has been observed in both the oral cavity and in extra oral sites. In this review, several studies were summarized showing a strong association between orodigestive cancers and poor oral health, presence of periodontitis-associated bacteria, tooth loss, and clinical signs of periodontitis. Proinflammatory pathways were also summarized. Such pathways are activated either by mono- or polymicrobial infections, resulting in an increase in the expression of proinflammatory molecules such as IL-6, IL-8, IL-1β, and TNF-α. In addition, it has been shown that several periodontitis-associated species induce the expression of genes related to cell proliferation, cell cycle, apoptosis, transport, and immune and inflammatory responses. Intriguingly, many of these pathways are linked to carcinogenesis. Among them, the activation of Toll-like receptors (TLRs) and antiapoptotic pathways (such as the PI3K/Akt, JAK/STAT, and MAPK pathways), the reduction of proapoptotic protein expression, the increase in cell migration and invasion, and the enhancement in metastasis are addressed. Considering that periodontitis is a polymicrobial disease, it is likely that mixed species promote carcinogenesis both in the oral cavity and in extra oral tissues and probably—as observed in periodontitis—synergistic and/or antagonistic interactions occur between microbes in the community. To date, a good amount of studies has allowed us to understand how monospecies infections activate pathways involved in tumorigenesis; however, more studies are needed to determine the combined effect of oral species in carcinogenesis.
1. Introduction
Periodontal diseases are dysbiotic conditions in the gingival margin, which are characterized by an imbalance between subgingival communities and the host immune response [1]. Such diseases include gingivitis, which is a reversible condition characterized by the inflammation of the gingiva driven by the combined effect of specific microbial taxa. If not treated, gingivitis could progress to periodontitis, characterized by the destruction of supporting tissues of the teeth. From health to gingivitis, to periodontitis, several ecological successions occur in the subgingival microbiome, leading to both an increased biomass and the establishment of distinct dysbiotic communities. Interestingly, not only local effects in the oral cavity have been associated with such disorders but also periodontitis has been largely considered as a risk factor for a number of both oral and systemic diseases [2–5]. Among these, orodigestive cancers are highly influenced by both a direct carcinogenic effect of periodontitis-associated bacteria in either oral cells or in other body sites and inflammatory mediators migrating from the oral cavity [6, 7]. Either way, there is extensive evidence showing that species such as Porphyromonas gingivalis (highly abundant and prevalent in periodontitis) and Fusobacterium nucleatum (closely interacting with periodontitis-associated species in the disease) directly activate transduction pathways leading to cell transformation [7–12]. Comparatively, less information exists about other periodontitis-associated bacteria.
However, although increasing evidence links periodontitis and carcinogenesis, the fact that periodontitis is a polymicrobial disease has not been well addressed in the context of cancer. This is especially relevant when evaluating the direct carcinogenic effect exerted by oral bacteria, since combined species act locally in oral cells and also migrate from the oral cavity. Thus, more studies evaluating how interbacterial interactions affect carcinogenesis process are needed.
2. Periodontal Diseases
Periodontal diseases are associated with chronic inflammation, which affects the supporting tissues of the teeth including the gums or gingival tissue, as well as the periodontal ligament and the alveolar bone in more severe forms of the diseases [13]. Gingivitis is a periodontal disease characterized by local inflammatory processes driven by subgingival bacteria that in most cases do not promote destruction of the tissues and can be reversible. However, clinically, it is considered as the starting point of other periodontal diseases, such as periodontitis [14]. Periodontitis is triggered by an imbalance between resident subgingival microbiota and the inflammatory response of the host that leads to destruction of the supporting tissues of the teeth, even producing the loss of teeth [13]. According to the World Health Organization, between 35% and 50% of the world population are affected by periodontitis [15]. In the United States, the prevalence of gingivitis in children aged between 3 and 11 years is 9-17%, while at puberty, prevalence rises to 70-90% [16] and corresponds to 47% of adult population [17].
2.1. Role of Subgingival Communities in the Etiology of Periodontal Diseases
Both gingivitis and periodontitis are driven by bacterial communities interacting with the host immune system and therefore contributing to the inflammation of tissues. Because of the relevance of the bacterial component, different theories have been proposed in order to establish the importance of these subgingival bacterial communities in the etiology of periodontitis. In 1954, it was proposed that the accumulation of microorganisms promotes the release of compounds that produce inflammation in the gingival tissue [18, 19]. This idea eventually evolved into researchers demonstrating that the colonization of certain anaerobic subgingival bacteria, including P. gingivalis, Treponema denticola, and Tannerella forsythia, promoted both the onset and the development of periodontitis [20]. However, different studies that sought to determine the composition of the bacterial community associated with periodontitis managed to determine that these bacteria were not only present in patients with periodontitis but also in periodontally healthy individuals [1].
This was a key point in supporting current theories establishing that it is not the colonization of specific bacteria what triggers the disease, but rather the changes in the relative abundances of specific taxa in the subgingival communities due to dysbiotic processes occurring in subgingival areas that would determine the development of periodontitis. In this context, Marsh [21] created the concept of “ecological catastrophe,” which establishes that the environmental and host factors, such as poor hygiene, inappropriate diets, and use of tobacco and drugs that produce side effects in the immune defense of the patient, select and enrich pathogenic bacteria, and a disease state. The authors described that an increase in bacterial plaque increases local inflammation, which in turn increases the flow of crevicular gingival fluid (CGF), produces bleeding, and provides proteinaceous nutrients, which increase the proliferation of Gram-negative anaerobes [21, 22].
This theory has been supported by several studies aimed at characterizing the microbiome of periodontally healthy individuals and patients with periodontitis [1, 23–25]. Diaz et al. [26] reviewed these studies concluding that different subgingival microbiomes are characteristic of healthy individuals, as well as patients with gingivitis and periodontitis. While most health-associated bacteria are early colonizers of the subgingival biofilm, periodontitis-associated bacteria are mainly late colonizers. In the periodontitis-associated group of bacteria, species such as Filifactor alocis, P. gingivalis, Porphyromonas endodontalis, T. forsythia, and T. denticola are found in all the 4 studies reviewed. P. gingivalis was proposed as a key player among such species (“keystone pathogen”), since Hajishengallis et al. [27] demonstrated that even when it is found in low abundance in healthy individuals, it can promote changes in homeostasis of the normal microbiota, remodeling it towards a harmful microbiota that promotes destruction of tissues and inflammation in in vivo models. This concept was refined by Hajishengallis himself in 2012, proposing the polymicrobial synergy and dysbiosis theory (PSD). This theory adds the fact that every component of a symbiotic and synergistic microbiota is relevant in the onset of the disease and not only the periodontitis-associated bacteria. Thus, the whole dysbiotic community will synergistically initiate processes of tissue inflammation, activate production of cytokines, and initiate the recruitment of immune cells [28].
Interestingly, besides having determined both periodontitis and health-associated bacteria, a third group called “core species,” which are equally prevalent and found in the same proportion both in health and periodontitis individuals, was characterized, being F. nucleatum the most abundant in this group [1, 24]. F. nucleatum plays a central role in the subgingival biofilm, since it physically interacts with other microorganisms in the subgingiva [29]: P. gingivalis [30], Aggregatibacter actinomycetemcomitans [31], Prevotella spp. [32], Streptococcus gordonii [33], Candida albicans [34], and others [29, 35]. Such close interactions with several species in the biofilm are reflected in the fact that F. nucleatum acts as a bridge attaching early colonizers like Streptococcus spp. and other facultative species and late colonizers such as P. gingivalis [36, 37]. This process is essential for the ecological successions that establish the subgingival plaque and determine the progression of periodontitis [35], in which thousands of species colonize the subgingival area in an ordered manner.
These successions include specific modification of the local environment in the biofilm [38, 39] which select specific groups of bacteria and eventually induce changes in the subgingival bacterial communities that lead to a dysbiotic community able to induce a deregulation of the host inflammatory response and eventually cause chronic inflammation.
2.2. Chronic Inflammation Driven by Periodontitis-Associated Bacteria
In the periodontal pocket, the first host responses to the dysbiotic subgingival community are characterized by the infiltration of natural killer (NK) cells, neutrophils, and granulocytes (polymorphonuclear cells) that promote the initial inflammatory response and the subsequent infiltration of lymphocytes to present antigens to dendritic cells [40]. The neutrophils are overwhelmed with the abundance and persistence of microorganisms, being destroyed or undergoing either apoptosis or necrosis as they interact with bacteria within the gingival crevice.
T cells promote a profile characterized by CD8+ and CD4+ cells that generate a proinflammatory medium rich in cytokines such as tumor necrosis factor alpha (TNF-α), interleukin- (IL-) 1, IL-4, IL-10, interferon-γ (IFN-γ), and transforming growth factor β (TGF-β) [41]. In addition, T CD4+ lymphocytes produce RANK-L, a cytokine that promotes bone resorption [42]. It was also described that T cells contribute to cell-mediated immune responses by stimulating T helper cells such as Th1, Th2, Th9, Th17, and Th22 and the deregulation of this response could be related to the appearance of the disease and its chronicity [43, 44]. On the other hand, B cells produce antibodies against the microorganisms present in the subgingival pocket in order to eliminate them and decrease the local inflammation [44].
In addition to the inflammatory mediators produced by the immune cells, the gingival epithelium also releases other cytokines such as IL-1, IL-8, and TNF-α, which in turn promotes the recruitment of macrophages [45]. Concordantly with these studies performed in vitro, in periodontitis tissue samples, an increase in mRNA of IL-1β, IL-6, IL8, and TNF-α, regulated upon activation normal T cell expressed and secreted (RANTES) and monocyte chemotactic protein-1 (MCP-1), was observed, compared to healthy gingiva [46]. In the same context, a higher expression of IL-1β was observed in gingival fluid from deeper sites of periodontitis patients [47].
As a consequence of this inflammatory response, ecological changes in the subgingival region occur, which contribute to the ecological successions in the subgingival area that are associated with periodontitis progression. Interestingly, some periodontitis-associated bacteria have been shown to contribute directly to the chronic inflammation by activating specific intracellular pathways.
Because of the polymicrobial nature of periodontitis and considering that interbacterial interactions occurring in the subgingival biofilm contribute to the disease, current models of periodontitis include the study of the effect of multiple species in the stimulation of immune response. Very recently, Herrero et al. [48] showed that the exposure of epithelial and fibroblast cultures to a dysbiotic biofilm increased the expression of IL-6, IL-8, IL-1β, TNF-α, and MMP-8. In the same context, other studies showed that epithelial cells produce higher cytokine levels when they are exposed to either monospecies or multispecies biofilms [49]. Interestingly, an increased expression of IL-8, C-X-C motif chemokine ligand 3 (CXCL-3), CXCL-1, IL-1, IL-6, colony-stimulating factor 2 (CSF2), and TNF-α was observed in cells stimulated with the multispecies biofilms. Similarly, polymicrobial infection (P. gingivalis, T. denticola, and T. forsythia) using a murine calvarial bone model affected the expression of several genes related to cell proliferation, cell cycle, apoptosis, transport, immune response, and inflammatory response. In the proinflammatory context, the cytokines that increased the most were IL-1, IL-6, and TNF-α, which are precisely those related to chronic inflammation and chronic bone damage [50].
Nonetheless, despite the fact that multispecies infection constitutes a more realistic model considering the polymicrobial etiology of the disease, many studies using planktonic monospecies bacteria have permitted to determine the contribution of key species to the inflammatory process. For example, studies using T. denticola monoinfections have shown that the bacterium can activate Toll-like receptor 5 (TLR5) through the flagellin, the main component of the bacterial flagellum. This interaction leads to an increase in IL-1β and TNF-α [51]. T. denticola can also suppress the action of antimicrobial peptides such as human β-defensin 3, regulating the signaling pathway activated by TLR2 [52]. Additionally, works by Tanabe et al. [53] demonstrated that T. denticola peptidoglycan induces the secretion of proinflammatory cytokines such as IL-8, IL-6, and TNF-α, in murine macrophages, stimulating the production of PGE2 and decreasing their viability. However, T. denticola can also counteract the increase of these cytokines, as it has been shown in a study conducted in peripheral blood mononuclear cells, where it was determined that T. denticola hydrolyzes IL-1β, IL-6, and TNF-α through the PrtP complex (dentilisin or chymotrypsin-like protease (CTLP)) [54].
On the other hand, the infection of mice with T. forsythia increased levels of IgG and IgM, both markers of immune response activation. Moreover, an increase in CD4+ T lymphocytes was shown [55]. Intriguingly, this bacterium has a glycosylated S layer [56], which is important for the mechanical stabilization and protection of the bacterium. A study by Settem et al. [57] showed that glycosylation of S layer of T. denticola can deregulate the immune response by preventing Th17 production, probably inhibiting the recruitment of neutrophils to the site of infection. This effect produces tissue and bone destruction.
A Gram-positive anaerobic bacterium that has been emerging as a periodontitis-associated species is F. alocis. Infection of gingival epithelial cells (GECs) by F. alocis stimulates the production of proinflammatory cytokines such as IL-1β, IL-6, and TNF-α [58]. This is important, since these cytokines are related to the stimulation of osteoclasts and bone resorption [58]. Moreover, these cytokines have been shown to increase in an in vivo model (mouse subcutaneous chamber model) and to increase the influx of neutrophils to the site of infection [59].
In spite of the growing evidence showing the relevance of a number of species in the progression of periodontitis, one of the most studied species is P. gingivalis. Through such studies, nowadays, we have a good understanding of its role in the pathogenesis of periodontitis. This bacterium is internalized by macrophages and is also able to induce its own internalization by GECs. Once the bacterium is inside the GECs, it can use the machinery of the host cell for its survival and persistence. For example, infected GECs activate antiapoptotic pathways, such as the JAK/STAT and phosphatidylinositol 3-kinase (PI3K)/Akt, which inhibit the intrinsic pathway of apoptosis probably to persist for longer periods. Both pathways have also been related to inflammation. Some cytokines such as IL-6, TNF-α, or IFN-γ function through the JAK/STAT pathway [60]; additionally, the JAK/STAT pathway activates NF-κB and stimulates TNF-α production [61]. The PI3K/Akt pathway, on the other hand, is involved in the increase of TLR4 mRNA, in response to bacterial lipopolysaccharide (LPS) [62]. Finally, phosphorylation of Akt and its consequent activation induces NF-κB, which increases the transcription of antiapoptotic genes [63].
Periodontitis-associated species seek to prolong bacterial growth within the infected cell and also evade the immune system. Once P. gingivalis is internalized, it is incorporated into early phagosomes, where it prevents fusion to the lysosome and therefore its degradation [64]. P. gingivalis secretes the nucleoside diphosphate kinase (NDK) enzyme that removes ATP through the P2X7 receptor. In macrophages, this receptor stimulates the production and secretion of IL-1β, the apoptosis of the host cell, and killing of bacteria [65].
Moreover, infection of human monocytic cell line with P. gingivalis activates NLRP3 and AIM2 inflammasome through caspase 1 activation, which produces the processing of pro-IL-1β to its active form IL-1β [66]. During periodontitis progression, tissue damage occurs both by the direct effect of bacterial virulence factors and the deregulation of the immune system response. P. gingivalis interacts with the GECs through the TLRs mediated by the recognition of P. gingivalis virulence factors such as fimbria and the LPS. It has been shown that this interaction increases the transcription of TLR2 and TLR4 in GECs [67]. Intriguingly, P. gingivalis can modify the lipid A region of its LPS by incorporating different units of acyl groups to its structure. A tetra-acylated structure of P. gingivalis lipid A is a TLR4 antagonist with anti-inflammatory potential [68]. However, the penta-acylated structure of P. gingivalis lipid A is a TLR4 agonist with proinflammatory potential [68] that activates the NF-κB and MAPK-p38 pathways [69]. Nevertheless, P. gingivalis has developed strategies to evade or delay the immune response. For example, within its virulence factors, it possesses gingipain proteases that degrade the CD14 protein (a coreceptor of TLR4 and TLR2), interfering with the optimal recognition of bacterial LPS [70].
P. gingivalis can also modify the expression of adhesion receptors—like E-selectin—for leukocyte adhesion and transmigration, preventing its upregulation. In this context, gingipain proteases produced by P. gingivalis degrade the intracellular adhesion molecule 1 (ICAM-1) in GECs, disrupting neutrophils-oral epithelial cell interaction [71]. These proteases affect also the integrity of the cytokines IL-6, IL-8, IL-12, and TNF-α, which are produced in response to the infection [72–75].
Interestingly, in addition to the inflammation induced by periodontitis-associated bacteria, some “core species” have also been linked to inflammation. For example, it has been demonstrated that F. nucleatum upregulates the production of MMP-13 and IL-8, through the MAPK/p38 pathway in epithelial cells [76]. Moreover, F. nucleatum increases IL-8 mRNA levels through the activation of NF-κB in human GECs [77].
Similar to P. gingivalis, F. nucleatum also activates NLRP3 inflammasome, inducing the releases of damage-associated molecular patterns (DAMPs) like high mobility group box 1 protein (HMGB1) and proteins that recruit and activate caspases (ASC), increasing the inflammation in GECs [78]. After infection, HMGB1 is released into the extracellular space, which is required for the activation of the inflammasome and the caspase 1 activation [79, 80]. On the other hand, ASC functions as an adapter of the NLRP3 inflammasome assembly and is secreted by macrophages during inflammation [81].
Limited data exist regarding the effect of combined subgingival species in carcinogenesis. Coinfection studies using F. nucleatum and P. gingivalis show that they induce a synergic virulence response in a mouse periodontitis model, with a stronger inflammatory response triggered by elevated levels of TNF-α, NF-κB, and interleukin IL-1β [82], as well as higher levels of attachment and invasion into host cells [83, 84].
3. Systemic Diseases Associated with Chronic Inflammation in Periodontitis
Although the inflammatory processes occur locally in the oral cavity, several studies have determined that the chronic inflammation during periodontal diseases or the dissemination of bacterial components could cause various extraoral diseases. Some of these diseases and a brief description of their associations with periodontal disease are summarized as follows:
Cardiovascular diseases: many studies have linked the presence of periodontal diseases with cardiovascular diseases [5, 85, 86]. Among them, Peng et al. [86] determined through a retrospective cohort study that periodontal therapy promoted a decreased risk of cardiovascular disease. Also, different meta-analyses have managed to link the presence of periodontal diseases with an increased risk of cardiovascular disease [85, 87]. Moreover, some periodontitis-associated species have been linked to such diseases. Thus, Damgaard et al. [88] linked the presence of IgG antibodies against P. gingivalis with the presence of cardiovascular disease in serum from 576 participants and Bale et al. [89] proposed that A. actinomycetemcomitans, P. gingivalis, T. forsythia, T. denticola, and F. nucleatum are related to higher risk of atherosclerosis. Interestingly, some cardiovascular diseases are related to chronic inflammation. Two of them, myocarditis and endocarditis, are diseases characterized by a high infiltration of lymphocytes and monocytes. P. gingivalis is proposed as an aggravator of autoimmune myocarditis in an in vivo model [90].
Rheumatoid arthritis (RA): RA is an autoimmune disease characterized by the thickening of the synovium, a tissue that exists inside the joints. IL-1 and TNF-α are highly related with the pathogenesis of RA, but other cytokines like IL-4 and IL-17 have also a role in this disease. Many studies confirm a relationship between periodontitis and RA [4, 91], like Mikuls et al. [92] who were able to determine that periodontitis and the presence of P. gingivalis is related to the self-activity, characteristic of RA. Additionally, both F. nucleatum and P. gingivalis are highly prevalent in patients with RA [93].
Cancer: it has been shown that patients affected by periodontal disease have a higher risk of suffering from some type of cancer [34]; specifically, a positive association between periodontal disease and orodigestive cancers (oral, esophageal, gastric, colonic, and pancreatic) has been well established [2, 3], as well as other types of cancers such as breast, prostate, and bladder [48, 94–96]. A deeper explanation of such associations and possible mechanisms involved in these associations will be addressed in following paragraphs.
4. Association between Periodontitis and Orodigestive Cancer
As stated above, multiple epidemiological studies showed a strong association between orodigestive cancers and poor oral health [97–102], periodontal diseases [103–106], tooth loss [98, 99, 101, 102, 106, 107], and periodontal diagnostic parameters such as clinical attachment loss (CAL) and alveolar bone loss [108, 109]. Additionally, patients showing gastric precancerous lesions were more likely to have higher percentages of sites with gingival bleeding [97, 110].
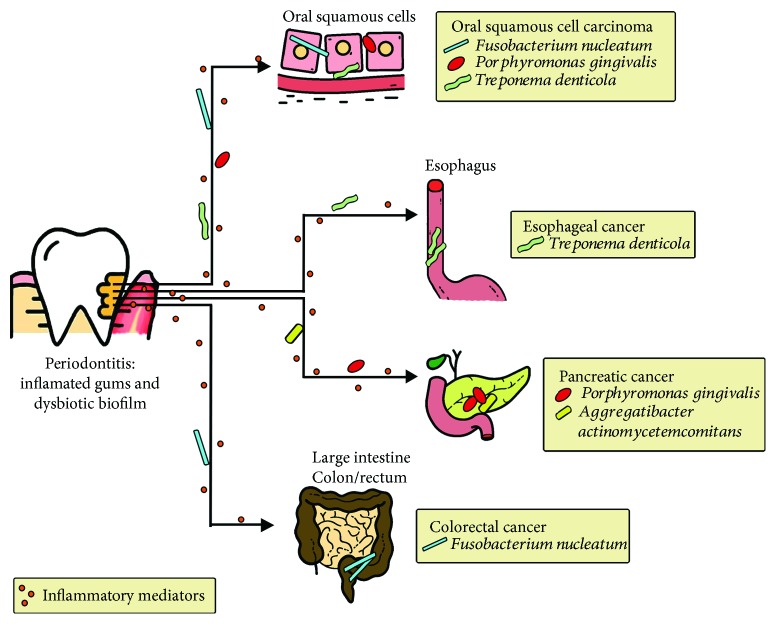
Together with the increasing evidence associating periodontal diseases with several types of extra oral cancer, the question of how these bacteria exert their effect in distal sites in the human body is gaining more and more attention. Thus, some types of cancer have associated carcinogenesis with the chronic inflammation generated in the oral cavity and the concomitant mobilization of inflammatory mediators to distal sites in the human body (Figure 1) [3, 111], while other studies have associated it with a direct carcinogenic effect mediated by periodontitis-associated bacterial species either directly in oral cells or by migrating from the oral cavity (Figure 1) [112]. Interestingly, despite the natural dissemination of oral bacteria due to swallowing of saliva, which contains a large number of bacteria, explaining therefore its involvement in orodigestive tract [113, 114], there is also evidence showing dissemination through the bloodstream (Figure 1) [115].
Figure 1.
Association of periodontal bacteria with orodigestive cancer. Periodontitis has been associated with orodigestive cancers through the chronic inflammation generated in the oral cavity and the concomitant mobilization of inflammatory mediators to distal sites in the human body, as well as a direct carcinogenic effect mediated by periodontitis-associated bacterial species either directly in oral cells or by migrating from the oral cavity.
Systemic spread of oral bacteria either after routine activities or dental procedures was early reported by Cobe [116]. Particularly, oral anaerobes are released to circulation after some daily activities, such as tooth brushing, flossing, and chewing [117], and also immediately after therapeutic oral procedures such as scaling and root planning [118]. Therefore, dental or oral surgery is considered to be a predisposing factor for anaerobes bacteremia in both adults and children [119, 120]. However, in periodontal disease, migration of bacteria from the oral cavity to other organs in the human body is likely to occur through the blood circulation probably because there is a 3-log increase in the biomass of the subgingival biofilm and the mean surface area where this biofilm is contacting the ulcerated gingiva is approximately 20 cm2 [1, 121], providing a portal of entry for oral bacteria into the vessels and thereby allowing them to spread to distant sites [122]. These bacteremias are usually polymicrobial, with higher numbers of Gram-negative bacilli and species of the genera Peptostreptococcus, Clostridium, Fusobacterium, among others [115].
As stated above, F. nucleatum is part of the subgingival microbiota and it is present in most subjects maintaining its proportion from health to disease, probably acting as a metabolic cornerstone for the whole community. Interestingly, extensive evidences associating bacteremia caused by F. nucleatum with underlying malignancy have been reported [123]. Moreover, comorbidity between F. nucleatum bacteremia and several types of cancer has been found in hospitalized patients [124–126]. Particularly, F. nucleatum is considered as a risk factor for colorectal cancer (CRC) (Figure 1) [7, 127, 128], as the bacterium is overrepresented in colorectal tumor tissues versus normal tissues in CRC patients [129–131]; moreover, higher loads of the bacterium have been found in CRC compared to premalignant lesions [127]. It is worth noting that as the bacterium is found together with other oral species in CRC such as Parvimonas micra, Peptostreptococcus stomatis, Gemella morbillorum, Porphyromonas spp, Leptotrichia spp., and Campylobacter spp., it strongly suggests that the source of the microbes is the oral cavity [130, 132–135]. More recently, F. nucleatum was also associated with other malignancies as oral cancer (Figure 1) [7], with higher levels of this species found in oral squamous cell carcinoma (OSCC) patients compared to controls [136, 137]. Similar to CRC, other periodontitis-associated taxa, such as Dialister spp., Peptostreptococcus spp., Filifactor spp., Treponema spp., and Parvimonas spp., were also enriched in these tumors [138]. This is interesting since a combined effect of such species could contribute to cell transformation.
Remarkably, the periodontitis-associated species P. gingivalis is the oral bacteria most commonly associated with cancers of the orodigestive tract and it probably has a positive effect in mortality [6, 139]. Among these cancers, P. gingivalis shows a strong correlation with OSCC [136], as well as with pancreatic cancer (Figure 1) [6, 140]. This species has been found in tumor tissues from patients with OSCC along with other oral anaerobes as species of the genera Veillonella, Fusobacterium, Prevotella, Actinomyces, and Clostridium [141], indicating that a combined effect of multiple bacterial species may be involved in carcinogenesis. Similar results have been observed in gingival squamous cell carcinoma where P. gingivalis is augmented compared to normal tissues [142], probably due to its invasive ability. In fact, tissue invasion is probably one of the significant ways of oral bacteria dissemination, since both F. nucleatum and P. gingivalis—the oral species mostly associated with orodigestive cancers—invade gingival tissues and have been found composing 15% to 40% of the total bacteria within the gingival tissue obtained from periodontal lesions [143]. The occurrence of both species in the tissue is likely to happen as a consequence of an intimate interaction between them in the oral cavity and probably also in extra oral sites.
Remarkably, the sole presence of a bacterium in tumorous tissue is not necessarily indicative of its role in the disease. A recent metatranscriptomic analysis showed that although both P. gingivalis and F. nucleatum were active in OSCC tumor sites compared to healthy control tumor-matched sites, only F. nucleatum showed a significant difference in transcriptional activity, as shown by linear discriminant analysis effect size (LefSe) analysis [144]. This indicates that either different species have a role in different stages of the tumorigenesis or that close interactions between microbial species in the tumoral tissues may modify the gene expression of the companions, as it has been shown in an in vitro multispecies community model [145, 146]. Interestingly, although it is not a periodontitis-associated species, F. nucleatum has been found to be transcriptionally active in different forms of periodontal diseases [147, 148]. Moreover, synergistic interactions between F. nucleatum and two periodontitis-associated bacteria, T. denticola and P. gingivalis, have been reported in chronic periodontitis [149].
This is interesting, since in addition to P. gingivalis, other periodontitis-associated taxa have been associated with orodigestive cancers. While carriage of A. actinomycetemcomitans correlates with higher risk of pancreatic cancer [150], T. denticola has been detected in both tongue squamous cell carcinoma [151] and esophageal cancer tissues (Figure 1) [152]. The question of how these species interact with each other in carcinogenesis has not been fully understood. It has neither been elucidated how migrating oral bacteria affect the local microbiome in distal sites and therefore alter host cell responses. For instance, Arimatsu et al. [153] showed that oral administration of P. gingivalis induces changes in the ileal microbiota in a mouse model, increasing systemic inflammation.
5. The Mechanism of Cancer Promotion by Periodontitis-Associated Bacteria
Although the exact mechanisms involved in cancer promotion by periodontal bacteria have not been completely elucidated, local inflammatory effects triggered by bacterial infection have been associated with cellular transformation [6]. Moreover, among all the subgingival species found in tumorous tissue, there is only information regarding carcinogenic mechanisms triggered by a few of them.
P. gingivalis was shown to activate carcinogenesis through several mechanisms (Figure 2). First, the bacterium has been associated directly with activation of oncogenic pathways, such as the promotion of survival in GECs through both the activation of the PI3K/Akt pathway and the inhibition of cytochrome c release [11], as well as with the reduction of the expression of proapoptotic proteins [10]. Additionally, P. gingivalis blocks apoptosis through the JAK/STAT pathway in GECs and therefore modulates the intrinsic cell death pathway and regulates the expression of several antiapoptotic proteins [154]. The LPS of P. gingivalis, in particular the O-antigen region, contributes to the apoptosis inhibition and induces proliferation in GECs [67]. This effect is associated with increased expression of TLR4 [67].
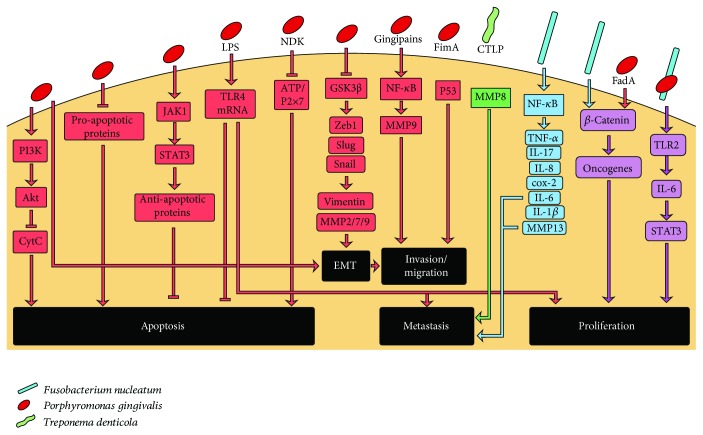
Figure 2.
Host response mechanisms of cellular transformation induced by periodontal bacteria. Inhibition of apoptosis, epithelial-mesenchymal transition (EMT), invasion and migration, metastasis, and proliferation are triggered through the activation of prooncogenic pathways by P. gingivalis (red arrows), T. denticola (purple arrows), F. nucleatum (yellow arrow), and P. gingivalis+F. nucleatum coinfection (orange arrows).
P. gingivalis was also shown to induce GECs migration in a manner dependent on the overexpression of Zeb1 [155], an activator of the epithelial-mesenchymal transition (EMT). Moreover, P. gingivalis increases proliferation and promotes invasion and migration in an in vitro model of persistent infection [9]. Likewise, P. gingivalis infection inhibits the activity of glycogen synthase kinase 3 (GSK3b), an important EMT regulator, in primary oral epithelial cells [156]. Additionally, other EMT-associated transcription factors, as well as mesenchymal intermediates, such as vimentin, MMP-2, MMP-7, and MMP-9, are increased and associated with higher levels of cell migration.
Several virulence factors are involved in the direct activation of inflammation and cell proliferation mediated by P. gingivalis [6]. Among them, nucleoside diphosphate kinase (NDK), FimA, and the LPS of P. gingivalis participate in the first stages of carcinogenesis, while gingipains and GroEL are associated with later stages. NDK inhibits proapoptotic mechanisms in oral epithelial cells by inhibiting the ATP/P2X7 cell death signaling [65, 157, 158]. FimA attenuates the host p53-mediated tumor suppression and cell cycle progression in oral epithelial cells [6, 67] and controls the epithelial–mesenchymal transition [155]. Gingipain proteases of P. gingivalis activate NF-κB and MMP-9 in oral squamous carcinoma cells, which is important for cancer cell invasion and metastasis [159, 160]. Finally, GroEL produced by P. gingivalis increases tumor volume and the mortality of mice implanted with the mouse colon carcinoma cell line (C26) [161]. Recently, Mfa1 fimbria was shown to induce oncogenic signaling, producing myeloid-derived dendritic suppressor cells (MDDSCs) from monocytes activating the pAKT1-pFOXO1 pathway through dendritic cell-specific intercellular adhesion molecule 3-grabbing non-integrin (DC-SIGN) receptor [162].
Although comparatively less information exists regarding carcinogenic mechanisms triggered by F. nucleatum, three virulence factors have been associated with CRC promotion: the adhesin FadA, the LPS, and the autotransporter protein Fap2 (Figure 2) [7]. FadA induces inflammation and activation of procarcinogenic pathways directly in colorectal cells, activating E-cadherin-β-catenin signaling [163]. The LPS of F. nucleatum induces the production of inflammatory cytokines both in the gingiva and in the colonic tissue [129, 164]. Consistently, increased expression of proinflammatory cytokine such as IL-6, IL-12, IL-17, and TNF-α has been found in F. nucleatum-enriched colorectal adenoma subjects compared to nonadenoma controls [165]. Finally, Fap2 decreases the cytotoxicity of immune cells, favoring cancer progression [166]. In vivo studies showed that F. nucleatum increases tumor multiplicity and recruitment of tumor-infiltrating immune cells in a mouse model of intestinal tumorigenesis [167]. In this model, F. nucleatum generates a proinflammatory microenvironment associated with an NF-κB-mediated response (COX-2, IL-1β, IL-6, IL-8, IL-10, and TNF-α) [167], which provides a critical link between inflammation and cancer [168] and is implicated in potentiating colorectal tumorigenesis in mice [167, 169]. In addition, F. nucleatum increases the proliferation and invasion ability of colonic epithelial cells, promoting EMT, activating NF-κB signaling, and increasing the production of IL-6, IL-1β, and MMP-13 [170].
Even less studies evaluated the association of other periodontitis-associated taxa with cancer, among them the contribution of T. denticola to carcinogenesis has recently been reported (Figure 2). This species is a highly invasive anaerobic bacteria and possesses a chymotrypsin-like proteinase (CTLP) as a major virulence factor. Recently, CTLP was detected within orodigestive tumor tissues including OSCC, tongue, tonsil, and esophagus [171]. Intriguingly, CTLP converts pro-MMP-8 and pro-MMP-9 to their active forms, which are associated with metastasis in tongue, esophageal, gastric, pancreatic, and CRC [8, 12, 172].
As mentioned above, systemic spread of periodontitis-associated bacteria is usually polymicrobial. In this context, although combined effect of periodontal bacteria is well established in the etiology of periodontitis, its contribution to cancer onset is less understood. Therefore, it is relevant to understand if these bacterial cooccurrences have synergistic or antagonist effect in respect to the activation of inflammatory pathways associated to cancer.
In this context, it has been shown that coinfection of oral epithelial cells with P. gingivalis and F. nucleatum triggers the TLR2 pathway resulting in IL-6 production and STAT3 activation, which in turn stimulate cell proliferation (Figure 2) [173]. In addition, infection of oral epithelial cells with cocultures of P. gingivalis and F. nucleatum induces a slight increase in cell migration [156]; however, the pathways that are altered and could explain this effect have not been defined.
6. Conclusion
Periodontitis is a dysbiotic disease, in which chronic inflammation is produced in response to a disease-associated multispecies bacterial community established in the subgingival area. The recruitment of immune cells and the production of several inflammatory mediators contribute to the tissue damage. Additionally, the direct effect of periodontitis-associated bacteria as well as other subgingival microorganisms equally prevalent both in healthy and diseased subjects “core species” contributes to the chronicity of the disease through the activation of specific inflammatory pathways.
Chronic inflammation has also been associated with several systemic diseases, like cancer. The literature demonstrates that either inflammatory mediators produced during periodontitis development could mediate carcinogenesis or periodontal bacteria can exert its effect directly in transforming cells. Interestingly, several oral bacteria, also found in high loads in the periodontal pocket, have been shown to activate inflammatory pathways associated with several stages of cellular transformation (Figure 2). Among them, these bacteria can induce NF-κB-mediated responses, promote cell survival, activate oncogenic pathways, reduce proapoptotic proteins expression, increase cell migration and invasion, increase the expression of EMT-associated proteins, enhance metastasis, etc. In spite of this knowledge, more studies are needed to elucidate the mechanisms triggered by other periodontal bacteria and also understand the tumorigenic effect of combined bacterial infections. Such studies are relevant because, although the combined effect of species such as P. gingivalis and F. nucleatum has been studied in the etiology of periodontitis, the consequences of its effect in carcinogenesis remain poorly understood. Moreover, since bacterial spreading to distant sites on the human body occurs in coexistence, it is relevant to know the synergistic or antagonistic effects that these interactions may have in oral and extra oral carcinogenesis.
Acknowledgments
Thanks to Mr. Juan Fernández from the Language and Translation Services of the Dentistry Faculty for kindly proofreading and checking the spelling and grammar of this article. This work was supported by grants from the FIOUCh no. 17/20 and CONICYT-FONDAP 15130011.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
- 1.Abusleme L., Dupuy A. K., Dutzan N., et al. The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. The ISME Journal. 2013;7(5):1016–1025. doi: 10.1038/ismej.2012.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Corbella S., Veronesi P., Galimberti V., Weinstein R., del Fabbro M., Francetti L. Is periodontitis a risk indicator for cancer? A meta-analysis. PLoS One. 2018;13(4, article e0195683) doi: 10.1371/journal.pone.0195683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fitzpatrick S. G., Katz J. The association between periodontal disease and cancer: a review of the literature. Journal of Dentistry. 2010;38(2):83–95. doi: 10.1016/j.jdent.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 4.Koziel J., Mydel P., Potempa J. The link between periodontal disease and rheumatoid arthritis: an updated review. Current Rheumatology Reports. 2014;16(3):p. 408. doi: 10.1007/s11926-014-0408-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Southerland J. H., Taylor G. W., Moss K., Beck J. D., Offenbacher S. Commonality in chronic inflammatory diseases: periodontitis, diabetes, and coronary artery disease. Periodontology 2000. 2006;40(1):130–143. doi: 10.1111/j.1600-0757.2005.00138.x. [DOI] [PubMed] [Google Scholar]
- 6.Atanasova K. R., Yilmaz O. Looking in the Porphyromonas gingivalis cabinet of curiosities: the microbium, the host and cancer association. Molecular Oral Microbiology. 2014;29(2):55–66. doi: 10.1111/omi.12047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gholizadeh P., Eslami H., Kafil H. S. Carcinogenesis mechanisms of Fusobacterium nucleatum. Biomedicine & Pharmacotherapy. 2017;89:918–925. doi: 10.1016/j.biopha.2017.02.102. [DOI] [PubMed] [Google Scholar]
- 8.Aparna M., Rao L., Kunhikatta V., Radhakrishnan R. The role of MMP-2 and MMP-9 as prognostic markers in the early stages of tongue squamous cell carcinoma. Journal of Oral Pathology & Medicine. 2015;44(5):345–352. doi: 10.1111/jop.12245. [DOI] [PubMed] [Google Scholar]
- 9.Geng F., Liu J., Guo Y., et al. Persistent exposure to Porphyromonas gingivalis promotes proliferative and invasion capabilities, and tumorigenic properties of human immortalized oral epithelial cells. Frontiers in Cellular and Infection Microbiology. 2017;7:p. 57. doi: 10.3389/fcimb.2017.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yao L., Jermanus C., Barbetta B., et al. Porphyromonas gingivalis infection sequesters pro-apoptotic Bad through Akt in primary gingival epithelial cells. Molecular Oral Microbiology. 2010;25(2):89–101. doi: 10.1111/j.2041-1014.2010.00569.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yilmaz O., Jungas T., Verbeke P., Ojcius D. M. Activation of the phosphatidylinositol 3-kinase/Akt pathway contributes to survival of primary epithelial cells infected with the periodontal pathogen Porphyromonas gingivalis. Infection and Immunity. 2004;72(7):3743–3751. doi: 10.1128/IAI.72.7.3743-3751.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeng R., Duan L., Kong Y., et al. Clinicopathological and prognostic role of MMP-9 in esophageal squamous cell carcinoma: a meta-analysis. Chinese Journal of Cancer Research. 2013;25(6):637–645. doi: 10.3978/j.issn.1000-9604.2013.11.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi J. I., Seymour G. J. Vaccines against periodontitis: a forward-looking review. Journal of Periodontal & Implant Science. 2010;40(4):153–163. doi: 10.5051/jpis.2010.40.4.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kinane D. F., Podmore M., Ebersole J. Etiopathogenesis of periodontitis in children and adolescents. Periodontology 2000. 2001;26(1):54–91. doi: 10.1034/j.1600-0757.2001.2260104.x. [DOI] [PubMed] [Google Scholar]
- 15.Petersen P. E., Ogawa H. The global burden of periodontal disease: towards integration with chronic disease prevention and control. Periodontology 2000. 2012;60(1):15–39. doi: 10.1111/j.1600-0757.2011.00425.x. [DOI] [PubMed] [Google Scholar]
- 16.Holm-Pedersen P., Walls A., Ship J. A. Textbook of Geriatric Dentistry. Third Edition. Wiley Blackwell; 2015. [Google Scholar]
- 17.Eke P. I., Dye B. A., Wei L., et al. Prevalence of periodontitis in adults in the United States: 2009 and 2010. Journal of Dental Research. 2012;91(10):914–920. doi: 10.1177/0022034512457373. [DOI] [PubMed] [Google Scholar]
- 18.Schultz-Haudt S., Bibby B. G., Bruce M. A. Tissue-destructive products of gingival bacteria from nonspecific gingivitis. Journal of Dental Research. 1954;33(5):624–631. doi: 10.1177/00220345540330050601. [DOI] [PubMed] [Google Scholar]
- 19.Schultz-Haudt S., Bruce M. A., Bibby B. G. Bacterial factors in nonspecific gingivitis. Journal of Dental Research. 1954;33(4):454–458. doi: 10.1177/00220345540330040301. [DOI] [PubMed] [Google Scholar]
- 20.Socransky S. S., Haffajee A. D., Smith C., et al. Use of checkerboard DNA-DNA hybridization to study complex microbial ecosystems. Oral Microbiology and Immunology. 2004;19(6):352–362. doi: 10.1111/j.1399-302x.2004.00168.x. [DOI] [PubMed] [Google Scholar]
- 21.Marsh P. D. Are dental diseases examples of ecological catastrophes? Microbiology. 2003;149(2):279–294. doi: 10.1099/mic.0.26082-0. [DOI] [PubMed] [Google Scholar]
- 22.Marsh P. D., Head D. A., Devine D. A. Ecological approaches to oral biofilms: control without killing. Caries Research. 2015;49(1):46–54. doi: 10.1159/000377732. [DOI] [PubMed] [Google Scholar]
- 23.Griffen A. L., Beall C. J., Campbell J. H., et al. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. The ISME Journal. 2012;6(6):1176–1185. doi: 10.1038/ismej.2011.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hong B. Y., Furtado Araujo M. V., Strausbaugh L. D., Terzi E., Ioannidou E., Diaz P. I. Microbiome profiles in periodontitis in relation to host and disease characteristics. PLoS One. 2015;10(5, article e0127077) doi: 10.1371/journal.pone.0127077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirst M. E., Li E. C., Alfant B., et al. Dysbiosis and alterations in predicted functions of the subgingival microbiome in chronic periodontitis. Applied and Environmental Microbiology. 2015;81(2):783–793. doi: 10.1128/AEM.02712-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diaz P. I., Hoare A., Hong B. Y. Subgingival microbiome shifts and community dynamics in periodontal diseases. Journal of the California Dental Association. 2016;44(7):421–435. [PubMed] [Google Scholar]
- 27.Hajishengallis G., Liang S., Payne M. A., et al. Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host & Microbe. 2011;10(5):497–506. doi: 10.1016/j.chom.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hajishengallis G., Lamont R. J. Beyond the red complex and into more complexity: the polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Molecular Oral Microbiology. 2012;27(6):409–419. doi: 10.1111/j.2041-1014.2012.00663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kolenbrander P. E., Andersen R. N., Moore L. V. Coaggregation of Fusobacterium nucleatum, Selenomonas flueggei, Selenomonas infelix, Selenomonas noxia, and Selenomonas sputigena with strains from 11 genera of oral bacteria. Infection and Immunity. 1989;57(10):3194–3203. doi: 10.1128/iai.57.10.3194-3203.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rickard A. H., Gilbert P., High N. J., Kolenbrander P. E., Handley P. S. Bacterial coaggregation: an integral process in the development of multi-species biofilms. Trends in Microbiology. 2003;11(2):94–100. doi: 10.1016/S0966-842X(02)00034-3. [DOI] [PubMed] [Google Scholar]
- 31.Karched M., Bhardwaj R. G., Asikainen S. E. Coaggregation and biofilm growth of Granulicatella spp. with Fusobacterium nucleatum and Aggregatibacter actinomycetemcomitans. BMC Microbiology. 2015;15(1):p. 114. doi: 10.1186/s12866-015-0439-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okuda T., Kokubu E., Kawana T., Saito A., Okuda K., Ishihara K. Synergy in biofilm formation between Fusobacterium nucleatum and Prevotella species. Anaerobe. 2012;18(1):110–116. doi: 10.1016/j.anaerobe.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 33.Mutha N. V. R., Mohammed W. K., Krasnogor N., Tan G. Y. A., Choo S. W., Jakubovics N. S. Transcriptional responses of Streptococcus gordonii and Fusobacterium nucleatum to coaggregation. Molecular Oral Microbiology. 2018;33(6):450–464. doi: 10.1111/omi.12248. [DOI] [PubMed] [Google Scholar]
- 34.Wu T., Cen L., Kaplan C., et al. Cellular components mediating coadherence of Candida albicans and Fusobacterium nucleatum. Journal of Dental Research. 2015;94(10):1432–1438. doi: 10.1177/0022034515593706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bradshaw D. J., Marsh P. D., Watson G. K., Allison C. Role of Fusobacterium nucleatum and coaggregation in anaerobe survival in planktonic and biofilm oral microbial communities during aeration. Infection and Immunity. 1998;66(10):4729–4732. doi: 10.1128/iai.66.10.4729-4732.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kolenbrander P. E., London J. Adhere today, here tomorrow: oral bacterial adherence. Journal of Bacteriology. 1993;175(11):3247–3252. doi: 10.1128/jb.175.11.3247-3252.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kolenbrander P. E., Palmer R. J., Rickard A. H., Jakubovics N. S., Chalmers N. I., Diaz P. I. Bacterial interactions and successions during plaque development. Periodontology 2000. 2006;42(1):47–79. doi: 10.1111/j.1600-0757.2006.00187.x. [DOI] [PubMed] [Google Scholar]
- 38.Takahashl N., Saito K., Schachtele C. F., Yamada T. Acid tolerance and acid-neutralizing activity of Porphyromonas gingivalis, Prevotella intermedia and Fusobacterium nucleatum. Oral Microbiology and Immunology. 1997;12(6):323–328. doi: 10.1111/j.1399-302X.1997.tb00733.x. [DOI] [PubMed] [Google Scholar]
- 39.Bowen W. H., Burne R. A., Wu H., Koo H. Oral biofilms: pathogens, matrix, and polymicrobial interactions in microenvironments. Trends in Microbiology. 2018;26(3):229–242. doi: 10.1016/j.tim.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Benakanakere M., Kinane D. F. Innate cellular responses to the periodontal biofilm. Frontiers of Oral Biology. 2012;15:41–55. doi: 10.1159/000329670. [DOI] [PubMed] [Google Scholar]
- 41.Graves D. Cytokines that promote periodontal tissue destruction. Journal of Periodontology. 2008;79(8s):1585–1591. doi: 10.1902/jop.2008.080183. [DOI] [PubMed] [Google Scholar]
- 42.Vernal R., Dutzan N., Hernández M., et al. High expression levels of receptor activator of nuclear factor-kappa B ligand associated with human chronic periodontitis are mainly secreted by CD4+ T lymphocytes. Journal of Periodontology. 2006;77(10):1772–1780. doi: 10.1902/jop.2006.050376. [DOI] [PubMed] [Google Scholar]
- 43.Aranha A. M. F., Repeke C. E., Garlet T. P., et al. Evidence supporting a protective role for th9 and th22 cytokines in human and experimental periapical lesions. Journal of Endodontia. 2013;39(1):83–87. doi: 10.1016/j.joen.2012.10.015. [DOI] [PubMed] [Google Scholar]
- 44.Gemmell E., Seymour G. J. Immunoregulatory control of Th1/Th2 cytokine profiles in periodontal disease. Periodontology 2000. 2004;35(1):21–41. doi: 10.1111/j.0906-6713.2004.003557.x. [DOI] [PubMed] [Google Scholar]
- 45.Noh M. K., Jung M., Kim S. H., et al. Assessment of IL-6, IL-8 and TNF-α levels in the gingival tissue of patients with periodontitis. Experimental and Therapeutic Medicine. 2013;6(3):847–851. doi: 10.3892/etm.2013.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davanian H., Stranneheim H., Båge T., et al. Gene expression profiles in paired gingival biopsies from periodontitis-affected and healthy tissues revealed by massively parallel sequencing. PLoS One. 2012;7(9, article e46440) doi: 10.1371/journal.pone.0046440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lomba K. S. B., de Souza Breves Beiler T. F. C., Sete M. R. C., Pires F. R., da Silva Figueredo C. M. Use of minimally invasive gingival biopsies in the study of inflammatory mediators expression and their correlation with gingival fluid in patients with chronic periodontitis. Indian Journal of Dental Research. 2015;26(2):126–130. doi: 10.4103/0970-9290.159134. [DOI] [PubMed] [Google Scholar]
- 48.Herrero E. R., Fernandes S., Verspecht T., et al. Dysbiotic biofilms deregulate the periodontal inflammatory response. Journal of Dental Research. 2018;97(5):547–555. doi: 10.1177/0022034517752675. [DOI] [PubMed] [Google Scholar]
- 49.Ramage G., Lappin D. F., Millhouse E., et al. The epithelial cell response to health and disease associated oral biofilm models. Journal of Periodontal Research. 2017;52(3):325–333. doi: 10.1111/jre.12395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bakthavatchalu V., Meka A., Mans J. J., et al. Polymicrobial periodontal pathogen transcriptomes in calvarial bone and soft tissue. Molecular Oral Microbiology. 2011;26(5):303–320. doi: 10.1111/j.2041-1014.2011.00619.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beklen A., Sorsa T., Konttinen Y. T. Toll-like receptors 2 and 5 in human gingival epithelial cells co-operate with T-cell cytokine interleukin-17. Oral Microbiology and Immunology. 2009;24(1):38–42. doi: 10.1111/j.1399-302X.2008.00473.x. [DOI] [PubMed] [Google Scholar]
- 52.Shin J. E., Kim Y. S., Oh J. E., Min B. M., Choi Y. Treponema denticola suppresses expression of human β-defensin-3 in gingival epithelial cells through inhibition of the toll-like receptor 2 axis. Infection and Immunity. 2010;78(2):672–679. doi: 10.1128/IAI.00808-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tanabe S. I., Bodet C., Grenier D. Treponema denticola peptidoglycan induces the production of inflammatory mediators and matrix metalloproteinase 9 in macrophage-like cells. Journal of Periodontal Research. 2009;44(4):503–510. doi: 10.1111/j.1600-0765.2008.01141.x. [DOI] [PubMed] [Google Scholar]
- 54.Miyamoto M., Ishihara K., Okuda K. The Treponema denticola surface protease dentilisin degrades interleukin-1β (IL-1β), IL-6, and tumor necrosis factor alpha. Infection and Immunity. 2006;74(4):2462–2467. doi: 10.1128/IAI.74.4.2462-2467.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chukkapalli S. S., Rivera-Kweh M. F., Velsko I. M., et al. Chronic oral infection with major periodontal bacteria Tannerella forsythia modulates systemic atherosclerosis risk factors and inflammatory markers. Pathogens and Disease. 2015;73(3) doi: 10.1093/femspd/ftv009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee S. W., Sabet M., Um H. S., Yang J., Kim H. C., Zhu W. Identification and characterization of the genes encoding a unique surface (S-) layer of Tannerella forsythia. Gene. 2006;371(1):102–111. doi: 10.1016/j.gene.2005.11.027. [DOI] [PubMed] [Google Scholar]
- 57.Settem R. P., Honma K., Nakajima T., et al. A bacterial glycan core linked to surface (S)-layer proteins modulates host immunity through Th17 suppression. Mucosal Immunology. 2013;6(2):415–426. doi: 10.1038/mi.2012.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moffatt C. E., Whitmore S. E., Griffen A. L., Leys E. J., Lamont R. J. Filifactor alocis interactions with gingival epithelial cells. Molecular Oral Microbiology. 2011;26(6):365–373. doi: 10.1111/j.2041-1014.2011.00624.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang Q., Jotwani R., le J., et al. Filifactor alocis infection and inflammatory responses in the mouse subcutaneous chamber model. Infection and Immunity. 2014;82(3):1205–1212. doi: 10.1128/IAI.01434-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shouda T., Yoshida T., Hanada T., et al. Induction of the cytokine signal regulator SOCS3/CIS3 as a therapeutic strategy for treating inflammatory arthritis. The Journal of Clinical Investigation. 2001;108(12):1781–1788. doi: 10.1172/JCI13568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ahmad S. F., Ansari M. A., Zoheir K. M. A., et al. Regulation of TNF-α and NF-κB activation through the JAK/STAT signaling pathway downstream of histamine 4 receptor in a rat model of LPS-induced joint inflammation. Immunobiology. 2015;220(7):889–898. doi: 10.1016/j.imbio.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 62.Jiang D., Li D., Cao L., et al. Positive feedback regulation of proliferation in vascular smooth muscle cells stimulated by lipopolysaccharide is mediated through the TLR 4/Rac1/Akt pathway. PLoS One. 2014;9(3, article e92398) doi: 10.1371/journal.pone.0092398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lieberthal W., Levine J. S. The role of the mammalian target of rapamycin (mTOR) in renal disease. Journal of the American Society of Nephrology. 2009;20(12):2493–2502. doi: 10.1681/ASN.2008111186. [DOI] [PubMed] [Google Scholar]
- 64.Kozarov E. Bacterial invasion of vascular cell types: vascular infectology and atherogenesis. Future Cardiology. 2012;8(1):123–138. doi: 10.2217/fca.11.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Choi C. H., Spooner R., DeGuzman J., Koutouzis T., Ojcius D. M., Yilmaz Ö. Porphyromonas gingivalis-nucleoside-diphosphate-kinase inhibits ATP-induced reactive-oxygen-species via P2X7 receptor/NADPH-oxidase signalling and contributes to persistence. Cellular Microbiology. 2013;15(6):961–976. doi: 10.1111/cmi.12089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Park E., Na H. S., Song Y. R., Shin S. Y., Kim Y. M., Chung J. Activation of NLRP3 and AIM2 inflammasomes by Porphyromonas gingivalis infection. Infection and Immunity. 2014;82(1):112–123. doi: 10.1128/IAI.00862-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Soto C., Bugueño I., Hoare A., et al. The Porphyromonas gingivalis O antigen is required for inhibition of apoptosis in gingival epithelial cells following bacterial infection. Journal of Periodontal Research. 2016;51(4):518–528. doi: 10.1111/jre.12331. [DOI] [PubMed] [Google Scholar]
- 68.Dixon D. R., Darveau R. P. Lipopolysaccharide heterogeneity: innate host responses to bacterial modification of lipid A structure. Journal of Dental Research. 2005;84(7):584–595. doi: 10.1177/154405910508400702. [DOI] [PubMed] [Google Scholar]
- 69.Herath T. D. K., Darveau R. P., Seneviratne C. J., Wang C. Y., Wang Y., Jin L. Tetra- and penta-acylated lipid A structures of Porphyromonas gingivalis LPS differentially activate TLR4-mediated NF-κB signal transduction cascade and immuno-inflammatory response in human gingival fibroblasts. PLoS One. 2013;8(3, article e58496) doi: 10.1371/journal.pone.0058496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sugawara S., Nemoto E., Tada H., Miyake K., Imamura T., Takada H. Proteolysis of human monocyte CD14 by cysteine proteinases (gingipains) from Porphyromonas gingivalis leading to lipopolysaccharide hyporesponsiveness. The Journal of Immunology. 2000;165(1):411–418. doi: 10.4049/jimmunol.165.1.411. [DOI] [PubMed] [Google Scholar]
- 71.Tada H., Sugawara S., Nemoto E., et al. Proteolysis of ICAM-1 on human oral epithelial cells by gingipains. Journal of Dental Research. 2003;82(10):796–801. doi: 10.1177/154405910308201007. [DOI] [PubMed] [Google Scholar]
- 72.Banbula A., Bugno M., Kuster A., Heinrich P. C., Travis J., Potempa J. Rapid and efficient inactivation of IL-6 gingipains, lysine- and arginine-specific proteinases from Porphyromonas gingivalis. Biochemical and Biophysical Research Communications. 1999;261(3):598–602. doi: 10.1006/bbrc.1999.1075. [DOI] [PubMed] [Google Scholar]
- 73.Mikolajczyk-Pawlinska J., Travis J., Potempa J. Modulation of interleukin-8 activity by gingipains from Porphyromonas gingivalis: implications for pathogenicity of periodontal disease. FEBS Letters. 1998;440(3):282–286. doi: 10.1016/S0014-5793(98)01461-6. [DOI] [PubMed] [Google Scholar]
- 74.Yun P. L. W., DeCarlo A. A., Collyer C., Hunter N. Modulation of an interleukin-12 and gamma interferon synergistic feedback regulatory cycle of T-cell and monocyte cocultures by Porphyromonas gingivalis lipopolysaccharide in the absence or presence of cysteine proteinases. Infection and Immunity. 2002;70(10):5695–5705. doi: 10.1128/IAI.70.10.5695-5705.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Calkins C. C., Platt K., Potempa J., Travis J. Inactivation of tumor necrosis factor-alpha by proteinases (gingipains) from the periodontal pathogen, Porphyromonas gingivalis. Implications of immune evasion. Journal of Biological Chemistry. 1998;273(12):6611–6614. doi: 10.1074/jbc.273.12.6611. [DOI] [PubMed] [Google Scholar]
- 76.Krisanaprakornkit S., Kimball J. R., Weinberg A., Darveau R. P., Bainbridge B. W., Dale B. A. Inducible expression of human β-defensin 2 by Fusobacterium nucleatum in oral epithelial cells: multiple signaling pathways and role of commensal bacteria in innate immunity and the epithelial barrier. Infection and Immunity. 2000;68(5):2907–2915. doi: 10.1128/IAI.68.5.2907-2915.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huang G. T.-J., Zhang H. B., Dang H. N., Haake S. K. Differential regulation of cytokine genes in gingival epithelial cells challenged by Fusobacterium nucleatum and Porphyromonas gingivalis. Microbial Pathogenesis. 2004;37(6):303–312. doi: 10.1016/j.micpath.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 78.Bui F. Q., Johnson L., Roberts J. A., et al. Fusobacterium nucleatum infection of gingival epithelial cells leads to NLRP3 inflammasome-dependent secretion of IL-1β and the danger signals ASC and HMGB1. Cellular Microbiology. 2016;18(7):970–981. doi: 10.1111/cmi.12560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Andersson U., Tracey K. J. HMGB1 is a therapeutic target for sterile inflammation and infection. Annual Review of Immunology. 2011;29(1):139–162. doi: 10.1146/annurev-immunol-030409-101323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lamkanfi M., Sarkar A., Vande Walle L., et al. Inflammasome-dependent release of the alarmin HMGB1 in endotoxemia. Journal of Immunology. 2010;185(7):4385–4392. doi: 10.4049/jimmunol.1000803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Franklin B. S., Bossaller L., de Nardo D., et al. The adaptor ASC has extracellular and ‘prionoid’ activities that propagate inflammation. Nature Immunology. 2014;15(8):727–737. doi: 10.1038/ni.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Polak D., Wilensky A., Shapira L., et al. Mouse model of experimental periodontitis induced by Porphyromonas gingivalis/Fusobacterium nucleatum infection: bone loss and host response. Journal of Clinical Periodontology. 2009;36(5):406–410. doi: 10.1111/j.1600-051X.2009.01393.x. [DOI] [PubMed] [Google Scholar]
- 83.Ahn S. H., Song J. E., Kim S., et al. NOX1/2 activation in human gingival fibroblasts by Fusobacterium nucleatum facilitates attachment of Porphyromonas gingivalis. Archives of Microbiology. 2016;198(6):573–583. doi: 10.1007/s00203-016-1223-7. [DOI] [PubMed] [Google Scholar]
- 84.Li Y., Guo H., Wang X., Lu Y., Yang C., Yang P. Coinfection with Fusobacterium nucleatum can enhance the attachment and invasion of Porphyromonas gingivalis or Aggregatibacter actinomycetemcomitans to human gingival epithelial cells. Archives of Oral Biology. 2015;60(9):1387–1393. doi: 10.1016/j.archoralbio.2015.06.017. [DOI] [PubMed] [Google Scholar]
- 85.Janket S. J., Baird A. E., Chuang S. K., Jones J. A. Meta-analysis of periodontal disease and risk of coronary heart disease and stroke. Oral Surgery, Oral Medicine, Oral Pathology, Oral Radiology, and Endodontics. 2003;95(5):559–569. doi: 10.1067/moe.2003.107. [DOI] [PubMed] [Google Scholar]
- 86.Peng C. H., Yang Y. S., Chan K. C., Kornelius E., Chiou J. Y., Huang C. N. Periodontal treatment and the risks of cardiovascular disease in patients with type 2 diabetes: a retrospective cohort study. Internal Medicine. 2017;56(9):1015–1021. doi: 10.2169/internalmedicine.56.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mustapha I. Z., Debrey S., Oladubu M., Ugarte R. Markers of systemic bacterial exposure in periodontal disease and cardiovascular disease risk: a systematic review and meta-analysis. Journal of Periodontology. 2007;78(12):2289–2302. doi: 10.1902/jop.2007.070140. [DOI] [PubMed] [Google Scholar]
- 88.Damgaard C., Reinholdt J., Enevold C., Fiehn N. E., Nielsen C. H., Holmstrup P. Immunoglobulin G antibodies against Porphyromonas gingivalis or Aggregatibacter actinomycetemcomitans in cardiovascular disease and periodontitis. Journal of Oral Microbiology. 2017;9(1, article 1374154) doi: 10.1080/20002297.2017.1374154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bale B. F., Doneen A. L., Vigerust D. J. High-risk periodontal pathogens contribute to the pathogenesis of atherosclerosis. Postgraduate Medical Journal. 2017;93(1098):215–220. doi: 10.1136/postgradmedj-2016-134279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ashigaki N., Suzuki J. I., Aoyama N., et al. The periodontal pathogen Aggregatibacter actinomycetemcomitans affects experimental autoimmune myocarditis in mice. International Heart Journal. 2013;54(6):412–416. doi: 10.1536/ihj.54.412. [DOI] [PubMed] [Google Scholar]
- 91.Detert J., Pischon N., Burmester G. R., Buttgereit F. The association between rheumatoid arthritis and periodontal disease. Arthritis Research & Therapy. 2010;12(5):p. 218. doi: 10.1186/ar3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mikuls T. R., Payne J. B., Yu F., et al. Periodontitis and Porphyromonas gingivalis in patients with rheumatoid arthritis. Arthritis & Rhematology. 2014;66(5):1090–1100. doi: 10.1002/art.38348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schmickler J., Rupprecht A., Patschan S., et al. Cross-sectional evaluation of periodontal status and microbiologic and rheumatoid parameters in a large cohort of patients with rheumatoid arthritis. Journal of Periodontology. 2017;88(4):368–379. doi: 10.1902/jop.2016.160355. [DOI] [PubMed] [Google Scholar]
- 94.Dizdar O., Hayran M., Guven D. C., et al. Increased cancer risk in patients with periodontitis. Current Medical Research and Opinion. 2017;33(12):2195–2200. doi: 10.1080/03007995.2017.1354829. [DOI] [PubMed] [Google Scholar]
- 95.Sfreddo C. S., Maier J., de David S. C., Susin C., Moreira C. H. C. Periodontitis and breast cancer: a case-control study. Community Dentistry and Oral Epidemiology. 2017;45(6):545–551. doi: 10.1111/cdoe.12318. [DOI] [PubMed] [Google Scholar]
- 96.Xie W. Z., Jin Y. H., Leng W. D., Wang X. H., Zeng X. T., BPSC investigators Periodontal disease and risk of bladder cancer: a meta-analysis of 298476 participants. Frontiers in Physiology. 2018;9:p. 979. doi: 10.3389/fphys.2018.00979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Salazar C. R., Francois F., Li Y., et al. Association between oral health and gastric precancerous lesions. Carcinogenesis. 2012;33(2):399–403. doi: 10.1093/carcin/bgr284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shakeri R., Malekzadeh R., Etemadi A., et al. Association of tooth loss and oral hygiene with risk of gastric adenocarcinoma. Cancer Prevention Research. 2013;6(5):477–482. doi: 10.1158/1940-6207.CAPR-12-0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Garrote L. F., Herrero R., Reyes R. M. O., et al. Risk factors for cancer of the oral cavity and oro-pharynx in Cuba. British Journal of Cancer. 2001;85(1):46–54. doi: 10.1054/bjoc.2000.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Marshall J. R., Graham S., Haughey B. P., et al. Smoking, alcohol, dentition and diet in the epidemiology of oral cancer. European Journal of Cancer Part B: Oral Oncology. 1992;28(1):9–15. doi: 10.1016/0964-1955(92)90005-L. [DOI] [PubMed] [Google Scholar]
- 101.Rosenquist K., Wennerberg J., Schildt E. B., Bladström A., Göran Hansson B., Andersson G. Oral status, oral infections and some lifestyle factors as risk factors for oral and oropharyngeal squamous cell carcinoma. A population-based case-control study in southern Sweden. Acta Oto-Laryngologica. 2005;125(12):1327–1336. doi: 10.1080/00016480510012273. [DOI] [PubMed] [Google Scholar]
- 102.Zheng T., Boyle P., Hu H., et al. Dentition, oral hygiene, and risk of oral cancer: a case-control study in Beijing, People’s Republic of China. Cancer Causes & Control. 1990;1(3):235–241. doi: 10.1007/BF00117475. [DOI] [PubMed] [Google Scholar]
- 103.Lee D., Jung K. U., Kim H. O., Kim H., Chun H. K. Association between oral health and colorectal adenoma in a screening population. Medicine. 2018;97(37, article e12244) doi: 10.1097/MD.0000000000012244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.de Moraes R. C., Dias F. L., da Silva Figueredo C. M., Fischer R. G. Association between chronic periodontitis and oral/oropharyngeal cancer. Brazilian Dental Journal. 2016;27(3):261–266. doi: 10.1590/0103-6440201600754. [DOI] [PubMed] [Google Scholar]
- 105.Chung S. D., Tsai M. C., Huang C. C., Kao L. T., Chen C. H. A population-based study on the associations between chronic periodontitis and the risk of cancer. International Journal of Clinical Oncology. 2016;21(2):219–223. doi: 10.1007/s10147-015-0884-6. [DOI] [PubMed] [Google Scholar]
- 106.Maisonneuve P., Amar S., Lowenfels A. B. Periodontal disease, edentulism, and pancreatic cancer: a meta-analysis. Annals of Oncology. 2017;28(5):985–995. doi: 10.1093/annonc/mdx019. [DOI] [PubMed] [Google Scholar]
- 107.Bundgaard T., Wildt J., Frydenberg M., Elbrond O., Nielsen J. E. Case-control study of squamous cell cancer of the oral cavity in Denmark. Cancer Causes & Control. 1995;6(1):57–67. doi: 10.1007/BF00051681. [DOI] [PubMed] [Google Scholar]
- 108.Tezal M., Grossi S. G., Genco R. J. Is periodontitis associated with oral neoplasms? Journal of Periodontology. 2005;76(3):406–410. doi: 10.1902/jop.2005.76.3.406. [DOI] [PubMed] [Google Scholar]
- 109.Tezal M., Sullivan M. A., Reid M. E., et al. Chronic periodontitis and the risk of tongue cancer. Archives of Otolaryngology – Head & Neck Surgery. 2007;133(5):450–454. doi: 10.1001/archotol.133.5.450. [DOI] [PubMed] [Google Scholar]
- 110.Sun J., Zhou M., Salazar C. R., et al. Chronic periodontal disease, periodontal pathogen colonization, and increased risk of precancerous gastric lesions. Journal of Periodontology. 2017;88(11):1124–1134. doi: 10.1902/jop.2017.160829. [DOI] [PubMed] [Google Scholar]
- 111.Yao Q. W., Zhou D. S., Peng H. J., Ji P., Liu D. S. Association of periodontal disease with oral cancer: a meta-analysis. Tumour Biology. 2014;35(7):7073–7077. doi: 10.1007/s13277-014-1951-8. [DOI] [PubMed] [Google Scholar]
- 112.Li X., Kolltveit K. M., Tronstad L., Olsen I. Systemic diseases caused by oral infection. Clinical Microbiology Reviews. 2000;13(4):547–558. doi: 10.1128/CMR.13.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Nasidze I., Li J., Quinque D., Tang K., Stoneking M. Global diversity in the human salivary microbiome. Genome Research. 2009;19(4):636–643. doi: 10.1101/gr.084616.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Socransky S. S., Haffajee A. D. Periodontal microbial ecology. Periodontology 2000. 2005;38(1):135–187. doi: 10.1111/j.1600-0757.2005.00107.x. [DOI] [PubMed] [Google Scholar]
- 115.Brook I. The role of anaerobic bacteria in bacteremia. Anaerobe. 2010;16(3):183–189. doi: 10.1016/j.anaerobe.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 116.Cobe H. M. Transitory bacteremia. Oral Surgery, Oral Medicine, and Oral Pathology. 1954;7(6):609–615. doi: 10.1016/0030-4220(54)90071-7. [DOI] [PubMed] [Google Scholar]
- 117.Tomás I., Diz P., Tobías A., Scully C., Donos N. Periodontal health status and bacteraemia from daily oral activities: systematic review/meta-analysis. Journal of Clinical Periodontology. 2012;39(3):213–228. doi: 10.1111/j.1600-051X.2011.01784.x. [DOI] [PubMed] [Google Scholar]
- 118.Lafaurie G. I., Mayorga-Fayad I., Torres M. F., et al. Periodontopathic microorganisms in peripheric blood after scaling and root planing. Journal of Clinical Periodontology. 2007;34(10):873–879. doi: 10.1111/j.1600-051X.2007.01125.x. [DOI] [PubMed] [Google Scholar]
- 119.Goldstein E. J. C. Anaerobic bacteremia. Clinical Infectious Diseases. 1996;23(Supplement 1):S97–101. doi: 10.1093/clinids/23.Supplement_1.S97. [DOI] [PubMed] [Google Scholar]
- 120.Finegold S. M. Anaerobic Bacteria in Human Disease. New York: Academic Press; 1977. [Google Scholar]
- 121.Hujoel P. P., White B. A., Garcia R. I., Listgarten M. A. The dentogingival epithelial surface area revisited. Journal of Periodontal Research. 2001;36(1):48–55. doi: 10.1034/j.1600-0765.2001.00011.x. [DOI] [PubMed] [Google Scholar]
- 122.Herzberg M. C., Weyer M. W. Dental plaque, platelets, and cardiovascular diseases. Annals of Periodontology. 1998;3(1):151–160. doi: 10.1902/annals.1998.3.1.151. [DOI] [PubMed] [Google Scholar]
- 123.Afra K., Laupland K., Leal J., Lloyd T., Gregson D. Incidence, risk factors, and outcomes of Fusobacterium species bacteremia. BMC Infectious Diseases. 2013;13(1):p. 264. doi: 10.1186/1471-2334-13-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yang C. C., Ye J. J., Hsu P. C., et al. Characteristics and outcomes of Fusobacterium nucleatum bacteremia—a 6-year experience at a tertiary care hospital in northern Taiwan. Diagnostic Microbiology and Infectious Disease. 2011;70(2):167–174. doi: 10.1016/j.diagmicrobio.2010.12.017. [DOI] [PubMed] [Google Scholar]
- 125.Denes E., Barraud O. Fusobacterium nucleatum infections: clinical spectrum and bacteriological features of 78 cases. Infection. 2016;44(4):475–481. doi: 10.1007/s15010-015-0871-x. [DOI] [PubMed] [Google Scholar]
- 126.Yusuf E., Wybo I., Pierard D. Case series of patients with Fusobacterium nucleatum bacteremia with emphasis on the presence of cancer. Anaerobe. 2016;39:1–3. doi: 10.1016/j.anaerobe.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 127.Flanagan L., Schmid J., Ebert M., et al. Fusobacterium nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome. European Journal of Clinical Microbiology & Infectious Diseases. 2014;33(8):1381–1390. doi: 10.1007/s10096-014-2081-3. [DOI] [PubMed] [Google Scholar]
- 128.Shang F. M., Liu H. L. Fusobacterium nucleatum and colorectal cancer: a review. World Journal of Gastrointestinal Oncology. 2018;10(3):71–81. doi: 10.4251/wjgo.v10.i3.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kostic A. D., Gevers D., Pedamallu C. S., et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Research. 2012;22(2):292–298. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Warren R. L., Freeman D. J., Pleasance S., et al. Co-occurrence of anaerobic bacteria in colorectal carcinomas. Microbiome. 2013;1(1):p. 16. doi: 10.1186/2049-2618-1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Mima K., Nishihara R., Qian Z. R., et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut. 2016;65(12):1973–1980. doi: 10.1136/gutjnl-2015-310101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Drewes J. L., White J. R., Dejea C. M., et al. High-resolution bacterial 16S rRNA gene profile meta-analysis and biofilm status reveal common colorectal cancer consortia. NPJ Biofilms and Microbiomes. 2017;3(1):p. 34. doi: 10.1038/s41522-017-0040-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Marchesi J. R., Dutilh B. E., Hall N., et al. Towards the human colorectal cancer microbiome. PLoS One. 2011;6(5, article e20447) doi: 10.1371/journal.pone.0020447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Chen W., Liu F., Ling Z., Tong X., Xiang C. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS One. 2012;7(6, article e39743) doi: 10.1371/journal.pone.0039743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Castellarin M., Warren R. L., Freeman J. D., et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Research. 2012;22(2):299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hooper S. J., Crean S. J., Fardy M. J., et al. A molecular analysis of the bacteria present within oral squamous cell carcinoma. Journal of Medical Microbiology. 2007;56(12):1651–1659. doi: 10.1099/jmm.0.46918-0. [DOI] [PubMed] [Google Scholar]
- 137.Pushalkar S., Ji X., Li Y., et al. Comparison of oral microbiota in tumor and non-tumor tissues of patients with oral squamous cell carcinoma. BMC Microbiology. 2012;12(1):p. 144. doi: 10.1186/1471-2180-12-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zhao H., Chu M., Huang Z., et al. Variations in oral microbiota associated with oral cancer. Scientific Reports. 2017;7(1):p. 11773. doi: 10.1038/s41598-017-11779-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ahn J., Segers S., Hayes R. B. Periodontal disease, Porphyromonas gingivalis serum antibody levels and orodigestive cancer mortality. Carcinogenesis. 2012;33(5):1055–1058. doi: 10.1093/carcin/bgs112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Michaud D. S. Role of bacterial infections in pancreatic cancer. Carcinogenesis. 2013;34(10):2193–2197. doi: 10.1093/carcin/bgt249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Nagy K. N., Sonkodi I., Szöke I., Nagy E., Newman H. N. The microflora associated with human oral carcinomas. Oral Oncology. 1998;34(4):304–308. doi: 10.1016/S1368-8375(98)80012-2. [DOI] [PubMed] [Google Scholar]
- 142.Katz J., Onate M. D., Pauley K. M., Bhattacharyya I., Cha S. Presence of Porphyromonas gingivalis in gingival squamous cell carcinoma. International Journal of Oral Science. 2011;3(4):209–215. doi: 10.4248/IJOS11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Baek K., Ji S., Choi Y. Complex intratissue microbiota forms biofilms in periodontal lesions. Journal of Dental Research. 2018;97(2):192–200. doi: 10.1177/0022034517732754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Yost S., Stashenko P., Choi Y., et al. Increased virulence of the oral microbiome in oral squamous cell carcinoma revealed by metatranscriptome analyses. International Journal of Oral Science. 2018;10(4):p. 32. doi: 10.1038/s41368-018-0037-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kuboniwa M., Hendrickson E. L., Xia Q., et al. Proteomics of Porphyromonas gingivalis within a model oral microbial community. BMC Microbiology. 2009;9(1):p. 98. doi: 10.1186/1471-2180-9-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Frias-Lopez J., Duran-Pinedo A. Effect of periodontal pathogens on the metatranscriptome of a healthy multispecies biofilm model. Journal of Bacteriology. 2012;194(8):2082–2095. doi: 10.1128/JB.06328-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Yost S., Duran-Pinedo A. E., Teles R., Krishnan K., Frias-Lopez J. Functional signatures of oral dysbiosis during periodontitis progression revealed by microbial metatranscriptome analysis. Genome Medicine. 2015;7(1):p. 27. doi: 10.1186/s13073-015-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Jorth P., Turner K. H., Gumus P., Nizam N., Buduneli N., Whiteley M. Metatranscriptomics of the human oral microbiome during health and disease. MBio. 2014;5(2):e01012–e01014. doi: 10.1128/mbio.01012-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Deng Z. L., Sztajer H., Jarek M., Bhuju S., Wagner-Döbler I. Worlds apart-transcriptome profiles of key oral microbes in the periodontal pocket compared to single laboratory culture reflect synergistic interactions. Frontiers in Microbiology. 2018;9:p. 124. doi: 10.3389/fmicb.2018.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Fan X., Alekseyenko A. V., Wu J., et al. Human oral microbiome and prospective risk for pancreatic cancer: a population-based nested case-control study. Gut. 2018;67(1):120–127. doi: 10.1136/gutjnl-2016-312580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Listyarifah D., Nieminen M. T., Mäkinen L. K., et al. Treponema denticola chymotrypsin-like proteinase is present in early-stage mobile tongue squamous cell carcinoma and related to the clinicopathological features. Journal of Oral Pathology & Medicine. 2018;47(8):764–772. doi: 10.1111/jop.12729. [DOI] [PubMed] [Google Scholar]
- 152.Narikiyo M., Tanabe C., Yamada Y., et al. Frequent and preferential infection of Treponema denticola, Streptococcus mitis, and Streptococcus anginosus in esophageal cancers. Cancer Science. 2004;95(7):569–574. doi: 10.1111/j.1349-7006.2004.tb02488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Arimatsu K., Yamada H., Miyazawa H., et al. Oral pathobiont induces systemic inflammation and metabolic changes associated with alteration of gut microbiota. Scientific Reports. 2014;4(1, article 4828) doi: 10.1038/srep04828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Mao S., Park Y., Hasegawa Y., et al. Intrinsic apoptotic pathways of gingival epithelial cells modulated by Porphyromonas gingivalis. Cellular Microbiology. 2007;9(8):1997–2007. doi: 10.1111/j.1462-5822.2007.00931.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Sztukowska M. N., Ojo A., Ahmed S., et al. Porphyromonas gingivalis initiates a mesenchymal-like transition through ZEB1 in gingival epithelial cells. Cellular Microbiology. 2016;18(6):844–858. doi: 10.1111/cmi.12554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Lee J., Roberts J. A. S., Atanasova K. R., Chowdhury N., Han K., Yilmaz Ö. Human primary epithelial cells acquire an epithelial-mesenchymal-transition phenotype during long-term infection by the oral opportunistic pathogen, Porphyromonas gingivalis. Frontiers in Cellular and Infection Microbiology. 2017;7:p. 493. doi: 10.3389/fcimb.2017.00493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Yilmaz O., Yao L., Maeda K., et al. ATP scavenging by the intracellular pathogen Porphyromonas gingivalis inhibits P2X7-mediated host-cell apoptosis. Cellular Microbiology. 2008;10(4):863–875. doi: 10.1111/j.1462-5822.2007.01089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Yilmaz Ã., Sater A. A., Yao L., Koutouzis T., Pettengill M., Ojcius D. M. ATP-dependent activation of an inflammasome in primary gingival epithelial cells infected by Porphyromonas gingivalis. Cellular Microbiology. 2010;12(2):188–198. doi: 10.1111/j.1462-5822.2009.01390.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Inaba H., Amano A., Lamont R. J., Murakami Y. Involvement of protease-activated receptor 4 in over-expression of matrix metalloproteinase 9 induced by Porphyromonas gingivalis. Medical Microbiology and Immunology. 2015;204(5):605–612. doi: 10.1007/s00430-015-0389-y. [DOI] [PubMed] [Google Scholar]
- 160.Inaba H., Sugita H., Kuboniwa M., et al. Porphyromonas gingivalis promotes invasion of oral squamous cell carcinoma through induction of pro MMP9 and its activation. Cellular Microbiology. 2014;16(1):131–145. doi: 10.1111/cmi.12211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Lin F. Y., Huang C. Y., Lu H. Y., et al. The GroEL protein of Porphyromonas gingivalis accelerates tumor growth by enhancing endothelial progenitor cell function and neovascularization. Molecular Oral Microbiology. 2015;30(3):198–216. doi: 10.1111/omi.12083. [DOI] [PubMed] [Google Scholar]
- 162.Arjunan P., Meghil M. M., Pi W., et al. Oral pathobiont activates anti-apoptotic pathway, promoting both immune suppression and oncogenic cell proliferation. Scientific Reports. 2018;8(1):p. 16607. doi: 10.1038/s41598-018-35126-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Rubinstein M. R., Wang X., Liu W., Hao Y., Cai G., Han Y. W. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host & Microbe. 2013;14(2):195–206. doi: 10.1016/j.chom.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Okahashi N., Koga T., Nishihara T., Fujiwara T., Hamada S. Immunobiological properties of lipopolysaccharides isolated from Fusobacterium nucleatum and F. necrophorum. Journal of General Microbiology. 1988;134(6):1707–1715. doi: 10.1099/00221287-134-6-1707. [DOI] [PubMed] [Google Scholar]
- 165.McCoy A. N., Araújo-Pérez F., Azcárate-Peril A., Yeh J. J., Sandler R. S., Keku T. O. Fusobacterium is associated with colorectal adenomas. PLoS One. 2013;8(1, article e53653) doi: 10.1371/journal.pone.0053653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Gur C., Ibrahim Y., Isaacson B., et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity. 2015;42(2):344–355. doi: 10.1016/j.immuni.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kostic A. D., Chun E., Robertson L., et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host & Microbe. 2013;14(2):207–215. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.DiDonato J. A., Mercurio F., Karin M. NF-κB and the link between inflammation and cancer. Immunological Reviews. 2012;246(1):379–400. doi: 10.1111/j.1600-065X.2012.01099.x. [DOI] [PubMed] [Google Scholar]
- 169.Allen-Vercoe E., Jobin C. Fusobacterium and Enterobacteriaceae: important players for CRC? Immunol Lett. 2014;162(2):54–61. doi: 10.1016/j.imlet.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Ma C. T., Luo H. S., Gao F., Tang Q. C., Chen W. Fusobacterium nucleatum promotes the progression of colorectal cancer by interacting with E-cadherin. Oncology Letters. 2018;16(2):2606–2612. doi: 10.3892/ol.2018.8947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Nieminen M. T., Listyarifah D., Hagström J., et al. Treponema denticola chymotrypsin-like proteinase may contribute to orodigestive carcinogenesis through immunomodulation. British Journal of Cancer. 2018;118(3):428–434. doi: 10.1038/bjc.2017.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Jakubowska K., Pryczynicz A., Januszewska J., et al. Expressions of matrix metalloproteinases 2, 7, and 9 in carcinogenesis of pancreatic ductal adenocarcinoma. Disease Markers. 2016;2016:7. doi: 10.1155/2016/9895721.9895721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Binder Gallimidi A., Fischman S., Revach B., et al. Periodontal pathogens Porphyromonas gingivalis and Fusobacterium nucleatum promote tumor progression in an oral-specific chemical carcinogenesis model. Oncotarget. 2015;6(26):22613–22623. doi: 10.18632/oncotarget.4209. [DOI] [PMC free article] [PubMed] [Google Scholar]