Abstract
DACH1 is the human homolog of the Drosophila dachshund gene, which is involved in the development of the eye, nervous system, and limbs in the fly. Here, we systematically investigate DACH1 expression patterns during human neurodevelopment, from 5 to 21 postconceptional weeks. By immunodetection analysis, we found that DACH1 is highly expressed in the proliferating neuroprogenitors of the developing cortical ventricular and subventricular regions, while it is absent in the more differentiated cortical plate. Single-cell global transcriptional analysis revealed that DACH1 is specifically enriched in neuroepithelial and ventricular radial glia cells of the developing human neocortex. Moreover, we describe a previously unreported DACH1 expression in the human striatum, in particular in the striatal medium spiny neurons. This finding qualifies DACH1 as a new striatal projection neuron marker, together with PPP1R1B, BCL11B, and EBF1. We finally compared DACH1 expression profile in human and mouse forebrain, where we observed spatio-temporal similarities in its expression pattern thus providing a precise developmental description of DACH1 in the 2 mammalian species.
Keywords: DACH1, human neocortex, human striatum, medium spiny neurons, neuroepithelial cells
Introduction
During human development, a complex interplay of dynamic processes operates within a genetically organized context in the forebrain primordium, which ultimately defines area-specific molecular expression domains underlying brain functions (Monuki and Walsh 2001; Rakic 2009; Lui et al. 2011; Silbereis et al. 2016). Recent studies have described the human forebrain transcriptome organization, delineating the spatio-temporal regulation of gene expression that specifies early neocortical and striatal progenitors and their progeny (Kang et al. 2011; Miller et al. 2014; Onorati et al. 2014). These efforts produced a plethora of differentially expressed genes (DEGs) and gene modules that distinguish the developing human striatum from the neocortex. In our previous study, we were able to define a precise spatio-temporal map of developmental factors involved in striatal and neocortical neuron determination and differentiation in humans. Among these relevant genes, we identified Dachshund family transcription factor 1 (DACH1) as a neocortical enriched DEG and bimodal expressed gene (Onorati et al. 2014). Then, by looking at the Human Brain Transcriptome Atlas, we found that DACH1, later on, decreases its expression in the neocortex but shows a stable expression in the striatum until adulthood (Kang et al. 2011). Based on these expression data, and since no data were available in human brain on the protein expression level, we carried out a detailed expression analysis of DACH1 during human cortical and striatal development to characterize its cell type specificity in these 2 brain regions.
The DACH1 homolog in Drosophila, dachshund, plays a pivotal role in the differentiation of the eye imaginal disc, in leg morphogenesis and in controlling neural differentiation in the mushroom bodies, a neural structure required for learning and memory in flies. Drosophila dachshund mutant flies lack eyes, have truncated limbs and exhibit brain abnormalities (Mardon et al. 1994; Kurusu et al. 2000; Martini et al. 2000).
In mouse, Dach1 encodes for a transcription factor expressed during development in restricted areas of the central nervous system (CNS), eye, neural crests, and limb buds (Mardon et al. 1994; Hammond et al. 1998; Caubit et al. 1999; Davis et al. 1999; Kozmik et al. 1999). Mouse Dach1-null homozygotes survive pregnancy but die within 24 h after birth (Davis et al. 2001; Backman et al. 2003). In the developing telencephalon, Dach1 mRNA shows strong expression in the neural stem/progenitor cells of the ventricular and subventricular zone (VZ and SVZ, respectively) of the neocortex, in medial and lateral ganglionic eminences (MGE and LGE, respectively), and hippocampus (Machon et al. 2002).
Recently, DACH1 expression was detected in the early fetal human neocortex by PCR (Straccia et al. 2015). Moreover, DACH1 was reported to mark in vitro-derived rosette-like neuroepithelial stem cells (Elkabetz et al. 2008) and to act as a tumor suppressor gene (Chu et al. 2014). In particular, its expression is altered in several cancers, including glioblastoma (Watanabe et al. 2011).
This work describes the dynamic expression profile of DACH1 in the human neocortex and striatum of samples aged 5–21 postconceptional weeks (pcw). We found that DACH1 is expressed in the VZ/SVZ regions of the developing human cerebral cortex at all the stages analyzed. Furthermore, by analyzing single-cell RNA-seq dataset on the human developing neocortex (Onorati et al. 2016), we identified DACH1 as a specific marker of neuroepithelial and ventricular radial glial cells (vRGCs). Starting from 10 pcw, we found a DACH1 protein expression in the human striatum that was not previously reported, to the best of our knowledge, suggesting a different role for DACH1 in 2 distinct regions and times, and qualifying DACH1 as a marker of striatal medium spiny projection neurons (MSNs).
Materials and Methods
Human Tissue
Deidentified postmortem human brain specimens were obtained from patients that requested pregnancy terminations and autopsy diagnostic procedures. All procedures were approved by the research ethical committees and research services division of the University of Cambridge and Addenbrooke’s Hospital in Cambridge (protocol 96/85, approved by Health Research Authority, Committee East of England—Cambridge Central in 1996 and with subsequent amendments, with the latest approved November 2017) and by the Ethics Committee of San Paolo Hospital, Milano (protocol 11 186, approved on 19 July 2013). The ethics were fully reviewed and approved in the UK, in accordance with the Human Tissue Act 2006. Both documents were submitted to the Ethics Committee of the University of Milano and ethical approval was obtained on 27 March 2013. Appropriate informed consent was obtained and all available nonidentifying information were recorded for each specimen. Tissue was handled in accordance with ethical guidelines and regulations for the research use of human brain tissue set forth by the National Institute of Health (NIH) (http://bioethics.od.nih.gov/humantissue.html) and the World Medical Association Declaration of Helsinki (http://www.wma.net/en/30publications/10policies/b3/index.html).
Definition of Human Developmental Periods
We used pcw to indicate the developmental age of the human embryo and fetus (also termed postfertilization or postovulatory age). For embryonic stages (up to 8 pcw), we based age on multiple physical features (e.g., somites). For fetal stages, we calculated the age by subtracting 2 weeks to the gestational age (calculated on the mother’s last menstruation), crown to rump length, anatomical landmarks, and by visual inspection, as previously described (Onorati et al. 2014).
According to Kang et al. (2011), we used the following developmental periods to describe the samples employed in this study:
Embryonic development, up to 8 pcw
Early fetal development, 8–13 pcw
Early midfetal development, 13–19 pcw
Late midfetal development, 19–24 pcw
Human Tissue Collection
This study was conducted with postmortem human brain specimens from tissues collected at the John van Geest Centre for Brain Repair, University of Cambridge, Cambridge, UK. Additional specimens were procured from the San Paolo Hospital, Milan, Italy. Details of the specimens analyzed are provided in Supplementary Table 1.
Brain Sampling for Immunostaining
As previously described (Onorati et al. 2014), specimens were chilled on ice during dissection and placed onto a chilled plate on ice. The entire brain (when possible) was removed and fixed for 2–4 days in 4% formaldehyde (FA) at 4 °C. Samples were then cryoprotected in 30% sucrose (in PBS), embedded in tissue-tek, stored at −80 °C and then cryosectioned (12–30 μm).
Immunofluorescence
Brain sections were rehydrated for 10 min with PBS, then permeabilized with 0.5% Triton X-100, and blocked with 5% FBS for 1 h at room temperature. Brain sections were incubated overnight at 4 °C with primary antibodies, listed in Supplementary Table 2. Alexa 488 or 568-conjugated secondary antibodies (Molecular Probes) were diluted 1:500 and mixed with DAPI (Molecular Probes, Invitrogen) to counterstain the nuclei. Sections were mounted with fluorescent mounting medium (Dako). Images were acquired with Leica DMI 6000B and DM4000B (Leica Microsystem) microscopes, analyzed with LAS-AF imaging software and then processed with Adobe Photoshop. Confocal images were acquired with a Leica TCS SP2 microscope (Leica Microsystem) and then processed with Adobe Photoshop.
Cell Counting
The positivity and specificity of each primary and secondary antibody used in this work was evaluated against a negative control intended as both nonexpressing human brain regions (e.g., spinal cord) and slices stained exclusively with secondary antibody. The majority of antibodies used were selected from the literature and previously tested on mouse and human brain. Automatic cell counting performed using Cell Profiler. Statistical tests were performed with PRISM software (GraphPad, ver 6). All results are expressed as mean ± SD.
In situ Hybridization
In situ hybridization experiments were performed as previously described (Onorati et al. 2014), on 15–25 μm thick brain cryosections upon postfixation in 4% FA. Sections were then washed 3 times in PBS and incubated with 0.5 mg/mL of proteinase K in 100 mM Tris-HCl (pH 8) and 50 mM EDTA for 10 min at 30 °C and then 15 min in 4% FA. Slices were washed 3 times in PBS before receiving H2O washes. Sections were incubated in Triethanolamine 0.1 M (pH 8) for 5 min, then 400 μL of acetic anhydride was added 2 times for 5 min each. Finally, sections were rinsed in H2O for 2 min and air-dried. Hybridization was performed overnight at 60 °C, incubating either sense or antisense P33 labeled riboprobes at a concentration of 6 × 106 counts per minute (cpm/slide). The following day, sections were rinsed in SSC 5× for 5 min then washed in formamide 50% SSC 2× for 30 min at 60 °C, before receiving Ribonuclease-A (Roche) treatment (20 mg/mL in 0.5 M NaCl, 10 mM Tris-HCl pH 8, and 5 mM EDTA 30 min at 37 °C). Sections were further washed in formamide 50% SSC 2× for 30 min at 60 °C and then rinsed 2 times in SSC 2×. Slides were dried by using ethanol series. NTB (Kodak) emulsion was applied in a dark room, according to manufacturer’s instructions. Sections were developed after 1 week and mounted with DPX (BDH) mounting solution. Human DACH1 probe was cloned by PCR from nucleotide 1545 to nucleotide 2450 according to Genbank information (NM_080 759.5). Mouse Dach1 probe was cloned by PCR from nucleotide 1668 to nucleotide 2606, according to Genbank information (NM_007 826.3). Bright field imaging was performed by Olympus BX52 and Leica Aperio AT2—digital scanner.
Primers used for making human DACH1 probe:
Forward: GTCGGACTGGAACTTCCTTTTATGATG
Reverse: GGGTCAGAGAGTCATTTAAGACCCTG
Probe length 904 from Genbank NM_080 759.5
Primers used for making mouse Dach1 probe:
Forward: GTTAGCCATCCTCTCAACCATCTGC
Reverse: TCATTTAAGACCCGGAGACTGTCCG
(Allen brain atlas probe).
Single-Cell RNA-seq Data Analysis
Single-cell RNA-seq datasets of the developing neocortex were retrieved from an earlier study (Onorati et al., 2016). Prenatal neocortical cells from 5 to 20 pcw were analyzed. Only cells that passed quality control according to Onorati et al. (2016) were used. Cell clustering was conducted by supervised hierarchical clustering with a solicited list of marker genes identifying major cell types in the neocortex. The R program, Monocle, was used to reconstruct cell lineage and pseudotime for the cells in the dataset using default parameters (Trapnell et al. 2014). Log2 transformed RPKM values were used as input. Violin plots were generated using R package ggplot2 and gene RPKM values were used. The analysis pipeline to identify genes correlated and anticorrelated with DACH1 was performed using the package described by Lun et al. (2016) on log2 RPKM values. Briefly, genes that had Spearman’s rho > 0 and a FDR < 0.05 with DACH1 were considered as correlated with DACH1, whereas genes with a Spearman’s rho < 0 and a FDR < 0.05 with DACH1 were considered as anticorrelated with DACH1.
Hub Gene Identification
Identification of hub genes was performed using the TopGO function of the Functional Gene Networks (FGNet), an R/Bioconductor package (Aibar et al. 2015) on genes that highly correlate with DACH1. Node size was set to 20 and the P-value threshold <0.05. The gene universe was defined as all expressed genes that had 25 accumulative gene counts in all samples and RPKM >1 in at least 5 of the samples and only GO “Biological processes” were considered. Filter threshold for metagroups was set to be >40.
Results
DACH1 Shows a Peculiar Pattern in the Human VZ and SVZ
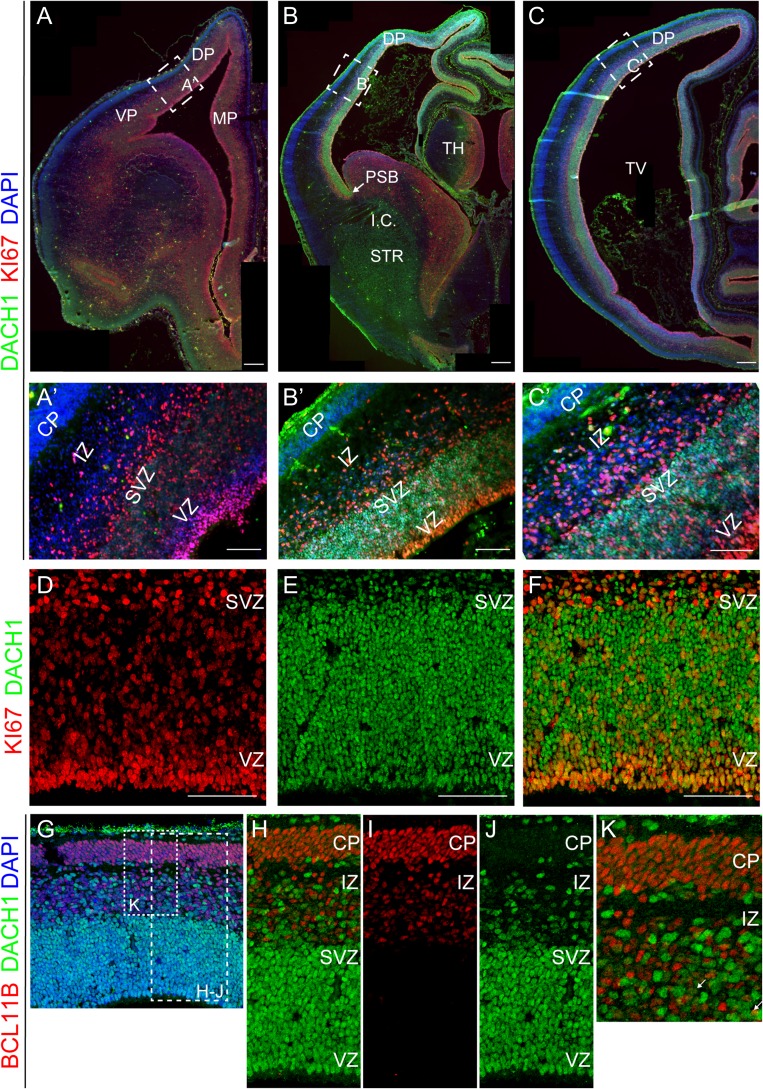
In order to define the regional expression of DACH1 in the cerebral cortex during human fetal development, we carried out an immunofluorescence analysis, first focusing our attention to a 10-pcw specimen, corresponding to early fetal development. This period is characterized by the consolidation of the cortical plate (CP), while the developing striatum starts to be separated into the caudate and putamen by the internal capsule. By immunostaining on whole human coronal brain sections, we found that DACH1 clearly showed a rostral to caudal expression pattern, with a very weak expression rostrally (Fig. 1A–C). At the level of the paracentral section, DACH1 expression was present in the VZ of the pallium, with a sharp edge at the level of the pallial–subpallial boundary (PSB; Fig. 1B, arrow). We performed a colabeling of DACH1 with KI67, a static marker of proliferative activity (Gerdes et al. 1983). DACH1 was expressed in the proliferative regions (VZ, SVZ) of the developing human neocortex (Fig. 1A–C’). Double immunofluorescence analysis at caudal levels clearly showed that many cells in the VZ/SVZ regions were proliferative as assessed by KI67 coexpression (Fig. 1D–F). In particular, we found that 48 ± 12.2% of total cells was DACH1+/KI67+ (Supplementary Table 3A). Finally, we compared DACH1 expression with that of BCL11B (also known as CTIP2), a transcription factor involved in the development of layer 5 projection neurons (Leid et al. 2004; Arlotta et al. 2005). DACH1 and BCL11B showed a mutually exclusive expression pattern, with DACH1 expressed mainly in the VZ/SVZ regions (Fig. 1G–K), and BCL11B mostly confined to the intermediate zone (IZ)/CP (Fig. 1G–K). We found only few cells coexpressing BCL11B and DACH1 (Fig. 1K, arrow).
Figure 1.
Expression pattern of DACH1 in the human fetal cerebral cortex at 10 pcw. (A–C) Rostro-caudal expression pattern for DACH1 compared with KI67+ proliferative cells. DACH1 shows a very low expression in rostral sections (frontal cortex) (A), and a higher expression when moving towards the paracentral (B) and occipital cortex (C). (A’–C’) Higher magnification of the boxes in A–C showing the strong DACH1 expression in more caudal sections. DACH1 is expressed in the ventricular/subventricular zone (VZ/SVZ) where KI67+ proliferative cells reside. In the cortical plate (CP) there is no discernible expression. (D–F) High magnification of DACH1 and KI67 expression in the dorsal pallium (DP) VZ/SVZ (caudal level) showing that they are coexpressed in these proliferative regions. The images are from the DP of a section adjacent to the one shown in (B). Some DACH1+ cells are KI67−, probably migrating neuroblasts. (G–K) DACH1 expression in the DP compared with BCL11B (CTIP2), a layer 5 marker. The 2 proteins are expressed in a complementary fashion. (H–J) High magnification of the region highlighted in the dashed box in (G), showing DACH1 expression in the VZ/SVZ and scattered in the intermediate zone (IZ), compared with BCL11B expression in the CP and IZ. (K) High magnification of the region highlighted in the dashed box in (G), showing the complementary expression pattern of DACH1 and BCL11B. Arrows in K point to occasional single cells coexpressing both BCL11B and DACH1. VP, ventral pallium; MP, medial pallium; TV, telencephalic ventricle; PSB, pallium–subpallium boundary; TH, thalamus; STR, striatum; I.C., internal capsule. Scale bars: 100 μm in A–C, 50 μm in A’–F.
DACH1 is Expressed in the VZ/SVZ Regions During Embryonic and Early Fetal Development Along the Medio-Lateral Axis
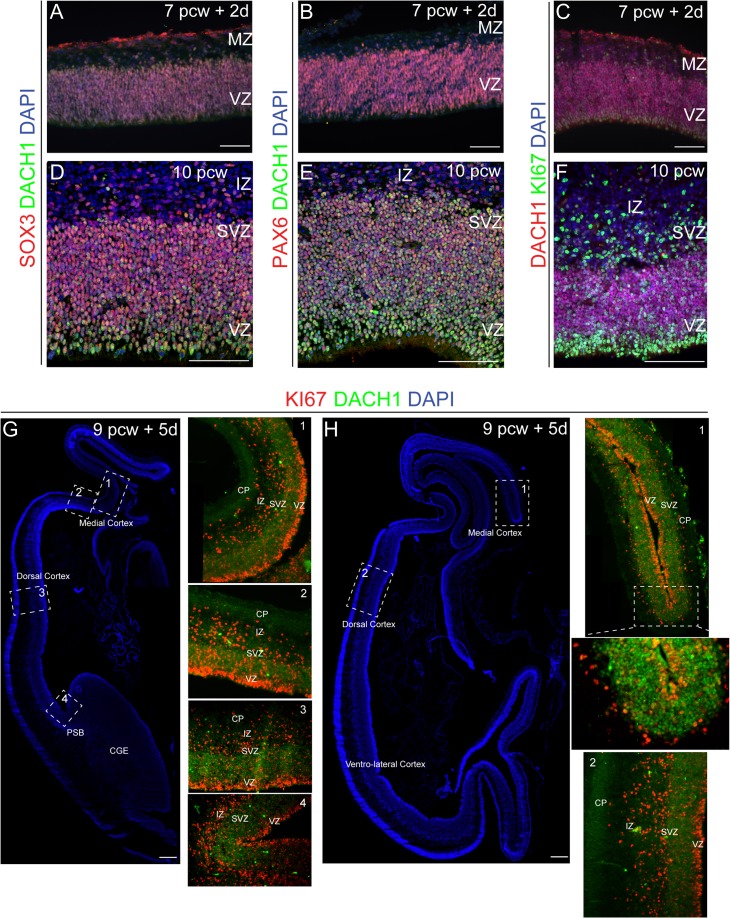
To determine whether DACH1 was also present at earlier stages of human development, we performed an immunodetection study on a 7-pcw + 2d embryonic specimen (Fig. 2A–C). We found that DACH1 was coexpressed in the VZ together with SOX3 (Fig. 2A), PAX6 (Fig. 2B), well-known neural progenitor markers, and KI67 (Fig. 2C). This pattern of expression was then conserved at 10 pcw (Fig. 2D–F) where 77 ± 4.1% of the cells was DACH1/SOX3 double-positive and 76 ± 4.0% was DACH1/PAX6 double-positive in the VZ/SVZ regions (Supplementary Table 3A).
Figure 2.
Spatial-temporal expression pattern of DACH1 in the developing human cerebral cortex. (A–C) Double immunodetection of DACH1 with SOX3 (A), PAX6 (B), and KI67 (C) at 7 pcw + 2d showing the coexpression of the 3 VZ markers with DACH1. (D–F) At 10 pcw DACH1 shows a similar coexpression pattern with SOX3, PAX6, and KI67. (G,H) DACH1 expression pattern in medio-lateral cortex at 9 pcw + 5d. The regions in the dashed box represent the area magnified in the boxes. DACH1 signal is localized in the VZ/SVZ, with no detectable signal in the CP. This pattern is conserved in more caudal sections (H). MZ, marginal zone; CP, cortical plate; IZ, intermediate zone; VZ/SVZ; ventricular/subventricular zone; PSB, pallium–subpallium boundary; CGE, caudal ganglionic eminence. Scale bars: 50 μm in A–F, 100 μm in G,H.
Next, we decided to investigate the medio-lateral expression pattern of DACH1 (Fig. 2G,H), using a (9 pcw + 5 d) human fetal brain specimen in order to substantiate the expression analysis described above. We decided to focus our attention on the rostro-caudal levels analyzed in Figure 1B,C where DACH1 expression was stronger. DACH1 signal was localized with similar levels along the medial-lateral axis (Fig. 2G, and boxes 1–4) in the VZ/SVZ regions, with no detectable signal in the CP. This pattern was conserved also in more caudal sections (Fig. 2H, and boxes 1–2).
Finally, we analyzed DACH1 expression at 21 pcw (Supplementary Fig. 1A–D). In this late midfetal specimen, DACH1 was localized in the cortical VZ, with fading expression in SVZ, IZ, subplate (SP), and CP (Supplementary Fig. 1A,A’). Supplementary Fig. 1A” shows the reduced DACH1 expression moving from the VZ to the SVZ.
We then performed in situ hybridization (ISH) for DACH1 on adjacent sections to study its weak expression in the CP (Supplementary Fig. 1B,B’). By using this more sensitive assay, we could detect the DACH1 transcript signal in the CP, although at low level. In the VZ/SVZ regions, also at this stage DACH1 colocalized with SOX3 (Supplementary Fig. 1C) and the RGC marker TFAP2C (also known as AP2γ) (Supplementary Fig. 1D).
Single-Cell Transcriptional Profiling Reveals DACH1 Expression in Neuroepithelial Cells and vRGCs
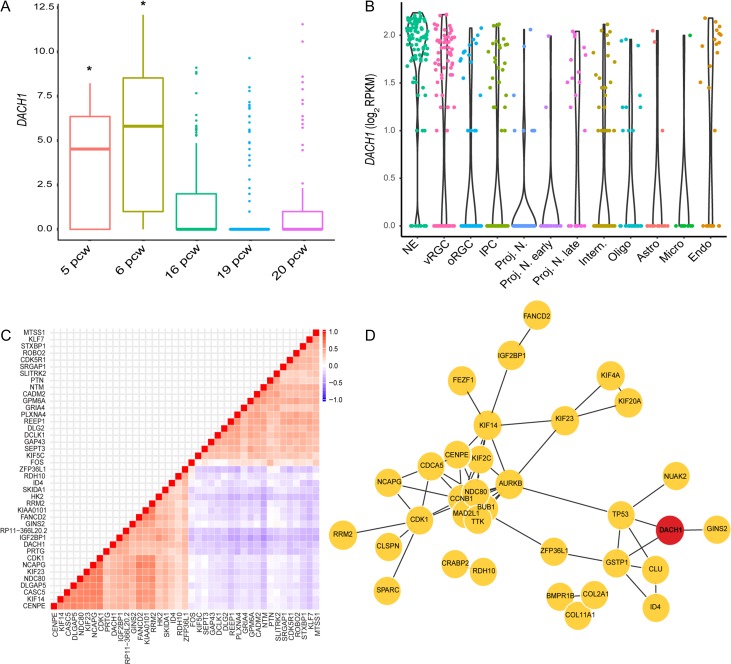
To fully comprehend DACH1 expression in time, and to pinpoint cell populations expressing this gene with a greater resolution, we decided to extend our investigation by analyzing the recently published single-cell RNA-seq dataset on the developing human neocortex (Onorati et al. 2016). Towards this aim, we initially clustered cells deriving from fetal samples from early embryonic to late midfetal periods (5–20 pcw) to verify the cellular identities arising in the developing neocortex. Supplementary Figure 2A shows the cellular subtypes identified by marker-driven clustering, where we could identify DACH1 as one of the players characterizing neuroepithelial and vRGCs. In particular, by pseudotemporal ordering of cells we observed that DACH1 has a significantly higher (P-adj. < 0.001) expression between 5 and 6 pcw compared with 16–20 pcw (Fig. 3A). Furthermore, DACH1 was specifically enriched (P-adj. < 0.001) in the cluster represented by neuroepithelial cells and vRGCs (Fig. 3B). The other cell populations of neural progenitors and neurons did not enrich for DACH1 (Fig. 3B). We then defined genes that have maximum correlation and anticorrelation pattern with DACH1 (Fig. 3C). We identified 61 genes that have a high correlation (FDR < 0.05, Supplementary Table 4) with DACH1, including the Cyclin Dependent Kinase (CDK1), the Centromere Protein E (CENPE), and the neuroprogenitor determinant ID4 (Fig. 3C). On the other hand, DACH1 was anticorrelated with synaptic and receptor genes, such as the glutamate ionotropic receptor GRIA4 and the plexin receptor PLXNA4 (Fig. 3C). Within the set of correlated genes, DACH1 emerged as a hub gene (Fig. 13D) in functional annotations like “Nuclear DNA replication”, “Regulation of DNA biosynthetic process”, “Neuronal stem cell population maintenance”, and “Cell proliferation in forebrain” (P < 0.05, Supplementary Table 5). This picture is in line with the nature of dividing neuroepithelial cells and vRGCs and with immunodetection results that showed DACH1 coexpressed with PAX6 and KI67 in the VZ (Fig. 1D–F).
Figure 3.
Single-cell transcriptional profiling of DACH1 expression in the developing human neocortex. (A) Boxplot showing pseudotemporal ordering of cells by DACH1 expression from 5 to 20 pcw, with a significant decrease in DACH1 expression during neocortical development. (*P-adj. < 0.001, one-way ANOVA with Tukey post hoc test). (B) Violin plots showing expression of DACH1 in different cell types of the developing neocortex. DACH1 has a significantly higher expression (P-adj. < 0.001, Wilcoxon test with Bonferroni correction for multiple comparisons) in neuroepithelial (NE) and ventricular radial glial cell (vRGCs) compared with the rest of the neural lineage (oRGCs, IPC, projection neurons, and interneurons). (C) 20 most correlated (red) and anticorrelated (blue) genes with DACH1 (ordered by false discovery rate). (D) Hub genes within genes with high correlation with DACH1. oRGC, outer radial glial cell; IPC, intermediate progenitor cell; Proj. N., projection neurons; Intern., interneurons; Oligo, oligodentrocytes; Astro, astrocytes; Micro, microglia; Endo, endothelial cells.
DACH1 is Expressed in the Human Fetal Striatum
We further studied DACH1 expression pattern during human brain development by taking into account the previously observed striatal expression (see Fig. 1B). First, we compared the DACH1 profile during early fetal development with FOXP1, FOXP2, and BCL11B, 3 transcription factors highly expressed during human striatum development and in particular in MSNs (Onorati et al. 2014) (Supplementary Fig. 3A–L). We found that DACH1 was similarly expressed in the developing striatum from 8 pcw to 10 pcw (Supplementary Fig. 3A–C).
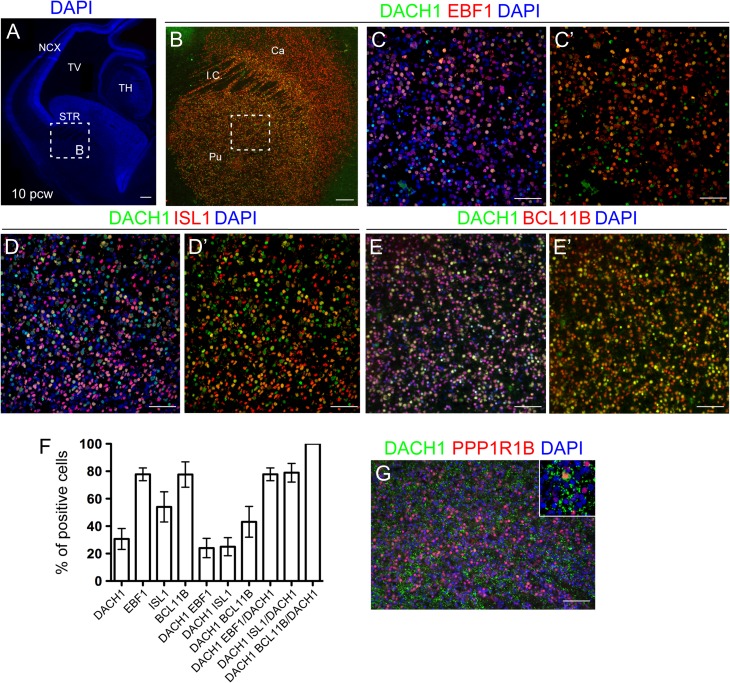
Next, we performed costainings with a panel of striatal markers at 10 pcw (Fig. 4A). Strong DACH1 signal was present in the striatum at this stage, coexpressed with EBF1 (Fig. 4B–C’), ISL1 (Fig. 4D,D’), and BCL11B (Fig. 4E,E’). We investigated coexpression patterns with different striatal markers at 10pcw where DACH1 was expressed in 30.6 ± 7.6% of total striatal cells (Fig. 4F). We quantified the number double-positive cells, finding 24 ± 7% of cells coexpressing DACH1 and EBF1, 25 ± 6.6% DACH1 and ISL1, and 43.1 ± 11.1% DACH1 and BCL11B. When the number of double-positive cells was normalized to the total number of DACH1+ cells, we found that 77.8 ± 4.6% of DACH1 expressed EBF1, 78.9 ± 6.7% expressed ISL1, and 100% expressed BCL11B (Fig. 4F and Supplementary Table 3B). DACH1 resulted also coexpressed with PPP1R1B (also known as DARPP-32), a mature striatal MSN marker (Onorati et al. 2014) (Fig. 4G).
Figure 4.
DACH1 expression in the developing human striatum. (A) 10 pcw human brain coronal hemisection. The region highlighted in the dashed box represents the striatal area analyzed in B. (B–C’) Coexpression analysis of DACH1 and EBF1, a striatal marker. The region in the dashed box in B represents the area magnified in C–E’. (D,D’) Coexpression analysis of DACH1 with the striatal marker ISL1. (E,E’) Coexpression analysis of DACH1 and the MSN marker BCL11B. (F) Histogram showing the percentage of cells coexpressing DACH1 and the indicated markers. All results are expressed as mean ± SD. (G) DACH1 coexpression with the MSN marker PPP1RB1 (see arrows in the inset). NCX, neocortex; TV, telencephalic ventricle; TH, thalamus; STR, striatum; I.C., internal capsule; Ca, caudate; Pu, putamen. Scale bars: 100 μm in A,B; 50 μm in C–E’, G.
We also evaluated DACH1 signal in the 21 pcw human striatum. DACH1 was localized in the mantle region together with EBF1 (Supplementary Fig. 4A,A’), ISL1 (Supplementary Fig. 4B,B’), BCL11B (Supplementary Fig. 4C,C’), and PPP1R1B (Supplementary Fig. 4D,D’) with similar coexpression patterns found at 10 pcw (Supplementary Table 3C). Double immunostaining analysis revealed that DACH1 was then expressed by SST+ interneurons but not by the NR2F2+ (also known as COUPTFII) or CALB2+ (also known as calretinin) interneurons (Supplementary Fig. 4E–G). Our finding of DACH1 being strongly expressed in the human striatum is further supported by the Human Brain Transcriptome Atlas (Kang et al. 2011) where DACH1 shows a stable expression until adulthood (Supplementary Fig. 5A).
We also suggest that DACH1 is the main player in human neurodevelopmental processes since its paralog DACH2 is not significantly expressed both at population and single-cell level in the human neocortex and striatum (Supplementary Fig. 6A,B).
DACH1 Expression is Conserved in the Mouse
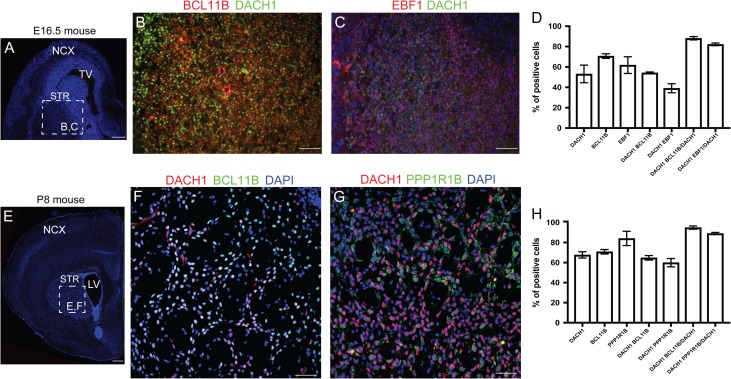
Finally, we performed a human versus mouse comparison by performing ISH and immunofluorescence analyses at different developmental time points in the mouse. We first found that at E13.5 (a stage comparable to the 10 pcw analyzed in Fig. 1) Dach1 was strongly expressed in the VZ/SVZ of the developing pallium (Supplementary Fig. 7A). At the end of the neurogenesis (E18.5), Dach1 was confined to the VZ/SVZ regions and virtually absent from the CP (Supplementary Fig. 7B–D). At E16.5 DACH1 showed a transitional expression pattern compared with E13.5 and E18.5, with high expression in the neocortical VZ/SVZ regions, together with PAX6, TFAP2C, and SOX3 (Supplementary Fig. 7E–G’). We also observed a strong expression in the striatum (Fig. 5A–H, Supplementary Fig. 7B) where 88 ± 1.63% of DACH1 cells coexpressed BCL11B and 82 ± 1.34% of the DACH1 cells were EBF1+ at E16.5 (Fig. 5A–D, Supplementary Table 3D). Then at P8 DACH1 marked MSNs together with BCL11B (94.98 ± 1.62%) and PPP1R1B (89 ± 1.10%) (Fig. 5D–H, Supplementary Table 3D).
Figure 5.
DACH1 expression pattern in the mouse striatum. (A) A representative E16.5 mouse brain coronal section. The region highlighted in the dashed box represents the striatal area analyzed in B,C. (B) Coimmunodetection analysis of DACH1 and BCL11B. (C) Coimmunodetection analysis of DACH1 and EBF1. (D) Histogram showing the percentage of cells coexpressing DACH1 together with BCL11B and EBF1. (E) A representative P8 mouse coronal brain section. The region highlighted in the dashed box represents the striatal area analyzed in E,F. (F) Coimmunodetection analysis showing DACH1 and BCL11B coexpression. (G) Coimmunodetection analysis of DACH1 and PPP1R1B. (H) Histogram showing the percentage of cells coexpressing DACH1 together with BCL11B and PPP1R1B. NCX, neocortex; TV, telencephalic ventricle; STR, striatum; LV, lateral ventricle. Scale bars: 100 μm in A,D; 50 μm in B,C,E,F.
Discussion
In this study, we demonstrate that DACH1 is expressed in neuroepithelial cells and vRGCs of the human forebrain from the earliest stage analyzed (5 pcw) and continues to be expressed until midfetal stages (21 pcw). Moreover, we describe a peculiar DACH1 expression in striatal MSNs, which is conserved in mouse. We show that DACH1 expression is temporally and spatially dynamic, suggesting multiple roles at different times and regions.
DACH1 is Expressed in Neocortical Neuroepithelial Cells and vRGCs
Single-cell transcriptional analysis showed that DACH1 expression significantly decreases between 5 and 20 pcw in proliferating neural progenitor populations in the neocortex. This was confirmed at protein level, as we observed a strong DACH1 signal in the embryonic neocortical VZ/SVZ, coexpressed with PAX6. This expression persists in the proliferative regions of the neocortex at least until 21 pcw, the latest developmental stage analyzed in this study. Single-cell transcriptomics revealed that DACH1 is found predominately in neuroepithelial cells and vRGCs starting from 5 pcw, corroborating the expression in 15–18 gestational week-old vRGCs shown by Pollen et al. (2015). Genes with high correlation with DACH1 show an enrichment in terms related to cell cycle and neural stem cell population maintenance, with most of the gene terms correlating to mechanisms involved in mitosis. Moreover, DACH1 results as a hub gene in these processes suggesting a potential role in regulating the cell cycle within dividing progenitors. Further strengthening this hypothesis is the fact that we also observed a strong DACH1 expression in the VZ of mice at E16.5. In line with these results, Dach1 expression is detected from E8.25 in the anterior neural ridge (Caubit et al. 1999) and persists at least until P21 in the ependymal and subependymal zones (Machon et al. 2002). DACH1 is considered one of the markers of neural rosette progenitors in vitro (Elkabetz et al. 2008). These observations, taken together, advocate for DACH1 having a fundamental role in neural progenitors during early stages of development, both in mouse and human.
A previous study examined Dach1 mutant mice with respect to proliferation defects at E17.5 (Backman et al. 2003), finding no impairments. However, they performed only a 2-h BrdU pulse at a single stage, thus probably missing more subtle defects that can be revealed by cumulative studies at different time points. DACH2 redundancy is unlikely because of its flat and lower expression in cerebral cortex proliferating zones (Supplementary Fig. 6A; Kang et al. 2011).
DACH1 has a Rostro-Caudal Expression Pattern in the Human Neocortex
Neocortical parcellation is an important developmental process that gives rise to 4 major areas: the primary visual (V1), somatosensory (S1), auditory (A1), and motor (M1) areas.
These areas are subdivisions that differ from one another by differences in cell composition, connections, and patterns of gene expression. The suggestion that intrinsic genetic mechanisms may control arealization was based on the observation of transcription factors with gradients of expression along the medio-lateral and rostro-caudal axes (Sur and Rubenstein 2005). In this study, we show that DACH1 at 10 pcw has a rostral-low to caudal-high expression pattern of gene expression, and thus it may have a role in correct cerebral cortical arealization similar to the transcription factors PAX6, EMX2, COUPTFI, and SP8 (Sur and Rubenstein 2005). Alternatively, DACH1 may have a dynamic expression pattern in different areas at different developmental ages.
A Secondary Site of DACH1 Expression, the Striatum
A few transcription factors, for example BCL11B and SP8, are expressed both in the neocortex and in the basal ganglia. In particular BCL11B is expressed, both in mouse (Arlotta et al. 2005, 2008) and in human (Onorati et al. 2014) in layer 5 projection neurons and in the MSNs of the striatum. In this study, we report that DACH1 has a similar dual expression. Its striatal expression was found both in mouse tissue (E13.5, E16.5, and E18.5) and in human specimens (10 pcw and 21 pcw). Interestingly, this striatal expression was not extensively reported before even in mouse. In Davis et al. (1999) they reported a generic ganglionic eminence expression at E12.5, while Caubit et al. (1999) reported an exclusive neocortical expression, even if their analysis was confined to the E12.5 stage. A more recent study examined Dach1 expression by ISH at E12.5, P0, and P21, but did not describe any expression in the striatum, except for faint LGE staining (Machon et al. 2002). We now show, using both ISH and immunostaining, that DACH1 is strongly expressed in the striatum in mouse and human, and that DACH1 expression is stable until adulthood (Supplementary Fig. 5A; Kang et al. 2011). Moreover, at 10 pcw, DACH1 was coexpressed with important mature striatal markers such as EBF1 (Garel et al. 1999; Onorati et al. 2014), ISL1 (Toresson et al. 2000; Onorati et al. 2014), BCL11B, and PPP1R1B.
Together, our findings suggest that DACH1 can have a dual role in the developing human telencephalon. The first one is in human cortical progenitors, at early developmental stages, as a consequence of its expression in neuroepithelial cells and vRGCs and not in cortical projection neurons or interneurons. Of interest, at 21 pcw it was also expressed in the LGE VZ, probably extending its role to other progenitor types. The second role is in the striatum, mostly in postmitotic striatal MSNs where it is coexpressed with well-known MSN markers. DACH1 is not the only gene that acts in 2 different contexts within the nervous system. Also, PAX6 and OTX2, known neural progenitor markers for the ventricular neuroepithelium of the developing CNS, are found in certain neuronal subpopulations in the adult brain (Stoykova et al. 2000, Omodei et al. 2008). In particular, PAX6 characterizes dopaminergic neurons in the glomerular layer of the olfactory bulb, granule neurons in the cerebellum and amacrine and ganglion cells of the retina (Stoykova et al. 2000). Instead, OTX2 is expressed in a neuronal subset within the ventral tegmental area (Di Salvio et al. 2010). In line, the dual role of DACH1 has been described in the literature with regards to promoting differentiation of the Drosophila eye and limb in postmitotic cells (Mardon et al. 1994; Hammond et al. 1998; Caubit et al. 1999; Davis et al. 1999; Kozmik et al. 1999) and in regulating proliferation and growth in cancer cells (Watanabe et al. 2011). In conclusion, here, we mirror this binary action in 2 different areas of the telencephalon: the neocortex and striatum. We describe its enrichment in neocortical progenitors in combination with genes involved in the cell cycle, together with its typifying role in defining MSNs in the developing striatum.
Supplementary Material
Footnotes
Conflict of Interest: None declared.
Supplementary Material
Supplementary material is available at Cerebral Cortex online.
Funding
This work was supported by NeuroStemcell (EU Seventh Framework Programme, grant agreement no. 222943), The Cure Huntington’s Disease Initiative (CHDI, USA, ID: A-4529), the Ministero dell’Istruzione dell’Università e della Ricerca (MIUR 2010JMMZLY_001, Italy), to E. Cattaneo; by NeuroStemcellRepair (EU Seventh Framework Programme, grant agreement no. 602278) to E. Cattaneo and R.A.B.; by Fondo per gli Investimenti della Ricerca di Base (FIRB, RBFR10A01S, Italy) to M.O.; and partially by TargetBrain (EU Seventh Framework Programme HEALTH-F2-2012–279 017) to G.M.; and NIH grants MH106934, NS095654, and MH110926 to N.S.
References
- Aibar S, Fontanillo C, Droste C, De Las Rivas J. 2015. Functional gene networks: R/Bioc package to generate and analyse gene networks derived from functional enrichment and clustering. Bioinformatics. 31:1686–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlotta P, Molyneaux BJ, Chen J, Inoue J, Kominami R, Macklis JD. 2005. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 45:207–221. [DOI] [PubMed] [Google Scholar]
- Arlotta P, Molyneaux BJ, Jabaudon D, Yoshida Y, Macklis JD. 2008. Ctip2 controls the differentiation of medium spiny neurons and the establishment of the cellular architecture of the striatum. J Neurosci. 28:622–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backman M, Machon O, Van Den Bout CJ, Krauss S. 2003. Targeted disruption of mouse Dach1 results in postnatal lethality. Dev Dyn. 226:139–144. [DOI] [PubMed] [Google Scholar]
- Caubit X, Thangarajah R, Theil T, Wirth J, Nothwang HG, Ruther U, Krauss S. 1999. Mouse Dac, a novel nuclear factor with homology to Drosophila dachshund shows a dynamic expression in the neural crest, the eye, the neocortex, and the limb bud. Dev Dyn. 214:66–80. [DOI] [PubMed] [Google Scholar]
- Chu Q, Han N, Yuan X, Nie X, Wu H, Chen Y, Guo M, Yu S, Wu K. 2014. DACH1 inhibits cyclin D1 expression, cellular proliferation and tumor growth of renal cancer cells. J Hematol Oncol. 7:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RJ, Shen W, Heanue TA, Mardon G. 1999. Mouse Dach, a homologue of Drosophila dachshund, is expressed in the developing retina, brain and limbs. Dev Genes Evol. 209:526–536. [DOI] [PubMed] [Google Scholar]
- Davis RJ, Shen W, Sandler YI, Amoui M, Purcell P, Maas R, Ou CN, Vogel H, Beaudet AL, Mardon G. 2001. Dach1 mutant mice bear no gross abnormalities in eye, limb, and brain development and exhibit postnatal lethality. Mol Cell Biol. 21:1484–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Salvio M, Di Giovannantonio LG, Acampora D, Prosperi R, Omodei D, Prakash N, Wurst W, Simeone A. 2010. Otx2 controls neuron subtype identity in ventral tegmental area and antagonizes vulnerability to MPTP. Nat Neurosci. 13(12):1481–1488. [DOI] [PubMed] [Google Scholar]
- Elkabetz Y, Panagiotakos G, Al Shamy G, Socci ND, Tabar V, Studer L. 2008. Human ES cell-derived neural rosettes reveal a functionally distinct early neural stem cell stage. Genes Dev. 22:152–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garel S, Marin F, Grosschedl R, Charnay P. 1999. Ebf1 controls early cell differentiation in the embryonic striatum. Development. 126:5285–5294. [DOI] [PubMed] [Google Scholar]
- Gerdes J, Schwab U, Lemke H, Stein H. 1983. Production of a mouse monoclonal antibody reactive with a human nuclear antigen associated with cell proliferation. Int J Canc Prev. 31:13–20. [DOI] [PubMed] [Google Scholar]
- Hammond KL, Hanson IM, Brown AG, Lettice LA, Hill RE. 1998. Mammalian and Drosophila dachshund genes are related to the Ski proto-oncogene and are expressed in eye and limb. Mech Dev. 74:121–131. [DOI] [PubMed] [Google Scholar]
- Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, Sousa AM, Pletikos M, Meyer KA, Sedmak G, et al. 2011. Spatio-temporal transcriptome of the human brain. Nature. 478:483–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozmik Z, Pfeffer P, Kralova J, Paces J, Paces V, Kalousova A, Cvekl A. 1999. Molecular cloning and expression of the human and mouse homologues of the Drosophila dachshund gene. Dev Genes Evol. 209:537–545. [DOI] [PubMed] [Google Scholar]
- Kurusu M, Nagao T, Walldorf U, Flister S, Gehring WJ, Furukubo-Tokunaga K. 2000. Genetic control of development of the mushroom bodies, the associative learning centers in the Drosophila brain, by the eyeless, twin of eyeless, and Dachshund genes. Proc Natl Acad Sci U S A. 97:2140–2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leid M, Ishmael JE, Avram D, Shepherd D, Fraulob V, Dolle P. 2004. CTIP1 and CTIP2 are differentially expressed during mouse embryogenesis. Gene Expr Patterns. 4:733–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui JH, Hansen DV, Kriegstein AR. 2011. Development and evolution of the human neocortex. Cell. 146:18–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lun AT, Bach K, Marioni JC. 2016. Pooling across cells to normalize single-cell RNA sequencing data with many zero counts. Genome Biol. 17:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machon O, van den Bout CJ, Backman M, Rosok O, Caubit X, Fromm SH, Geronimo B, Krauss S. 2002. Forebrain-specific promoter/enhancer D6 derived from the mouse Dach1 gene controls expression in neural stem cells. Neuroscience. 112:951–966. [DOI] [PubMed] [Google Scholar]
- Mardon G, Solomon NM, Rubin GM. 1994. Dachshund encodes a nuclear protein required for normal eye and leg development in Drosophila. Development. 120:3473–3486. [DOI] [PubMed] [Google Scholar]
- Martini SR, Roman G, Meuser S, Mardon G, Davis RL. 2000. The retinal determination gene, dachshund, is required for mushroom body cell differentiation. Development. 127:2663–2672. [DOI] [PubMed] [Google Scholar]
- Miller JA, Ding SL, Sunkin SM, Smith KA, Ng L, Szafer A, Ebbert A, Riley ZL, Royall JJ, Aiona K, et al. 2014. Transcriptional landscape of the prenatal human brain. Nature. 508:199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monuki ES, Walsh CA. 2001. Mechanisms of cerebral cortical patterning in mice and humans. Nat Neurosci. 4(Suppl):1199–1206. [DOI] [PubMed] [Google Scholar]
- Omodei D, Acampora D, Mancuso P, Prakash N, Giovanni Di Giovannantonio L, Wurst W, Simeone A. 2008. Anterior–posterior graded response to Otx2 controls proliferation and differentiation of dopaminergic progenitors in the ventral mesencephalon. Development. 135:3459–3470. [DOI] [PubMed] [Google Scholar]
- Onorati M, Castiglioni V, Biasci D, Cesana E, Menon R, Vuono R, Talpo F, Laguna Goya R, Lyons PA, Bulfamante GP, et al. 2014. Molecular and functional definition of the developing human striatum. Nat Neurosci. 17:1804–1815. [DOI] [PubMed] [Google Scholar]
- Onorati M, Li Z, Liu F, Sousa AMM, Nakagawa N, Li M, Dell’Anno MT, Gulden FO, Pochareddy S, Tebbenkamp ATN, et al. 2016. Zika virus disrupts phospho-TBK1 localization and mitosis in human neuroepithelial stem cells and radial glia. Cell Rep. 16:2576–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollen AA, Nowakowski TJ, Chen J, Retallack H, Sandoval-Espinosa C, Nicholas CR, Shuga J, Liu SJ, Oldham MC, Diaz A, et al. 2015. Molecular identity of human outer radial glia during cortical development. Cell. 163:55–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P. 2009. Evolution of the neocortex: a perspective from developmental biology. Nat Rev Neurosci. 10:724–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silbereis JC, Pochareddy S, Zhu Y, Li M, Sestan N. 2016. The cellular and molecular landscapes of the developing human central nervous system. Neuron. 89:248–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoykova A, Treichel D, Hallonet M, Gruss P. 2000. Pax6 modulates the dorsoventral patterning of the mammalian telencephalon. J Neurosci. 20(21):8042–8050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straccia M, Garcia-Diaz Barriga G, Sanders P, Bombau G, Carrere J, Mairal PB, Vinh NN, Yung S, Kelly CM, Svendsen CN, et al. 2015. Quantitative high-throughput gene expression profiling of human striatal development to screen stem cell-derived medium spiny neurons. Mol Ther Methods Clin Dev. 2:15030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sur M, Rubenstein JL. 2005. Patterning and plasticity of the cerebral cortex. Science. 310:805–810. [DOI] [PubMed] [Google Scholar]
- Toresson H, Potter SS, Campbell K. 2000. Genetic control of dorsal-ventral identity in the telencephalon: opposing roles for Pax6 and Gsh2. Development. 127:4361–4371. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, Rinn JL. 2014. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 32:381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe A, Ogiwara H, Ehata S, Mukasa A, Ishikawa S, Maeda D, Ueki K, Ino Y, Todo T, Yamada Y, et al. 2011. Homozygously deleted gene DACH1 regulates tumor-initiating activity of glioma cells. Proc Natl Acad Sci U S A. 108:12384–12389. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.