Abstract
Fragile X syndrome (FXS) is a neurodevelopmental disorder caused by mutations in the FMR1 gene. It is a leading monogenic cause of autism spectrum disorder and inherited intellectual disability and is often comorbid with attention deficits. Most FXS cases are due to an expansion of CGG repeats leading to suppressed expression of fragile X mental retardation protein (FMRP), an RNA-binding protein involved in mRNA metabolism. We found that the previously published Fmr1 knockout rat model of FXS expresses an Fmr1 transcript with an in-frame deletion of exon 8, which encodes for the K-homology (KH) RNA-binding domain, KH1. Notably, 3 pathogenic missense mutations associated with FXS lie in the KH domains. We observed that the deletion of exon 8 in rats leads to attention deficits and to alterations in transcriptional profiles within the medial prefrontal cortex (mPFC), which map to 2 weighted gene coexpression network modules. These modules are conserved in human frontal cortex and enriched for known FMRP targets. Hub genes in these modules represent potential therapeutic targets for FXS. Taken together, these findings indicate that attentional testing might be a reliable cross-species tool for investigating FXS and identify dysregulated conserved gene networks in a relevant brain region.
Keywords: 5-choice serial reaction time task, fragile X syndrome, medial prefrontal cortex, RNA sequencing
Introduction
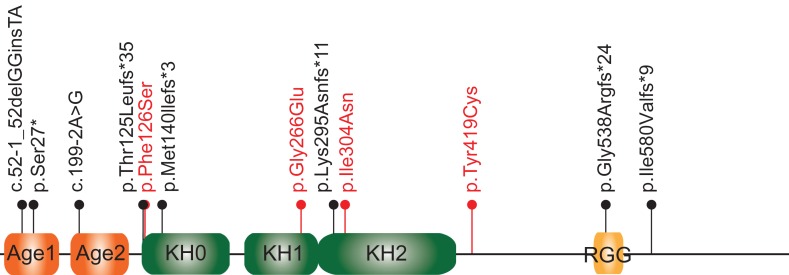
Fragile X syndrome (FXS) is a leading monogenic cause of autism spectrum disorder (ASD) and the most frequent known form of inherited intellectual disability (ID) (Bagni et al. 2012). FXS is caused by the loss of Fragile X Mental Retardation Protein (FMRP), which is encoded by the FMR1 gene on chromosome X (Bagni et al. 2012). FMRP is involved in the regulation of messenger RNA (mRNA) translation (Napoli et al. 2008; Darnell et al. 2011; De Rubeis et al. 2013), shuttling of mRNA to dendritic spines (Santoro et al. 2012), and stability of mRNA (Zalfa et al. 2007; Zhang et al. 2018). FMRP contains several RNA-binding domains, including 3 K-homology (KH) domains and an arginine–glycine box (RGG) (Vasilyev et al. 2015; D’Annessa et al. In Press). In most FXS cases, FMRP loss occurs when the unstable trinucleotide CGG repeat at the 5′ untranslated region of FMR1 expands to above 200 copies, resulting in the hypermethylation and transcriptional silencing of FMR1 (Richter et al. 2015). Notably, both point mutations (Suhl and Warren 2015) and deletions (Hammond et al. 1997; Coffee et al. 2008) within the FMR1 gene coding sequence have also been reported in a small number of individuals, including 3 missense mutations in the KH domains (Pozdnyakova and Regan 2005; Zang et al. 2009; Di Marino et al. 2014; Myrick et al. 2014) (Fig. 1).
Figure 1.
Likely pathogenic or pathogenic point mutations associated with FXS. Likely pathogenic or pathogenic mutations in the coding region of the FMR1 gene published (De Boulle et al. 1993; Lugenbeel et al. 1995; Pozdnyakova and Regan 2005; Zang et al. 2009; Collins et al. 2010; Gronskov et al. 2011; Di Marino et al. 2014; Handt et al. 2014; Myrick et al. 2014; Myrick, Deng, et al. 2015; Okray et al. 2015; Xiong et al. 2015; Quartier et al. 2017; Sitzmann et al. 2018) or deposited in ClinVar are summarized here. The FMRP domains are reported as described in Myrick, Hashimoto, et al. 2015. Mutations are indicated using the HGSV nomenclature. Reference sequences used are Q06787 for the protein and NM_002024.5 for the cDNA. The splice-site mutations are indicated by their splice-site nomenclature and localized to the position of the first amino acid predicted to be affected. For NM_002024.5:c.990+1 G > A and NM_002024.5:c.420–8 A > G (Quartier et al. 2017), we have indicated the amino acid change identified experimentally (p.Lys295Asnfs*11 and p.Met140llefs*3, respectively). All mutations are absent in gnomAD. Mutations p.Gly266Glu and p.Ile304Asn are supported by functional and/or structural analyses (Pozdnyakova and Regan 2005; Zang et al. 2009; Di Marino et al. 2014; Myrick et al. 2014). Age, Agenet-like domain (also known as Tudor domain); KH, K-homology domain; RGG; arginine–glycine-glycine box. Missense mutations are indicated in red; protein-truncating mutations are in black.
Attention deficits and hyperactivity are very common behavioral manifestations in FXS and are prevalent in both males and females (Gross, Hoffmann, et al. 2015). Attention in rodents requires an intact medial prefrontal cortex (mPFC) (Chudasama and Robbins 2006), which shares functions with prefrontal regions that are anatomically (Hoeft et al. 2010; Bray et al. 2011; Hallahan et al. 2011) and functionally (Menon et al. 2004; Hoeft et al. 2007) impaired in individuals with FXS. Despite the repeated implication of the mPFC as a nexus of cognitive dysfunction in FXS, it has been the focus of very few preclinical studies in animal models (Krueger et al. 2011; Sidorov et al. 2014; Gross, Raj, et al. 2015). In the current study, we used a rat model of FXS to study the involvement of the mPFC in FXS.
To analyze the role of Fmrp in the mPFC, we employed behavioral, molecular, and bioinformatic approaches using a recently described rat model with a 122 base-pair (bp) in-frame deletion in exon 8 that was previously shown to have enhanced protein synthesis, exaggerated Group 1 mGluR-dependent long-term depression (LTD), increased spine head width and spine density in the CA1 region of the hippocampus (Hamilton et al. 2014; Till et al. 2015), and macroorchidism (Hamilton et al. 2014). We found that this 122 bp deletion leads to skipping of exon 8 and the expression of a gene product that lacks the KH1 domain (Fmrp-ΔKH1). To study how this deletion affects visuospatial attention, which requires an intact mPFC (Muir et al. 1996; Chudasama et al. 2003), we used the 5-choice serial reaction time task (5-CSRTT) (Mar et al. 2013). In addition, we characterized the transcriptional profiles in the mPFC and identified discrete groups of conserved coregulated genes.
Materials and Methods
Generation of the Fmr1- Δ exon 8 Rat Model
The Fmr1-Δexon 8 rat model, previously reported as the Fmr1 knockout (KO) rat model (Engineer et al. 2014; Hamilton et al. 2014; Till et al. 2015; Kenkel et al. 2016; Berzhanskaya et al. 2017), was generated using zinc-finger nucleases (ZFN) in the outbred Sprague-Dawley background. The design and cloning of the ZFN, as well as the embryonic microinjection and screening for positive founder rats were performed by SAGE Labs (Boyertown, PA USA) as previously described (Kulinski et al. 2000). The best performing ZFN pair targeting the CATGAACAGTTTATCgtacgaGAAGATCTGATGGGT sequence, located between 18 686 and 18 721 bp in the Fmr1 gene (NCBI reference sequence NC_005120.4), was used for embryo microinjection. Positive Sprague-Dawley founder animals with a deletion in the Fmr1 gene were mated to produce F1 breeding pairs. Polymerase chain reaction (PCR) amplification at the target sites followed by sequencing analysis revealed the exact deletion of 122 bp at the junction of intron 7 and exon 8 (between 18 588 and 18 709 bp), as previously described by Hamilton et al. (2014).
Animal Breeding, Care, and Husbandry
This study used age-matched male and female littermate rats. To produce both male genotypes (Fmr1-Δexon 8+/y, Fmr1-Δexon 8−/y) and all 3 female genotypes (Fmr1-Δexon 8+/+, Fmr1-Δexon 8+/−, and Fmr1-Δexon 8−/−) we set up 2 pairs of breeders: Fmr1-Δexon 8+/y × Fmr1-Δexon 8+/−and Fmr1-Δexon 8−/y × Fmr1-Δexon 8+/−. Wild type (WT) and Fmr1-Δexon 8−/y male rats were used for both the RNA sequencing (RNAseq) analyses and the attentional task and WT, Fmr1-Δexon 8+/−, and Fmr1-Δexon 8−/− female rats were used for the attentional task. All rats were kept under veterinary supervision in a 12 h reverse light/dark cycle at 22 ± 2 °C. Animals were pair-caged with food and water available ad libitum. Rats tested on the 5-CSRTT were food restricted to 85% of their free-feeding weight. All animal procedures were approved by the Institutional Animal Care and Use Committee at the Icahn School of Medicine at Mount Sinai.
Testes Weight
Testes were dissected from 10-week-old male Fmr1-Δexon 8−/y rats (n = 19) and WT littermates (n = 19). After gonadal fat pads were removed and testes were weighed, testes:body weight ratios were calculated. The data was analyzed with a 2-tailed t-test.
Total Lysate Preparation
mPFC tissues were dissected from 8-week-old rats as previously described (Spijker 2011). Tissues were homogenized in 100 μL ice cold RIPA buffer supplemented with a 1:100 proteinase inhibitors cocktail (Thermo Scientific) and a 1:100 phosphatase inhibitors cocktail (Thermo Scientific), using a Teflon-glass homogenizer. The homogenate was centrifuged at 12 000 × g for 20 min at 4 °C. The recovered supernatant was centrifuged again at 12 000 rpm for 20 min at 4 °C. The protein concentration in the final recovered supernatant was determined using the BCA protein assay (Pierce).
Structural Data
FMR1 orthologs from 58 species, including Drosophila melanogaster, 2 Enterogona (Chordata: Tunicata), 12 fishes, Xenopus tropicalis, 2 reptiles, 5 birds, and 35 mammals, were extracted from Ensembl and aligned using Alvis v. 0.1 software. The X-ray structure of the human FMRP KH1-KH2 domains (PDBID = 2QND) was generated using Pymol v1.7.2.1.
Immunoblotting
Immunoblotting was performed using a standard protocol (Bozdagi et al. 2010). Briefly, 10 μg of each protein lysate were loaded to a 4–12% SDS-polyacrylamide gel electrophoresis (PAGE gel, Invitrogen; Carlsbad, CA USA), which was transferred to polyvinylidene fluoride membrane for immunoblotting. For Fmrp detection, we used the anti-Fmrp (G468) antibodies targeted against the C-terminus of the Fmrp (1:1000, Cell Signaling) and the anti-Fmrp (F3930) antibodies targeted against the N-terminus of Fmrp (1:1000, Sigma Aldrich). Anti beta III tubulin antibodies (1:2000, Abcam; ab18207) were used to quantify the beta III tubulin level, used for normalization. Horseradish peroxidase (HRP)-conjugated antirabbit (1:5000) and HRP-conjugated antimouse antibodies (1:5000) were purchased from Jackson ImmunoResearch Laboratories (West Grove, PA USA) and used as secondary antibodies. ECL substrate (Pierce; Thermo Scientific, Rockford, IL) or SuperSignal West Femto (Thermo Scientific, Rockford, IL) substrates were used to produce the signal that was detected on a G:Box Chemi-XT4 GENESys imager (Syngene; Cambridge, UK). Blots were quantified using ImageJ (Schneider et al. 2012).
Immunoprecipitation Followed by Immunoblotting Analysis
Prefrontal cortex were dissected from 8-week old WT and Fmr1-Δexon 8−/y rats and homogenized in lysis buffer (100 mM NaCl, 10 mM MgCl2, 10 mM Tris–HCl, 1 mM diothiolhreitol, 1% Triton X-100, (1:100) proteinase inhibitors cocktail (Thermo Scientific) and (1:100) phosphatase inhibitors cocktail (Thermo Scientific)), using an electric tissue homogenizer (Napoli et al. 2008). Samples were incubated on ice for 5 min and centrifuged at 12 000 × g for 8 min at 4 °C. The recovered supernatant was centrifuged again at 12 000 × g for 8 min at 4 °C. The protein concentration in the final recovered supernatant was determined using the BCA protein assay (Pierce). A 800 μg of protein extract was used for Fmrp immunoprecipitation experiments. Fmrp was immunoprecipitated based on the previously established protocol (Brown et al. 2001). Briefly, Fmrp was immunoprecipitated with 6.24 μg 7G1-1 Fmr1 monoclonal antibody conjugated to 1.5 mg of Protein A Dynabeads (Invitrogen). The same amount of monoclonal mouse IgG2B (R&D Systems) was used as a control. The immunoprecipitates as well as 20 μg of prefrontal cortex protein lysate from WT and Fmr1-Δexon 8−/y rats were loaded onto 4–12% SDS-polyacrylamide gel electrophoresis (PAGE gel, Invitrogen; Carlsbad, CA) and ran for 3 h at 200 V, 60 mA to allow an optimal separation of the bands around 75 MW followed by transfer onto PVDF membranes (Invitrogen) using XCell II Blot Module system (Thermo Scientific). Membranes were immunoblotted with anti-Fmrp antibodies (1:1000, N-Terminal, Sigma Aldrich F3930-25UL). Subsequently, membranes were incubated with appropriate antirabbit HRP-conjugated secondary antibodies (1:5000, Jackson ImmunoResearch Laboratories). Images were developed using West Femto (Thermo Scientific).
PCR Amplification on Genomic DNA Followed by Sanger Sequencing
Tail samples were collected from WT and Fmr1-Δexon 8−/y rats and DNA was extracted using the QIAamp DNA Mini Kit (Qiagen) according to the manufacturer’s instructions. 25 ng of each sample was PCR-amplified using the Fmr1-G-F and Fmr1-G-R primers. PCR products were loaded onto an agarose gel and pure bands from each of the WT and Fmr1-Δexon 8−/y samples were sliced from the gel and cleaned using the QIAquick Gel Extraction Kit (Qiagen). Purified samples were sent to GENEWIZ for Sanger sequencing using both of the Fmr1-G-F and Fmr1-G-R primers. The location and sequence of the primers are described in Supplementary Table S1.
Reverse Transcriptase PCR (RT-PCR) Followed by Sanger Sequencing
mPFC tissues were dissected from 8-week old WT and Fmr1-Δexon 8−/y male rats as previously described (Spijker 2011). RNA was extracted using the RNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. A 2 μg of RNA was then used to prepare cDNA, using the SuperScript II Reverse Transcriptase Kit (Invitrogen) and following the manufacturer’s instructions. A 100 ng of each sample was PCR-amplified using the F-Fmr1, which aligns to exons 6/7 junction and the R-Fmr1, which aligns to exons 9/10 junction. PCR products were loaded on an agarose gel and pure bands from each of the WT and Fmr1-Δexon 8−/y samples were sliced from the gel and cleaned using the QIAquick Gel Extraction Kit (Qiagen). Purified samples were sent to GENEWIZ for Sanger sequencing using both of the F-Fmr1 and R-Fmr1 primers. The location and sequence of the primers are described in Supplementary Table S1.
Open Field Test
Rats were exposed to a brightly lit novel 90 cm × 90 cm environment during their light-cycle for one hour. All horizontal movements were automatically tracked by Noldus Ethovision system and samples were analyzed in 10-min bins. After a significant effect of time and no significant effect of genotype were discovered in a first batch of animals (WT: n = 6, Fmr1-Δexon 8−/y: n = 7), the experiment was repeated and the same effect was replicated in a second batch (WT: n = 7, Fmr1-Δexon 8−/y: n = 5). The data was analyzed with the SPSS statistical package, version 23 (IBM SPSS Statistics, Armonk, North Castle, NY) using repeated measures ANOVA where time was the within-subjects factor, genotype was the between-subjects factor, and batch was a covariate.
5-CSRTT
The 5-CSRTT was carried out as we have previously described (Harony-Nicolas et al. 2017), with slight modifications due to performance. Briefly, training on the 5-CSRTT began when the rats were 8-week old and after they were habituated to being handled and food deprived to achieve ~85% of free feeding weight. Rats were first trained to touch the location of an illuminated white square presented at 1 of 5 locations on a Bussey-Saksida capacitive touchscreen system (Lafayette Instrument Company; Lafayette, IN) (Mar et al. 2013) using ABET II Software for Touch Screens. If a capacitive screen touch occurred at the illuminated location during the stimulus presentation, sucrose (valve open for 250 ms) was delivered in the reward receptacle located across the chamber from the touchscreen. Training occurred in stages, where the light stimulus duration decreased from 60 to 30 to 20 to 10 to 5 to 2.5 s. Rats advanced schedules once criterion performance was met. Criteria for progression were an accuracy rate higher than 80% (accuracy rate = number of correct trials/total trials) and an omission rate lower than 20% (omission rate = number of omitted trials/total trials). Trials where the rat made a correct response after the time allotted were termed “late responses.” Once criterion was reached with a stimulus duration of 2.5 s, training was recorded as complete. Four separate batches of animals were trained on the 5-CSRTT, totaling 12 WT male, 15 Fmr1-Δexon 8−/y male, 11 WT female, 13 Fmr1-Δexon 8+/− female, and 12 Fmr1-Δexon 8−/− female rats, where the experimenter was blind to subject genotype. Five Fmr1-Δexon 8−/y male, 1 WT female, 1 Fmr1-Δexon 8+/− female, and 2 Fmr1-Δexon 8−/− female rats that either did not reach criterion on a stimulus duration of 5 s after 30 sessions or on a stimulus duration of 2.5 s after 45 sessions were removed from analysis. When subsets of 10 animals per group were randomly sampled from the male and female WT and Fmr1-Δexon 8 dataset 10 000 times and analyzed via a 2-way ANOVA (genotype × sex), a significant main effect of genotype (P < 0.05) on the % omissions measure at a 2.5 s stimulus duration was observed 92% of the time, suggesting that there was reproducibility of a particularly significant finding regardless of batch. Therefore, we focused our analysis on the pooled data. Because Fmr1 is X-linked, there was an imbalance in the number of genotypes available between males (2) and females (3). Thus, the analysis included 2 separate comparisons: 1) WT male and female rats were compared with Fmr1-Δexon 8−/y male and Fmr1-Δexon 8−/− female rats and 2) WT female rats were compared with Fmr1-Δexon 8+/−and Fmr1-Δexon 8−/− female rats. Data analysis of training data was comprised of linear mixed-effects modeling (LMM) where sex, genotype, and stimulus duration were fixed factors and rat was nested into batch as a random factor using custom scripts written in the R statistical programming environment (R Development Core Team, 2006).
RNA Isolation, Library Preparation, and Data Availability
RNA was extracted from mPFC tissues from 8-week old WT (n = 12) and Fmr1-Δexon 8−/y (n = 12) littermate rats using the RNeasy Mini Kit (Qiagen), according to the manufacturer’s instructions. Subsequently, the quantity of all purified RNA samples was measured on a nanodrop (2.07 ± 0.01 A260/280; 2.11 ± 0.19 A260/230) and the quality and integrity were measured with the Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA). All RNA integrity numbers were greater than 9 (9.6 ± 0.3). Next, 1 μg of total RNA was used for the preparation of the RNAseq library using the Illumina Genome Analyzer IIx TruSeq mRNA Seq Kit supplied by Illumina (Cat number: RS-122-2001). A poly-A-based mRNA enrichment step was carried out and cDNA was synthesized and used for library preparation using the Illumina TruSeqTM RNA sample preparation kit as previously described (Tariq et al. 2011), except for the following step: adapter-ligated DNA fragments were size-selected by gel-free size selection using appropriate concentration of SPRI AMPure beads to get an average 200 bp peak size in adapter ligated DNA. The size selected adapter-ligated DNA fragments were amplified by LM-PCR. Then, Illumina recommended 6 bp barcode bases were introduced at one end of the adapters during PCR amplification step. The amplified PCR products were then purified with SPRI AMPure XP magnetic beads to get the final RNAseq library, which was used for high-throughput RNAseq.
All samples were sequenced on the Illumina Genome Analyzer IIx. A total of 40 million 100 bp paired-end sequences were used to reliably assess expression for each sample. Overall, the design of the experiment was as follows: 12 barcoded samples/per brain region were used, of which 6 of each genotype were pooled and loaded onto 2 lanes so that each sample was spread across 2 lanes in order to minimize confounds associated with lane effects. These raw RNAseq fastq data have been submitted to Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE126057.
Short Read Mapping and Quantification of Gene Expression
All high-quality short reads were mapped to the rat reference genome rn4 using the STAR Aligner v2.4.0g1 (Dobin et al. 2013) with 2-pass mapping strategy (--twopassMode Basic). RNAseq read and alignment quality was checked with the FastQC and RNA-seqQC tools, respectively. Uniquely mapped reads with overlapping genes were counted with featureCounts v1.4.4 (Liao et al. 2014) parameters (featureCounts -T 10 -p -t exon -g gene_id).
RNAseq Data Preprocessing
Raw count data measured 16 499 transcripts across all samples (12 WT and 12 Fmr1-Δexon 8−/y rats). Nonspecific filtering required more than 2 counts per million (cpm) in at least 12 samples and retrained 14 745 transcripts. Filtered raw count data was subjected to conditional quantile normalization (CQN) (Hansen et al. 2012) to remove systematic bias introduced by GC-content and correct for global distortions, resulting in a normally distributed data matrix. Normalized data were inspected for outlying samples using unsupervised clustering of samples (Pearson’s correlation coefficient and average distance metric) and principal component analysis to identify outliers outside 2 standard deviations from these grand averages. Based on these metrics, 2 outliers were removed from these data (WT = 2). Rattus ENSEMBL symbols were converted to HGNC symbols, then converted to human orthologues using Ensemble biomart conversion (http://www.ensembl.org/biomart). In order to form the bases for cross-species comparisons (rat, human), one large reference transcriptome of the mPFC in WT rats was constructed by integrating RNAseq gene expression data from an additional 24 WT rats (Ntot = 35). These data were processed in an identical fashion as described above, and were generated in 3 batches (i.e., different processing dates). Combat batch correction (Leek et al. 2012) was applied to resolve systematic sources of variability across batches.
Finally, to estimate the relative frequencies of brain cell types for each sample, Cibersort deconvolution analysis was applied (https://cibersort.stanford.edu/). Cibersort (Newman et al. 2015) relies on known cell subset specific marker genes and applies linear support vector regression to estimate the relative frequencies of cell types from bulk tissue. A priori defined brain cell type specific RNAseq expression markers (Zhang et al. 2014) were used as a signature matrix to obtain estimates for neurons, oligodendrocytes and astrocytes in all samples.
Visualization of the Fmr1 Deletion and Splice Junctions
To enable in-depth visualization of the deletion in exon 8 of the Fmr1 gene, we used depth of coverage plots from the Integrated Genome Browser (IGB) (http://bioviz.org/igb/). The Rattus norvegicus reference genome version Nov_2004 was used as a reference genome, and 2 pooled BAM alignment files were used as input, one for each genotype (WT and Fmr1-Δexon 8−/y). Subsequently, to visualize predicted splice junctions in the Fmr1 gene, we used the Integrative Genome Viewer (IGV) (http://software.broadinstitute.org/software/igv/). Similarly, the Rattus norvegicus reference genome version Nov_2004 was selected, and sorted BAM files from each rat were loaded separately (10 WT and 12 Fmr1-Δexon 8−/y) and then pooled across genotypes to show the total number for each genotype.
Differential Gene Expression Analysis
Differential gene expression (DGE) signatures between WT and Fmr1-Δexon 8−/y rats were identified using moderated t-tests in the limma package (Ritchie et al. 2015). The covariates RIN, parents, age, and date of dissection were included in the models to adjust for their potential confounding influence on gene expression between-group main effects. P-value significance was set to an FDR-corrected P-value of < 0.05. This assumption was later relaxed to an FDR-corrected P-value of < 0.1 to yield a sufficient number of genes for down-stream network enrichment analyses.
Weighted Gene Coexpression Network Analysis
Weighted gene coexpression network analysis (WGCNA) (Langfelder and Horvath 2008) was used to build coexpression networks. To construct each network, the absolute values of Pearson correlation coefficients were calculated for all possible gene pairs and resulting values were transformed with an exponential weight (β) so that the final matrices followed an approximate scale-free topology (R2 ≥ 0.80). The dynamic tree-cut algorithm was used to detect network modules with a minimum module size set to 50 and cut tree height set to 0.99. These parameters were used to construct 3 separate networks. First, we built one large reference transcriptome network using all available mPFC samples from WT rats (n = 35, genes = 14 745, β = 10). Nonspecific filtering required more than 2 counts per million (cpm) in at least 12 samples (1 batch) and retrained 14 552 transcripts. The resulting WT modules were assessed for enrichment for Fmr1-Δexon 8−/y DGE signatures, FMRP targets, CNS cell type specificity, genetic risk loci for neurodevelopmental disorders and gene coexpression modules implicated in ASD cases, as described below. We then sought to determine whether any WT modules displaying significant enrichment for Fmr1-Δexon 8−/y DGE signatures and FMRP targets were also preserved in human cortex samples. To this end, we collected previously published healthy unaffected human cortical (BA 9/41) gene expression data (Voineagu et al. 2011) (RIN, 7.5 ± 0.6; Age, 33.25 ± 13.09; PMI, 23.58 ± 6.3; Sex, 16 M/1 F) and restricted our analysis to HGNC gene symbols that were commonly expressed in both rat and human data (genes = 6 926). Using this subset of genes, a second WT network was constructed (β = 10) and a separate, third network was constructed for human cortical gene expression (β = 15). Our module preservation analysis sought to determine whether any fundamental differences exist in the underlying gene coregulatory patterns, as being preserved or disrupted, in WT rats as compared with humans, and vice versa. For these analyses, module preservation was assessed using a permutation-based preservation statistic, Zsummary, implemented within WGCNA with 500 random permutations of the data (Langfelder et al. 2011). Zsummary takes into account the overlap in module membership as well as the density and connectivity patterns of genes within modules. Zsummary score < 2 indicates no evidence of preservation, 2 < Zsummary < 10 implies weak preservation and Zsummary > 10 suggests strong preservation.
Functional Annotation and Protein Interaction Networks
Gene modules and DGE signatures with an FDR-corrected P-value < 0.1 and an absolute log fold-change > 0.10 were subjected to functional annotation. First, the ToppFunn module (Chen et al. 2009) of ToppGene Suite software was used to assess enrichment of Gene Ontology (GO) terms specific to biological processes and molecular factors using a one-tailed hypergeometric distribution with family-wise FDR at 5%. Second, gene modules implicated in the neurobiology of FMR1 were used to build direct protein–protein interaction (PPI) networks, which can reveal key genes and transcription factors mediating the regulation of multiple target genes. PPIs were obtained from the STRING database (Franceschini et al. 2013) with a signature query of the reported module gene list. STRING implements a scoring scheme to report the confidence level for each direct PPI (low confidence: < 0.4; medium: 0.4–0.7; high: >0.7). We used a combined STRING score of >0.7 and reported only the highest confidence interactions. We further used STRING to test whether the number of observed PPIs were significantly more than expected by chance using a nontrivial random background model (that is, null model). For visualization, the STRING network was imported into CytoScape (Shannon et al. 2003).
Module Overlap and User-Defined List Enrichment Analyses
DGE signatures and WT networks were annotated as described above. In addition, cell type enrichment was performed by cross-referencing gene modules with previously defined lists of genes known to be preferentially expressed in different brain cell types (Cahoy et al. 2008; Zeisel et al. 2015). Neurodevelopmental genetic risk loci were curated from human genetic studies of ASD (Xu et al. 2012; De Rubeis et al. 2014; Sanders et al. 2015), ID (Parikshak et al. 2013), schizophrenia (SCZ) (Fromer et al. 2016), and a list of well-known FMRP target genes (Darnell et al. 2011). Over-representation analysis of these gene sets within DGE signatures and WT transcriptome modules was analyzed using a one-sided Fishers exact test to assess the statistical significance. All P-values, from all gene sets and modules, were adjusted for multiple testing using Bonferroni procedure. We required an adjusted P-value < 0.05 to claim that a gene set is enriched within a user-defined list of genes. All user-defined lists can be found in Supplementary Data 1.
Quantitative RT-PCR
RNA was prepared, as described above, from a new cohort of WT and Fmr1-Δexon 8−/y littermate rats (n = 7/genotype). A 1 μg cDNA was synthesized from RNA samples using the SuperScript II Reverse Transcriptase Kit (Invitrogen). The universal probe library (UPL) system (Roche) was used to perform RT-PCR. Two reference genes (Rplp0 and Gapdh) were used for normalization. The relative expression levels were calculated using qBase software (Hellemans et al. 2007), now available from Biogazelle (Ghent, Belgium). Primers for each gene were designed using ProbeFinder Software (Roche). The location and sequences of the primers and the UPL probe numbers are listed in Supplementary Table S1.
Results
Validation of an Fmr1-Δexon 8 Rat Model of FXS
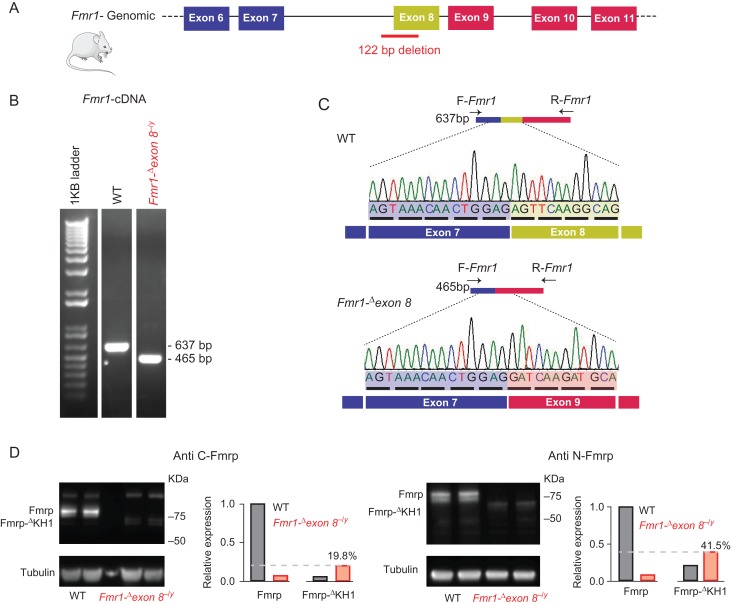
ZFN targeting Fmr1 were used to introduce a 122 bp genomic deletion in the gene in order to develop a rat model of FXS, which has been published on previously (Engineer et al. 2014; Hamilton et al. 2014; Till et al. 2015; Kenkel et al. 2016; Berzhanskaya et al. 2017) and referred to here as the Fmr1-Δexon 8 rat (Fig. 2A). We used both PCR and genomic Sanger sequencing to confirm the 122 bp deletion between base-pairs 18 588 and 18 709 (NCBI reference sequence: NC_005120.4), spanning part of intron 7 and exon 8 of the Fmr1 gene (Supplementary Fig. S1A–C). To examine the effect of the genomic deletion on Fmr1 mRNA, we amplified the coding sequence between exons 7 and 9 using RT-PCR (Fig. 2B) followed by Sanger sequencing (Fig. 2C). This analysis revealed that in the Fmr1-Δexon 8−/y rat at least a portion of mRNAs show a skipping of exon 8, which results in an in-frame deletion (Fig. 2C). Using Western blot analysis with antibodies directed against the C-terminus or the N-terminus of Fmrp, we detected the full-length Fmrp (~75KDa) in WT, but not in Fmr1-Δexon 8−/y rats, where we detected a band at a lower molecular weight (~70KDa) (Fig. 2D). Immunoprecipitation using a monoclonal antibody against the N-terminus of Fmrp and detection with both N-terminus and C-terminus directed antibodies (Supplementary Fig. S2A) recovered the ~75KDa band in WT rats and the 70KDa band in Fmr1-Δexon 8−/y rats. The sequencing results, the immunoprecipitation results, and the ~70 kDa molecular weight are compatible with an Fmr1-Δexon 8−/y rat in-frame deletion of the 57 amino acids encoded by exon 8 and composing the KH1 domain of Fmrp (Supplementary Fig. S2B). Notably, the ~70KDa band had decreased expression relative to Fmrp in WT rats (19.8–41.5%), indicating that although the in-frame deletion did not cause Fmr1-Δexon 8 mRNA degradation, it might have resulted in reduced translational efficiency or most likely reduced stability of the resultant protein.
Figure 2.
Skipping of exon 8 of the Fmr1 gene in the Fmr1-Δexon 8 rat model (previously presented as Fmr1 KO). (A) A schematic representation of exons 6–11 of the Fmr1 gene and the position of the genomic 122 bp deletion spanning intron 7 and exon 8 in the Fmr1-Δexon 8 rat model. (B) RT-PCR analysis primed with a forward primer (F-Fmr1), designed to align to exon junction 6/7, and a reverse primer (R-Fmr1), designed to align to exon junction 9/10 (as described in C and detailed in Supplementary Table S1). (C) Sanger sequencing of the RT-PCR products of WT and Fmr1-Δexon 8−/y rats, primed with the F-Fmr1 primer. Sequencing results confirm the skipping of exon 8 in Fmr1-Δexon 8−/y rats, keeping the frame intact. (D) Immunoblotting and quantification of Fmrp and FmrpΔKH1 protein levels in WT and Fmr1-Δexon 8−/y mPFC samples, using anti-C-terminus or anti-N-terminus Fmrp antibodies.
The FMRP KH1 domain is structurally organized by 3 antiparallel β-strands and 3 α-helices (β1-α1-α2-β2-β′-α′ configuration) with a GxxG loop between α1 and α2 forming a cleft for the RNA binding (Supplementary Fig. S2C). As shown by the sequence conservation obtained from alignment of FMR1 orthologs across 58 species (Valverde et al. 2008), the KH1 domain has very strong evolutionary conservation (Supplementary Fig. S2A). Interestingly, amongst the point mutations in FMR1 associated with FXS, there are 3 missense and 2 frameshift mutations in the KH domains, including p.Gly266Glu, which lies in the KH1 domain (Myrick et al. 2014) (Fig. 1). The phenotype associated with this mutation includes the characteristic dysmorphic facial features of FXS, macroorchidism, and ID in comorbidity with ASD and Attention-Deficit/Hyperactivity Disorder (ADHD) (Myrick et al. 2014).
The rat with the deletion of exon 8 shares some phenotypes with the Fmr1 KO mouse (Huber et al. 2002; Qin et al. 2005; Dolen et al. 2007; Osterweil et al. 2010; Wijetunge et al. 2014), including enhanced protein synthesis, alterations in mGluR-dependent LTD, and deficits in spine density and morphology (Hamilton et al. 2014; Till et al. 2015). We also observed a significant increase in testes:body weight ratio in the Fmr1-Δexon 8−/y rats compared with their WT littermates (Supplementary Fig. S3; 2-tailed t-test, P = 0.019), replicating the macroorchidism phenotype reported in the rat (Hamilton et al. 2014) and in mouse models for FXS (Bakker et al. 1994). In contrast to the findings from the Fmr1 KO mouse model (Baker et al. 2010; Kazdoba et al. 2014; Sorensen et al. 2015), however, the rat model does not appear to have increased locomotion in the open field test (Hamilton et al. 2014; Till et al. 2015). We replicated this lack of hyperactivity in the open field test, observing a significant effect of time (Supplementary Fig. S4; repeated measures ANOVA for time, P < 0.0001), where animals slowed down over the course of the trial, but no interaction between time and genotype. We focused our studies on attention, another key cognitive behavior that is impaired in FXS and that is dependent upon a brain region clinically implicated in FXS, the mPFC.
Fmr1- Δ exon 8 Rats Have Deficits in Sustained Attention
We assessed visuospatial attention with the 5-CSRTT. In this task, rats must respond quickly with a nose poke to briefly presented light stimuli after a 5-s delay (Fig. 3A) (Mar et al. 2013). ADHD is comorbid with FXS in both males and females (Gross, Hoffmann, et al. 2015). To compare across sexes, we tested Fmr1-Δexon 8−/y and Fmr1-Δexon 8−/−, which have comparable genetic vulnerability, and Fmr1-Δexon 8+/− to probe for a dose effect. This type of study cannot be carried out in humans due to the fact that homozygous mutations in females with FXS do not exist. Rats were trained in stages where the duration of the light stimulus was incrementally decreased from 60 to 30 to 20 to 10 to 5 to 2.5 s and were progressed to the next stage once performance criterion were met (≥80% accuracy and ≤20% omissions). Decreasing stimulus durations increases demands on sustained attention because briefer stimuli require more attentional effort in order to continue to detect and respond to them successfully.
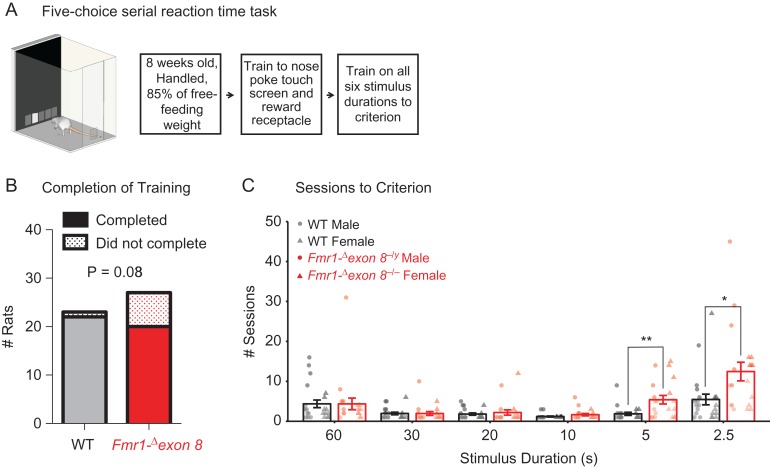
Figure 3.
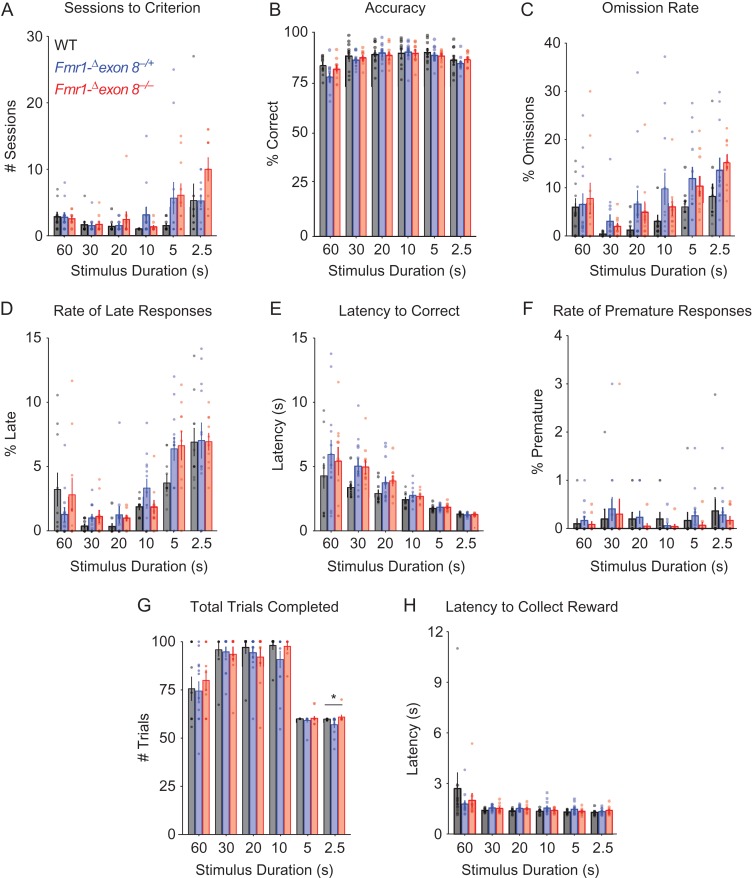
Completion of the 5-choice serial reaction time task (5-CSRTT) by male and female Fmr1-Δexon 8 (Fmr1-Δexon 8−/yand Fmr1-Δexon 8−/−) rats and WT littermates. (A) Schematic of the 5-CSRTT apparatus and training timeline. (B) The number of rats that completed training to criterion at a 2.5 s stimulus duration and the reported Phi value from a 2 by 2 contingency table that shows a significant association between genotype and completion of training. (C) The mean number of sessions rats required to reach criterion across the 6 5-CSRTT training stages ± SEM (WT, n = 22; Fmr1-Δexon 8, n = 20), black = WT, red = Fmr1-Δexon 8, circles = males, triangles = females, **P < 0.01, *P < 0.05.
Two separate analyses were performed: 1) male and female WT rats were compared with male Fmr1-Δexon 8−/y and female Fmr1-Δexon 8−/− rats and 2) female WT rats were compared with Fmr1-Δexon 8+/− and Fmr1-Δexon 8−/− rats (see Methods). Whereas all but one of the WT controls were able to meet criterion on the most difficult stage (stimulus duration of 2.5 s) and were therefore able to complete training, 5 male Fmr1-Δexon 8−/y, 1 female Fmr1-Δexon 8+/−, and 2 female Fmr1-Δexon 8−/− rats were unable to complete training. When we analyzed male and female WT rats compared with Fmr1-Δexon 8−/y and Fmr1-Δexon 8−/− rats, there was a significant association between genotype and completion of training (Fig. 3B; 2 × 2 contingency table for genotype × completion of training, Phi = 0.038). There was, however, no association between sex and completion of training (Phi = 0.599). Furthermore, there was no interaction between sex and genotype on attentional performance across training in WT and Fmr1-Δexon 8 rats (Supplementary Table S2), suggesting that, much like patients with FXS, both male and female Fmr1-Δexon 8 rats behave similarly. Therefore, data from both sexes were pooled for visual representation and presented in 2 groups: WT and Fmr1-Δexon 8.
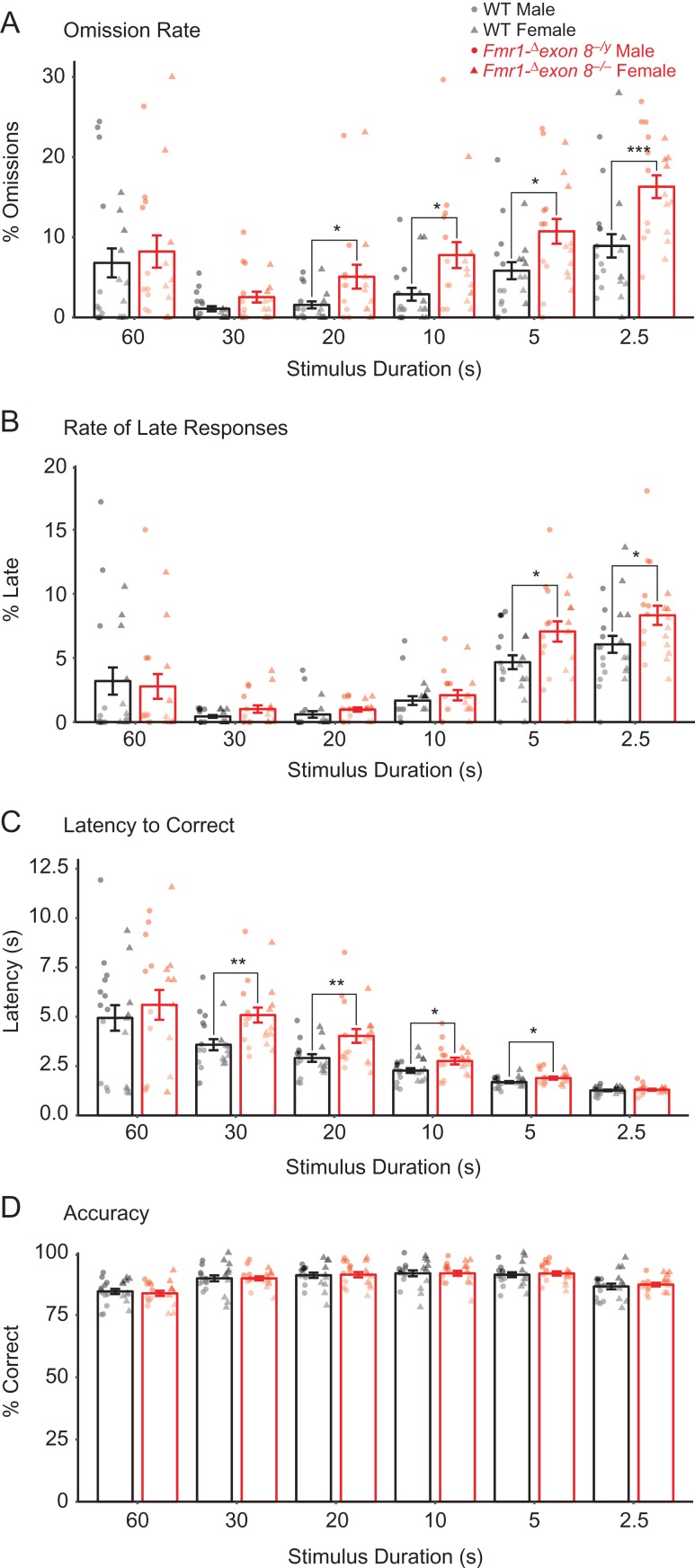
By examining the performance of all rats that completed training, we found that Fmr1-Δexon 8 rats took more sessions to reach criterion in the final 2 stages (where the stimulus durations were 5 s and 2.5 s) compared with WT littermates (Fig. 3C; linear mixed-effects model (LMM), for genotype × schedule across training P = 0.001, for genotype at 5 s, P = 0.003, 2.5 s, P = 0.015). We also found that the decline in performance was paralleled by an increase in omission rates (Fig. 4A; LMM for genotype across training, P = 0.001), where the Fmr1-Δexon 8 rats omitted more often than WT littermates at stimulus durations shorter than 30 s (Fig. 4A; LMM for genotype at 20 s, P = 0.038, 10 s, P = 0.011, 5 s, P = 0.02, 2.5 s, P = 0.001). Some of the responses that we previously believed were omissions were in fact correct responses that were performed after the time allotted. We refer to these as “late responses.” We also found that the Fmr1-Δexon 8 rats performed more of these late responses than WT littermates (Fig. 4B; LMM for genotype across training, P = 0.009) at the shortest stimulus durations (Fig. 4B; LMM for genotype at 5 s, P = 0.012, 2.5 s, P = 0.018). When the Fmr1-Δexon 8 rats did make the correct choice in the time allotted, they took longer to respond than WT littermates (Fig. 4C; LMM for genotype across training, P = 2.01 × 10−33, at 30 s, P = 0.003, 20 s, P = 0.009, 10 s, P = 0.026, 5 s, P = 0.032). Altogether, these findings indicate impaired sustained attention in male and female Fmr1-Δexon 8 rats across training.
Figure 4.
Attentional performance on the 5-CSRTT by male and female Fmr1-Δexon 8 (Fmr1-Δexon 8−/yand Fmr1-Δexon 8−/−) rats and WT littermates across training. Across six 5-CSRTT training stages, bars represent (A) mean percentage of trials that were omitted, (B) mean percentage of trials with a late response, (C) mean latency to perform a correct response, and (D) mean accuracy (number of correct responses/number of total responses) ± SEM (WT, n = 22; Fmr1-Δexon 8, n = 20), black = WT, red = Fmr1-Δexon 8, circles = males, triangles = females, ***P < 0.001, **P < 0.01, *P < 0.05.
Importantly, these deficits were not attributed to impairments in learning or sensory perception because 1) accuracy remained unaffected across all training sessions for each stimulus duration (Fig. 4D; LMM for stimulus duration × genotype, P = 0.463, and genotype, P = 0.924) and 2) the increased omission rates only appeared at the 20 s stimulus duration and onward. Furthermore, the deficits were not due to decreased motivation for food or motor deficits because the latency of Fmr1-Δexon 8 rats to collect reward after a correct response was comparable to WT rats (Supplementary Fig. S5A; LMM for stimulus duration × genotype, P = 0.671, and genotype, P = 0.931) and the average total amount of trials completed did not differ by genotype (Supplementary Fig. S5B; LMM for stimulus duration × genotype, P = 0.755, and genotype, P = 0.385). Additionally, the rate of premature responses in Fmr1-Δexon 8 rats was equal to WT littermates and both were low overall, suggesting that impulsivity was not a factor in the delay in reaching criterion (Supplementary Fig. S5C; LMM for stimulus duration × genotype, P = 0.926, and genotype, P = 0.613).
Notably, Sprague Dawley rats can typically perform the task with a stimulus duration of 1 s or less (Auclair et al. 2009; Harony-Nicolas et al. 2017). Furthermore, often training on the 5-CSRTT is followed by testing of baseline performance at the shortest stimulus duration obtainable and then challenge trial testing where the task parameters are manipulated (Robbins 2002; Semenova 2012; Boutros et al. 2017). In Fmr1-Δexon 8 rats, sustained attention deficits appeared during training and the rats were not able to perform the task at stimulus durations shorter than 2.5 s, suggesting a relatively severe deficit in sustained attention that did not allow for further testing on more demanding tasks.
We did not find sex differences amongst Fmr1-Δexon 8 rats; however, males, regardless of genotype, took longer to collect reward than females (Supplementary Fig. S6A; LMM for sex × schedule, P = 0.024, for sex at 30 s, P = 0.022, 20 s, P = 0.01, 10 s, P = 0.005, 5 s, P = 0.024). Also, females, regardless of genotype, made more perseverative responses after a correct choice at 60, 30, 5, and 2.5 s (Supplementary Fig. S6B; LMM for sex, P = 0.001, at 60 s, P = 0.012, 30 s, P = 0.021, 5 s, P = 0.023, 2.5 s, P = 0.004). To our knowledge, this is the first study to report sex differences in perseverative responses and reward collection latency in rats on the 5-CSRTT. Both might be explained by overall greater activity in females, which are also more active than males in the open field test (Archer 1975). In our second analysis, we found that female Fmr1-Δexon 8+/− rats, which have variable expression of FMRP with the in-frame deletion due to random X chromosome inactivation, did not have significant deficits in any of these measurements compared with their WT and Fmr1-Δexon 8−/− sex-matched littermates (Fig. 5).
Figure 5.
Attentional performance of female WT, Fmr1-Δexon 8+/−, and Fmr1-Δexon 8−/− rats on the 5-CSRTT. Across six 5-CSRTT training stages, bars indicate (A) mean number of sessions required to reach criterion, (B) mean accuracy (number of correct responses/number of total responses), (C) mean percentage of trials that were omitted, (D) mean percentage of trials with a late response, (E) mean latency to perform a correct response, (F) mean percentage of trials with a premature response, (G) mean number of total trials completed each session, and (H) mean latency to collect a reward ± SEM (WT, n = 10; Fmr1-Δexon 8+/−, n = 12; Fmr1-Δexon 8−/−, n = 10), black = WT, blue = Fmr1-Δexon 8+/−, red = Fmr1-Δexon 8−/−, *P < 0.05.
In summary, these results indicate that male and female Fmr1-Δexon 8 rats have impairments that are specific to sustained attention, which is also commonly disrupted in individuals with FXS (Cornish et al. 2001).
Changes to the mPFC Gene Expression Profile in the Fmr1-Δexon 8 Rat Model of FXS
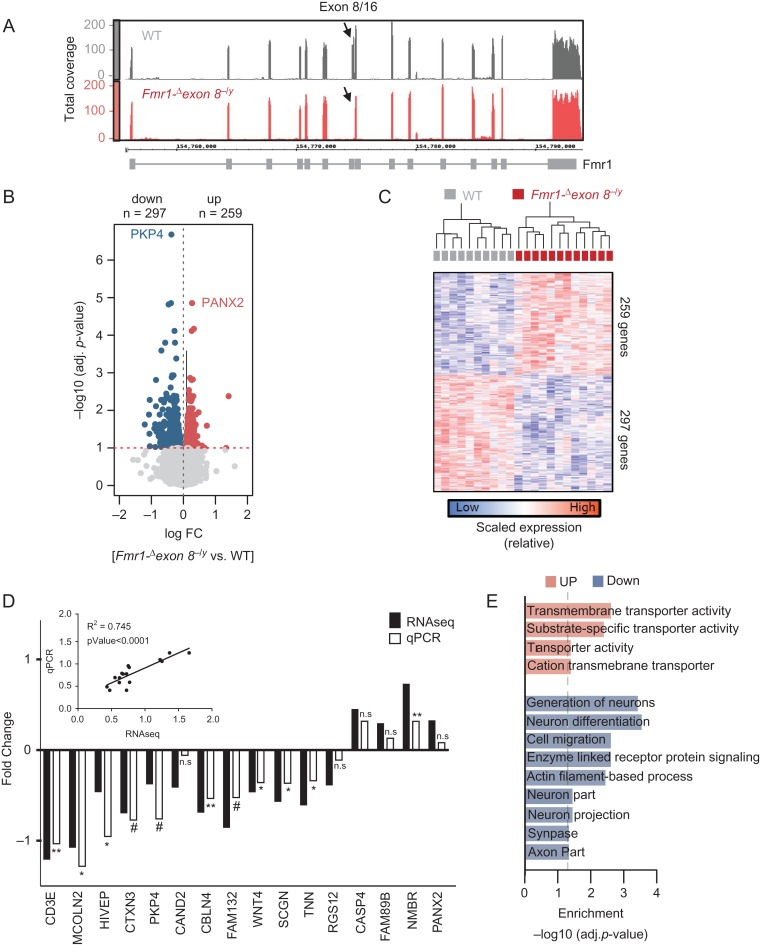
To identify gene expression differences associated with Fmr1-exon 8 deletion in a brain region largely responsible for sustained attention, we applied transcriptome-wide RNAseq and measured global gene expression profiles in mPFC samples of WT and Fmr1-Δexon 8−/y rats following the analytic pipeline and data preprocessing described in Supplementary Figures S7 and S8 (see Materials and Methods). We first confirmed, based on the RNAseq data and using the IGV and the IGB, that the deletion in Fmr1 leads to exon 8 skipping (Supplementary Fig. S10). Exon 8 is in fact not detected in Fmr1-Δexon 8−/y rats (Fig. 6A). Similarly, we found that the levels of RNA transcripts aligning to Fmr1 exon sequences (except for exon 8) are comparable between WT and Fmr1-Δexon 8−/y samples, indicating that the decreased level of the protein observed in our immunoblotting analysis is not due to reduced mRNA levels. Subsequently, we sought to identify DGE signatures and found 259 up- and 297 down-regulated genes in Fmr1-Δexon 8−/y rats compared with WT (using False Discovery Rate [FDR] < 0.1) (Fig. 6B, Supplementary Data 2). Notably, these genes were mainly associated with differences in genotype and not with any other factor, including differences in parents, RIN values, age, date of dissection, or estimated cell type proportions (Supplementary Fig. S8H). Consistent with this result, DGE signatures largely separated the 2 genotypes (Fig. 6C).
Figure 6.
Comparative RNAseq analysis between Fmr1-Δexon 8−/y and WT rats. (A) Depth of RNAseq coverage (y-axis) plots across all 16 exons of the FMR1 gene (x-axis) using the Integrative Genome Browser (IGB) viewer. Plots represent pooled coverage across all animals for each genotype (WT = 10, Fmr1-Δexon 8−/y = 12). (B) Volcano plot comparing the extent of FDR q-value significance (y-axis) and log fold change (x-axis) for differential gene expression (DGE) between Fmr1-Δexon 8−/y and WT rats. (C) DGE signatures segregate Fmr1-Δexon 8−/y and WT samples. Normalized editing levels (z-scores) were used in hierarchical clustering. Each row corresponds to one gene and each column corresponds to one sample. (D) RT-PCR validation of 16 genes identified to be differentially expressed by our RNAseq analysis. Validation was done on an independent set of Fmr1-Δexon 8−/y and WT littermate mPFC samples (n = 7/genotype). Eight genes showed statistically significant changes (one tail t-test, P < 0.05) and 3 showed a trend towards significance (one tail t-test, P < 0.1). Inset shows the significant correlation between the absolute values of the RNAseq and RT-PCR mean fold changes. (E) Functional annotation of DGE signatures, parsed by up- and down-regulated genes.
To validate our findings in an independent cohort of rats (n = 7/genotype), we used RT-PCR on a cross section of the differentially expressed genes (DEGs), including both up- and down-regulated genes, for a total of 16 DEGs. Analysis of the correlation between the 2 studies (i.e., absolute values of the RNAseq and RT-PCR mean fold changes) showed a significant correlation (R2 = 0.74, P < 0.0001) (Fig. 6D). Moreover, 8 out of the 16 DEGs showed statistically significant changes (one tail t-test, P < 0.05) and 3 showed a trend towards significance (one tail t-test, P < 0.1) (Fig. 6D).
Next, to gain biological insights into the function of the DEGs, we performed GO enrichment analysis (Fig. 6E). We found that up-regulated genes were enriched in biological processes that included transmembrane transporter activity (q-value FDR B&H < 4.02×10−02). In parallel, down-regulated genes were enriched for 1) cellular components, including neuron part (q-value = 2.45 × 10−02), neuron projection (q-value = 2.45E-02), synapse (q-value = 2.85 × 10−02), and axon part (q-value = 3.74 × 10−02) and 2) biological processes including generation, differentiation, and migration of neurons (q-value = 4.57 × 10−02), enzyme linked receptor protein signaling pathway (q-value = 4.57 × 10−02), and actin filament based processes (q-value = 4.57 × 10−02) (Supplementary Data 2).
We then tested whether the Fmr1-Δexon 8−/y DEGs were enriched for Fmrp targets (Darnell et al. 2011) or risk genes for ASD (Xu et al. 2012; De Rubeis et al. 2014; Sanders et al. 2015), ID (Parikshak et al. 2013), or genes found with de novo mutations in SCZ (Fromer et al. 2016). We found that the down-regulated genes in Fmr1-Δexon 8−/y rats show significant overlap with Fmrp targets (∩ = 33, FDR P = 0.0002) and SCZ genes (∩ = 7, FDR P = 0.03) (Supplementary Fig. S10).
Rat mPFC Gene Networks, Preserved in Human Frontal Cortex, are Altered in Fmr1-Δexon 8 Rats
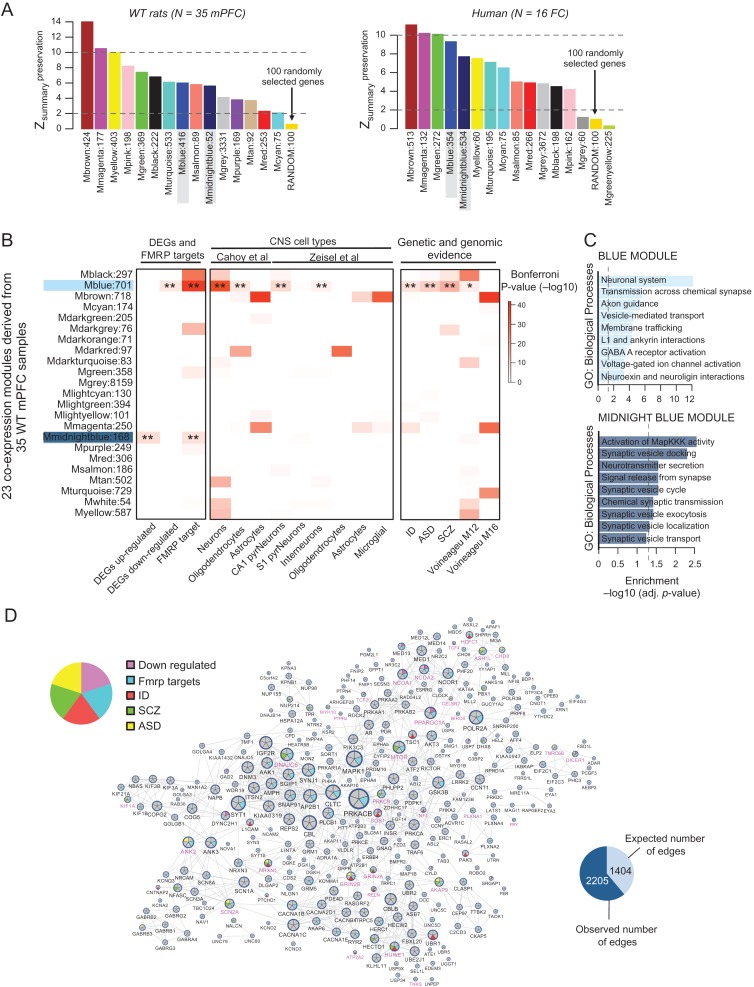
Next, we asked whether mPFC gene networks in WT rats are conserved in human and if any of the conserved networks were especially vulnerable to the effects of Fmr1-Δexon 8 deletion. To address these questions, we first built a reference WT coexpression network by combining mPFC RNAseq data across 35 WT rats, matched for age and sex, and using WGCNA (see Materials and Methods). Because these 35 samples were prepared in 3 different batches, we first used the Combat batch correction (Leek et al. 2012) to resolve any systematic sources of variability (Supplementary Fig. S11) before performing our conetwork analysis. Our WGCNA identified 23 modules specific to the mPFC of WT rats (Supplementary Data 1). Next, we determined whether the coexpression patterns of these 23 modules were preserved in the human brain. For this purpose, we created separate transcriptional networks from previously published human cortex tissue (BA 9/41) sampled from control individuals (Voineagu et al. 2011) in order to systematically explore potential species similarities and differences. Interspecies coexpression preservation has been shown to prioritize disease gene selection under genetic disease loci and to categorize the function of poorly characterized genes better than coexpression in a single species (Miller et al. 2010; Mueller et al. 2017). This approach is sensitive to detecting fundamental differences in the underlying gene coregulatory patterns between WT rats and healthy humans, and vice versa, as being preserved or disrupted. Using a permutation-based preservation statistic (Zsummary) with 500 random permutations, we observed strong to moderate preservation between the 2 species (all network modules displayed a Zsummary score > 2, which was higher than a random sampling of 100 genes), indicating similar levels of gene coregulation between rat mPFC and human frontal cortex (Fig. 7A).
Figure 7.
mPFC gene coexpression networks of WT rats and preservation with human frontal cortex coexpression networks. WGCNA identified 23 modules across 35 WT mPFC samples. (A) Permutation-based preservation statistic (Zsummary) scores (y-axis) were generated with 500 random permutations across all WT modules (x-axis), comparing WT rat mPFC (left) and healthy human frontal cortex (right) preservation to 100 randomly selected genes. Zsummary < 2 indicates no evidence of preservation, 2 < Zsummary < 10 implies moderate preservation, and Zsummary > 10 suggests strong preservation. These results indicate that the blue and midnightblue modules, which are significantly enriched for differentially expressed genes, are indeed preserved. (B) All modules were assessed for enrichment of Fmr1-Δexon 8−/y DGE signatures and Fmrp targets, CNS cell type specific signatures, and genetic risk loci and genomic evidence of neurodevelopmental disorders. Over-representation analysis of these gene sets within DGE signatures and WT transcriptome modules was analyzed using a one-sided Fishers exact test to assess the statistical significance. All P-values, from all gene sets and modules, were adjusted for multiple testing using Bonferroni procedure. (C) Functional annotation of the blue (top) and midnightblue (bottom) modules. (D) Protein interaction network for blue module genes shows a significant over-representation of high-confidence direct protein interactions, beyond what was expected by chance (P < 0.0001). Hub genes include numerous Fmrp target genes and several ID, SCZ, and ASD genetic risk loci. Genes in pink lettering were differentially expressed in Fmr1-Δexon 8−/y rats.
To assess whether WT rat modules were vulnerable to the Fmr1-Δexon 8 deletion, we tested for enrichment of the Fmr1-Δexon 8−/y DEGs. Of the 23 identified modules, one module (blue) contained a strong, significant over-representation for Fmr1-Δexon 8−/y down-regulated genes and FMRP targets, and another module (midnightblue) was enriched for Fmr1-Δexon 8 up-regulated genes and FMRP targets (Fig. 7B). The blue module also contained a significant enrichment for neuronal cell type markers and genes implicated in ASD, ID and SCZ (Fig. 7B, Supplementary Data 1). Functional annotation of the blue module revealed enrichment primarily associated with synaptic signaling, gated channel activity, and neuronal system-related terms (Fig. 7C). The midnightblue module did not display any cell type specificity nor any enrichment for risk genes. Functional annotation of the midnightblue module revealed functional terms implicating MapKKK activity, synaptic vesicle docking, and neurotransmitter secretion (Fig. 7C).
Subsequently, we tested whether genes that are coexpressed together in the blue module indeed interact with each other at the protein level. A significant over-representation of high-confidence direct protein interactions was identified in the blue module, beyond what was expected by chance (P < 0.0001) (Fig. 7D). Hub genes in the blue module include numerous FMRP target genes including MTOR, ANK2, ANK3, SCN2A, GRIN2A, RELN, and NRXN1 (Darnell et al. 2011).
Discussion
This study is the first to uncover that the “Fmr1 KO rat” is not a null KO of Fmr1, but instead results in a gene product with a loss of exon 8, which encodes a domain within Fmr1 that is responsible for RNA-binding, the KH1 domain (Burd and Dreyfuss 1994). Notably, a point mutation was reported in the Fmrp-KH1 domain of an individual with FXS and was shown to cause deficits in mRNA binding, polyribosome association, and mGluR-mediated trafficking of AMPA receptors (Myrick et al. 2014). Further, disrupted function of the KH1 domain was sufficient to cause the classic symptoms of FXS that usually follow from silencing of the entire FMR1 gene, including attention deficits. Similarly, we find that the Fmr1-Δexon 8 rats have impairments in sustained attention that parallel those reported in individuals with FXS (Cornish et al. 2001). In addition, we find alterations in their mPFC transcriptional profiles, which is of potential translational value for subjects with FXS. It is important to note that the deletion in exon 8 in this rat model both caused a depletion of the KH1 domain and led to a decrease in Fmrp expression. Therefore, we cannot discriminate whether the observed phenotype is specifically due to a loss of function of the KH1 domain or simply low expression of Fmrp.
The basis of the deficit in sustained attention could be ascribed to functional impairments in the mPFC of Fmr1-Δexon 8 rats. Dysregulated sustained attention has been shown to follow manipulations of mPFC activity in rats that have previously acquired the task. Lister hooded rats that underwent treatment with an immunotoxin to deplete cholinergic function in the nucleus basalis magnocellularis of the basal forebrain, which sends cholinergic projections to the medial frontal cortex, had increased omissions and no difference in accuracy (Risbrough et al. 2002). Increased omissions and response latency were also observed in rats with lesions to mPFC or imbalanced inhibition/disinhibition in mPFC (Muir et al. 1996; Pezze et al. 2014). The performance of Fmr1-Δexon 8 rats during the acquisition of the 5-CSRTT mirrors the performance of rats that underwent specific manipulations of mPFC activity, indicating that the mPFC is implicated in the manifestation of these attentional deficits in the Fmr1-Δexon 8 rat model and could be a result of an insult to the mPFC by the Fmr1 mutation during early developmental stages. Similar findings were reported in the Fmr1 KO mice, where deficits in acquisition of the 5-CSRTT were accompanied by alterations in prefrontal synaptic composition and neural activity (Krueger et al. 2011). We recently reported a similar deficit in sustained attention and impairment in mPFC synaptic plasticity in a Shank3-deficient rat (Harony-Nicolas et al. 2017), suggesting convergent findings across models of ASD and ID disorders that are often comorbid with ADHD. Taken together, these findings indicate that the mPFC warrants further study as the basis of cognitive impairment in this Fmr1-Δexon 8 rat model of FXS.
Our approach to address this was to probe the molecular profile of the mPFC following the loss of exon 8 using RNAseq analysis. We observed hundreds of dysregulated genes (FDR 10%) associated with Fmr1-Δexon 8. Up-regulated genes were enriched in biological processes that included transmembrane transporter activity. Genes within this GO category included several solute carrier proteins including a member of the Na+/H+ exchanger superfamily, SLC9A9. This family of exchangers controls ion transport across membranes, which is essential for regulating cellular pH and electrical excitability that is known to be affected in FXS (Kondapalli et al. 2014). SLC9A9 is highly expressed in the brain and mutations in the encoding gene have been associated with ASD (Prasad et al. 2017), ADHD (de Silva et al. 2003; Brookes et al. 2006; Lasky-Su et al. 2008), and epilepsy, which are all prevalent in FXS (Kondapalli et al. 2014).
Down-regulated genes were enriched for neural and synaptic components and for biological processes including generation, differentiation, and migration of neurons and actin filament-based processes. Impaired actin cytoskeletal function has consistently been reported in FXS models and is thought to underlie the abnormal dendritic spine phenotype common to subjects with FXS (Maurin et al. 2014). It is possible that the observed down-regulation at the transcriptional level is a secondary effect of increased translation caused by the mutation and/or reduced Fmrp levels. Alternatively, this down-regulation could instead be a direct consequence caused by the loss of Fmrp. For example, Fmrp has been shown to regulate mRNA stability (Zalfa et al. 2007; Zhang et al. 2018). The decreased levels of transcription observed could reflect the destabilization of mRNA molecules in the absence of Fmrp. Future studies are needed to further examine the mechanisms by which a loss of Fmrp leads to the observed transcriptional alterations.
Results derived from our reference WT transcriptome coexpression network echo our DGE findings and further refine the biological processes involved in Fmr1-Δexon 8. Functional annotation of the blue module, which we found to be conserved in human PFC coexpression networks and to be significantly enriched for down-regulated Fmr1-Δexon 8-related genes, known Fmrp targets, neuronal cell type signatures, and genes implicated in ID, ASD and SCZ, revealed enriched GO terms including neuronal system, axon guidance, and neurexin (an Fmrp target (Darnell et al. 2011)) and neuroligin interactions. These biological processes were previously reported to be dysregulated in a transcriptomic study of the cerebellum of Fmr1 KO mice (Kong et al. 2014) and in functional studies of Fmrp (Tucker et al. 2006; Bhakar et al. 2012). Amongst the hub genes in the blue module are numerous FMRP target genes and several ASD risk genes, including MTOR, ANK2, ANK3, TSC1, SCN2A, GRIN2A, RELN and NRXN1. Interestingly, all of these genes are implicated in neurodevelopmental disorders. ANK2, SCN2A, and NRXN1 are top risk genes for ASD (Sanders et al. 2015) and the others are associated with neurodevelopmental syndromes (e.g., MTOR in Smith-Kingsmore syndrome) (MIM 616638), ANK3 in an autosomal recessive ID syndrome (MIM 615493), GRIN2A in a form of focal epilepsy and speech disorder with or without ID (MIM 245570), RELN in a lissencephaly syndrome (MIM 257320), and NRXN1 in Pitt-Hopkins-like syndrome (MIM 614325). This module also includes the TSC1 gene, which is associated with Tuberous Sclerosis (MIM 191100).
In summary, we have shown here that a specific deletion of exon 8 in Fmr1 is sufficient to cause FXS-like phenotypes in rat. The behavioral task we employed provides a tool to screen potential therapeutic candidates for efficacy in treating a highly common cognitive deficit observed in these rats that is also seen in both males and females diagnosed with FXS: dysregulated attention, which is associated with mPFC dysfunction. In addition, the results from our RNAseq analysis of the mPFC supply multiple potential treatment avenues to explore. Now that these deficits are elucidated in the Fmr1-Δexon 8 rat, we can begin to uncover their underlying circuit mechanisms by probing the mPFC with in vivo imaging and electrophysiology.
Supplementary Material
Footnotes
Carla E.M. Golden is a Seaver Graduate Fellow and Michael S. Breen is a Seaver Postdoctoral Fellow. We thank Eilam Doron, who contributed to this work, and The Mount Sinai Genomics Core Facility for their services in carrying out the Next Generation RNA sequencings. Conflict of Interest: None declared.
Funding
Seaver Foundation (to J.D.B., H.H.N., S.D.R., C.E.M.G., and M.S.B.); Autism Speaks (to J.D.B.); Autism Science Foundation (Grant number 17-001 to M.S.B.); and the National Institute of Mental Health (Grant number F31 MH115656-01 to C.E.M.G., and 5R01MH101584-05 to J.D.B.).
References
- Archer J. 1975. Rodent sex differences in emotional and related behavior. Behav Biol. 14:451–479. [DOI] [PubMed] [Google Scholar]
- Auclair AL, Besnard J, Newman-Tancredi A, Depoortere R. 2009. The five choice serial reaction time task: comparison between Sprague-Dawley and Long-Evans rats on acquisition of task, and sensitivity to phencyclidine. Pharmacol Biochem Behav. 92:363–369. [DOI] [PubMed] [Google Scholar]
- Bagni C, Tassone F, Neri G, Hagerman R. 2012. Fragile X syndrome: causes, diagnosis, mechanisms, and therapeutics. J Clin Invest. 122:4314–4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker KB, Wray SP, Ritter R, Mason S, Lanthorn TH, Savelieva KV. 2010. Male and female Fmr1 knockout mice on C57 albino background exhibit spatial learning and memory impairments. Genes Brain Behav. 9:562–574. [DOI] [PubMed] [Google Scholar]
- Bakker CE, Verheij C, Willemsen R, Helm R, Oerlemans F, Vermey M, Bygrave A, Hoogeveen AT, Oostra BA, Reyniers E, et al. , The Dutch-Belgian Fragile X Consortium . 1994. Fmr1 knockout mice: a model to study fragile X mental retardation. Cell. 78:23–33. [PubMed] [Google Scholar]
- Berzhanskaya J, Phillips MA, Gorin A, Lai C, Shen J, Colonnese MT. 2017. Disrupted cortical state regulation in a rat model of fragile X syndrome. Cereb Cortex. 27:1386–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhakar AL, Dolen G, Bear MF. 2012. The pathophysiology of fragile X (and what it teaches us about synapses). Annu Rev Neurosci. 35:417–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutros N, Der-Avakian A, Markou A, Semenova S. 2017. Effects of early life stress and adolescent ethanol exposure on adult cognitive performance in the 5-choice serial reaction time task in Wistar male rats. Psychopharmacology (Berl). 234:1549–1556. [DOI] [PubMed] [Google Scholar]
- Bozdagi O, Sakurai T, Papapetrou D, Wang X, Dickstein DL, Takahashi N, Kajiwara Y, Yang M, Katz AM, Scattoni ML, et al. 2010. Haploinsufficiency of the autism-associated Shank3 gene leads to deficits in synaptic function, social interaction, and social communication. Mol Autism. 1:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray S, Hirt M, Jo B, Hall SS, Lightbody AA, Walter E, Chen K, Patnaik S, Reiss AL. 2011. Aberrant frontal lobe maturation in adolescents with fragile X syndrome is related to delayed cognitive maturation. Biol Psychiatry. 70:852–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookes K, Xu X, Chen W, Zhou K, Neale B, Lowe N, Anney R, Franke B, Gill M, Ebstein R, et al. 2006. The analysis of 51 genes in DSM-IV combined type attention deficit hyperactivity disorder: association signals in DRD4, DAT1 and 16 other genes. Mol Psychiatry. 11:934–953. [DOI] [PubMed] [Google Scholar]
- Brown V, Jin P, Ceman S, Darnell JC, O’Donnell WT, Tenenbaum SA, Jin X, Feng Y, Wilkinson KD, Keene JD, et al. 2001. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 107:477–487. [DOI] [PubMed] [Google Scholar]
- Burd CG, Dreyfuss G. 1994. Conserved structures and diversity of functions of RNA-binding proteins. Science. 265:615–621. [DOI] [PubMed] [Google Scholar]
- Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, et al. 2008. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 28:264–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Bardes EE, Aronow BJ, Jegga AG. 2009. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 37:W305–W311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chudasama Y, Passetti F, Rhodes SE, Lopian D, Desai A, Robbins TW. 2003. Dissociable aspects of performance on the 5-choice serial reaction time task following lesions of the dorsal anterior cingulate, infralimbic and orbitofrontal cortex in the rat: differential effects on selectivity, impulsivity and compulsivity. Behav Brain Res. 146:105–119. [DOI] [PubMed] [Google Scholar]
- Chudasama Y, Robbins TW. 2006. Functions of frontostriatal systems in cognition: comparative neuropsychopharmacological studies in rats, monkeys and humans. Biol Psychol. 73:19–38. [DOI] [PubMed] [Google Scholar]
- Coffee B, Ikeda M, Budimirovic DB, Hjelm LN, Kaufmann WE, Warren ST. 2008. Mosaic FMR1 deletion causes fragile X syndrome and can lead to molecular misdiagnosis: a case report and review of the literature. Am J Med Genet A. 146A:1358–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins SC, Bray SM, Suhl JA, Cutler DJ, Coffee B, Zwick ME, Warren ST. 2010. Identification of novel FMR1 variants by massively parallel sequencing in developmentally delayed males. Am J Med Genet A. 152A:2512–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish KM, Munir F, Cross G. 2001. Differential impact of the FMR-1 full mutation on memory and attention functioning: a neuropsychological perspective. J Cogn Neurosci. 13:144–150. [DOI] [PubMed] [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, et al. 2011. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 146:247–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Boulle K, Verkerk AJ, Reyniers E, Vits L, Hendrickx J, Van Roy B, Van den Bos F, de Graaff E, Oostra BA, Willems PJ. 1993. A point mutation in the FMR-1 gene associated with fragile X mental retardation. Nat Genet. 3:31–35. [DOI] [PubMed] [Google Scholar]
- De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, Kou Y, Liu L, Fromer M, Walker S, et al. 2014. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 515:209–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rubeis S, Pasciuto E, Li KW, Fernandez E, Di Marino D, Buzzi A, Ostroff LE, Klann E, Zwartkruis FJ, Komiyama NH, et al. 2013. CYFIP1 coordinates mRNA translation and cytoskeleton remodeling to ensure proper dendritic spine formation. Neuron. 79:1169–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Silva MG, Elliott K, Dahl HH, Fitzpatrick E, Wilcox S, Delatycki M, Williamson R, Efron D, Lynch M, Forrest S. 2003. Disruption of a novel member of a sodium/hydrogen exchanger family and DOCK3 is associated with an attention deficit hyperactivity disorder-like phenotype. J Med Genet. 40:733–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Marino D, Achsel T, Lacoux C, Falconi M, Bagni C. 2014. Molecular dynamics simulations show how the FMRP Ile304Asn mutation destabilizes the KH2 domain structure and affects its function. J Biomol Struct Dyn. 32:337–350. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolen G, Osterweil E, Rao BS, Smith GB, Auerbach BD, Chattarji S, Bear MF. 2007. Correction of fragile X syndrome in mice. Neuron. 56:955–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Annessa I, Cicconardi F, Marino D In Press. Handling FMRP and its molecular partners: structural insights into fragile X syndrome. Prog Biophys Mol Biol. [DOI] [PubMed] [Google Scholar]
- Engineer CT, Centanni TM, Im KW, Rahebi KC, Buell EP, Kilgard MP. 2014. Degraded speech sound processing in a rat model of fragile X syndrome. Brain Res. 1564:72–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, et al. 2013. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 41:D808–D815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromer M, Roussos P, Sieberts SK, Johnson JS, Kavanagh DH, Perumal TM, Ruderfer DM, Oh EC, Topol A, Shah HR, et al. 2016. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat Neurosci. 19:1442–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronskov K, Brondum-Nielsen K, Dedic A, Hjalgrim H. 2011. A nonsense mutation in FMR1 causing fragile X syndrome. Eur J Hum Genet. 19:489–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross C, Hoffmann A, Bassell GJ, Berry-Kravis EM. 2015. a. Therapeutic strategies in fragile X syndrome: from bench to bedside and back. Neurotherapeutics. 12:584–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross C, Raj N, Molinaro G, Allen AG, Whyte AJ, Gibson JR, Huber KM, Gourley SL, Bassell GJ. 2015. b. Selective role of the catalytic PI3K subunit p110beta in impaired higher order cognition in fragile X syndrome. Cell Rep. 11:681–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallahan BP, Craig MC, Toal F, Daly EM, Moore CJ, Ambikapathy A, Robertson D, Murphy KC, Murphy DG. 2011. In vivo brain anatomy of adult males with Fragile X syndrome: an MRI study. Neuroimage. 54:16–24. [DOI] [PubMed] [Google Scholar]
- Hamilton SM, Green JR, Veeraragavan S, Yuva L, McCoy A, Wu Y, Warren J, Little L, Ji D, Cui X, et al. 2014. Fmr1 and Nlgn3 knockout rats: novel tools for investigating autism spectrum disorders. Behav Neurosci. 128:103–109. [DOI] [PubMed] [Google Scholar]
- Hammond LS, Macias MM, Tarleton JC, Shashidhar Pai G. 1997. Fragile X syndrome and deletions in FMR1: new case and review of the literature. Am J Med Genet. 72:430–434. [PubMed] [Google Scholar]
- Handt M, Epplen A, Hoffjan S, Mese K, Epplen JT, Dekomien G. 2014. Point mutation frequency in the FMR1 gene as revealed by fragile X syndrome screening. Mol Cell Probes. 28:279–283. [DOI] [PubMed] [Google Scholar]
- Hansen KD, Irizarry RA, Wu Z. 2012. Removing technical variability in RNA-seq data using conditional quantile normalization. Biostatistics. 13:204–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harony-Nicolas H, Kay M, Hoffmann JD, Klein ME, Bozdagi-Gunal O, Riad M, Daskalakis NP, Sonar S, Castillo PE, Hof PR, et al. 2017. Oxytocin improves behavioral and electrophysiological deficits in a novel Shank3-deficient rat. Elife. 6:e18904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. 2007. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 8:R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeft F, Carter JC, Lightbody AA, Cody Hazlett H, Piven J, Reiss AL. 2010. Region-specific alterations in brain development in one- to three-year-old boys with fragile X syndrome. Proc Natl Acad Sci USA. 107:9335–9339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeft F, Hernandez A, Parthasarathy S, Watson CL, Hall SS, Reiss AL. 2007. Fronto-striatal dysfunction and potential compensatory mechanisms in male adolescents with fragile X syndrome. Hum Brain Mapp. 28:543–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber KM, Gallagher SM, Warren ST, Bear MF. 2002. Altered synaptic plasticity in a mouse model of fragile X mental retardation. Proc Natl Acad Sci USA. 99:7746–7750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazdoba TM, Leach PT, Silverman JL, Crawley JN. 2014. Modeling fragile X syndrome in the Fmr1 knockout mouse. Intractable Rare Dis Res. 3:118–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenkel WM, Yee JR, Moore K, Madularu D, Kulkarni P, Gamber K, Nedelman M, Ferris CF. 2016. Functional magnetic resonance imaging in awake transgenic fragile X rats: evidence of dysregulation in reward processing in the mesolimbic/habenular neural circuit. Transl Psychiatry. 6:e763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondapalli KC, Prasad H, Rao R. 2014. An inside job: how endosomal Na(+)/H(+) exchangers link to autism and neurological disease. Front Cell Neurosci. 8:172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong SW, Sahin M, Collins CD, Wertz MH, Campbell MG, Leech JD, Krueger D, Bear MF, Kunkel LM, Kohane IS. 2014. Divergent dysregulation of gene expression in murine models of fragile X syndrome and tuberous sclerosis. Mol Autism. 5:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger DD, Osterweil EK, Chen SP, Tye LD, Bear MF. 2011. Cognitive dysfunction and prefrontal synaptic abnormalities in a mouse model of fragile X syndrome. Proc Natl Acad Sci USA. 108:2587–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulinski J, Besack D, Oleykowski CA, Godwin AK, Yeung AT. 2000. CEL I enzymatic mutation detection assay. Biotechniques. 29(44–46):48. [DOI] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. 2008. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 9:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Luo R, Oldham MC, Horvath S. 2011. Is my network module preserved and reproducible? PLoS Comput Biol. 7:e1001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasky-Su J, Neale BM, Franke B, Anney RJ, Zhou K, Maller JB, Vasquez AA, Chen W, Asherson P, Buitelaar J, et al. 2008. Genome-wide association scan of quantitative traits for attention deficit hyperactivity disorder identifies novel associations and confirms candidate gene associations. Am J Med Genet B Neuropsychiatr Genet. 147B:1345–1354. [DOI] [PubMed] [Google Scholar]
- Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. 2012. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 28:882–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. 2014. FeatureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 30:923–930. [DOI] [PubMed] [Google Scholar]
- Lugenbeel KA, Peier AM, Carson NL, Chudley AE, Nelson DL. 1995. Intragenic loss of function mutations demonstrate the primary role of FMR1 in fragile X syndrome. Nat Genet. 10:483–485. [DOI] [PubMed] [Google Scholar]
- Mar AC, Horner AE, Nilsson SR, Alsio J, Kent BA, Kim CH, Holmes A, Saksida LM, Bussey TJ. 2013. The touchscreen operant platform for assessing executive function in rats and mice. Nat Protoc. 8:1985–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurin T, Zongaro S, Bardoni B. 2014. Fragile X syndrome: from molecular pathology to therapy. Neurosci Biobehav Rev. 46(Pt 2):242–255. [DOI] [PubMed] [Google Scholar]
- Menon V, Leroux J, White CD, Reiss AL. 2004. Frontostriatal deficits in fragile X syndrome: relation to FMR1 gene expression. Proc Natl Acad Sci USA. 101:3615–3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JA, Horvath S, Geschwind DH. 2010. Divergence of human and mouse brain transcriptome highlights Alzheimer disease pathways. Proc Natl Acad Sci USA. 107:12698–12703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller AJ, Canty-Laird EG, Clegg PD, Tew SR. 2017. Cross-species gene modules emerge from a systems biology approach to osteoarthritis. NPJ Syst Biol Appl. 3:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muir JL, Everitt BJ, Robbins TW. 1996. The cerebral cortex of the rat and visual attentional function: dissociable effects of mediofrontal, cingulate, anterior dorsolateral, and parietal cortex lesions on a five-choice serial reaction time task. Cereb Cortex. 6:470–481. [DOI] [PubMed] [Google Scholar]
- Myrick LK, Deng PY, Hashimoto H, Oh YM, Cho Y, Poidevin MJ, Suhl JA, Visootsak J, Cavalli V, Jin P, et al. 2015. a. Independent role for presynaptic FMRP revealed by an FMR1 missense mutation associated with intellectual disability and seizures. Proc Natl Acad Sci USA. 112:949–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myrick LK, Hashimoto H, Cheng X, Warren ST. 2015. b. Human FMRP contains an integral tandem Agenet (Tudor) and KH motif in the amino terminal domain. Hum Mol Genet. 24:1733–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myrick LK, Nakamoto-Kinoshita M, Lindor NM, Kirmani S, Cheng X, Warren ST. 2014. Fragile X syndrome due to a missense mutation. Eur J Hum Genet. 22:1185–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli I, Mercaldo V, Boyl PP, Eleuteri B, Zalfa F, De Rubeis S, Di Marino D, Mohr E, Massimi M, Falconi M, et al. 2008. The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell. 134:1042–1054. [DOI] [PubMed] [Google Scholar]
- Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M, Alizadeh AA. 2015. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 12:453–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okray Z, de Esch CE, Van Esch H, Devriendt K, Claeys A, Yan J, Verbeeck J, Froyen G, Willemsen R, de Vrij FM, et al. 2015. A novel fragile X syndrome mutation reveals a conserved role for the carboxy-terminus in FMRP localization and function. EMBO Mol Med. 7:423–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterweil EK, Krueger DD, Reinhold K, Bear MF. 2010. Hypersensitivity to mGluR5 and ERK1/2 leads to excessive protein synthesis in the hippocampus of a mouse model of fragile X syndrome. J Neurosci. 30:15616–15627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikshak NN, Luo R, Zhang A, Won H, Lowe JK, Chandran V, Horvath S, Geschwind DH. 2013. Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell. 155:1008–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezze M, McGarrity S, Mason R, Fone KC, Bast T. 2014. Too little and too much: hypoactivation and disinhibition of medial prefrontal cortex cause attentional deficits. J Neurosci. 34:7931–7946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozdnyakova I, Regan L. 2005. New insights into Fragile X syndrome. Relating genotype to phenotype at the molecular level. FEBS J. 272:872–878. [DOI] [PubMed] [Google Scholar]
- Prasad H, Osei-Owusu J, Rao R. 2017. Functional analysis of Na(+)/H(+) exchanger 9 variants identified in patients with autism and epilepsy. Matters (Zur). 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin M, Kang J, Burlin TV, Jiang C, Smith CB. 2005. Postadolescent changes in regional cerebral protein synthesis: an in vivo study in the FMR1 null mouse. J Neurosci. 25:5087–5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quartier A, Poquet H, Gilbert-Dussardier B, Rossi M, Casteleyn AS, Portes VD, Feger C, Nourisson E, Kuentz P, Redin C, et al. 2017. Intragenic FMR1 disease-causing variants: a significant mutational mechanism leading to Fragile-X syndrome. Eur J Hum Genet. 25:423–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. 2006. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- Richter JD, Bassell GJ, Klann E. 2015. Dysregulation and restoration of translational homeostasis in fragile X syndrome. Nat Rev Neurosci. 16:595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risbrough V, Bontempi B, Menzaghi F. 2002. Selective immunolesioning of the basal forebrain cholinergic neurons in rats: effect on attention using the 5-choice serial reaction time task. Psychopharmacology (Berl). 164:71–81. [DOI] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins TW. 2002. The 5-choice serial reaction time task: behavioural pharmacology and functional neurochemistry. Psychopharmacology (Berl). 163:362–380. [DOI] [PubMed] [Google Scholar]
- Sanders SJ, He X, Willsey AJ, Ercan-Sencicek AG, Samocha KE, Cicek AE, Murtha MT, Bal VH, Bishop SL, Dong S, et al. 2015. Insights into autism spectrum disorder genomic architecture and biology from 71 risk loci. Neuron. 87:1215–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro MR, Bray SM, Warren ST. 2012. Molecular mechanisms of fragile X syndrome: a twenty-year perspective. Annu Rev Pathol. 7:219–245. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 9:671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenova S. 2012. Attention, impulsivity, and cognitive flexibility in adult male rats exposed to ethanol binge during adolescence as measured in the five-choice serial reaction time task: the effects of task and ethanol challenges. Psychopharmacology (Berl). 219:433–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13:2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidorov MS, Krueger DD, Taylor M, Gisin E, Osterweil EK, Bear MF. 2014. Extinction of an instrumental response: a cognitive behavioral assay in Fmr1 knockout mice. Genes Brain Behav. 13:451–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitzmann AF, Hagelstrom RT, Tassone F, Hagerman RJ, Butler MG. 2018. Rare FMR1 gene mutations causing fragile X syndrome: a review. Am J Med Genet A. 176:11–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen EM, Bertelsen F, Weikop P, Skovborg MM, Banke T, Drasbek KR, Scheel-Kruger J. 2015. Hyperactivity and lack of social discrimination in the adolescent Fmr1 knockout mouse. Behav Pharmacol. 26:733–740. [DOI] [PubMed] [Google Scholar]
- Spijker S. 2011. Dissection of rodent brain regions. Neuroproteomics. 57:13–26. [Google Scholar]
- Suhl JA, Warren ST. 2015. Single-nucleotide mutations in FMR1 reveal novel functions and regulatory mechanisms of the fragile X syndrome protein FMRP. J Exp Neurosci. 9:35–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tariq MA, Kim HJ, Jejelowo O, Pourmand N. 2011. Whole-transcriptome RNAseq analysis from minute amount of total RNA. Nucleic Acids Res. 39:e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till SM, Asiminas A, Jackson AD, Katsanevaki D, Barnes SA, Osterweil EK, Bear MF, Chattarji S, Wood ER, Wyllie DJA, et al. 2015. Conserved hippocampal cellular pathophysiology but distinct behavioural deficits in a new rat model of FXS. Hum Mol Genet. 24:5977–5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker B, Richards RI, Lardelli M. 2006. Contribution of mGluR and Fmr1 functional pathways to neurite morphogenesis, craniofacial development and fragile X syndrome. Hum Mol Genet. 15:3446–3458. [DOI] [PubMed] [Google Scholar]
- Valverde R, Edwards L, Regan L. 2008. Structure and function of KH domains. FEBS J. 275:2712–2726. [DOI] [PubMed] [Google Scholar]
- Vasilyev N, Polonskaia A, Darnell JC, Darnell RB, Patel DJ, Serganov A. 2015. Crystal structure reveals specific recognition of a G-quadruplex RNA by a beta-turn in the RGG motif of FMRP. Proc Natl Acad Sci USA. 112:E5391–E5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voineagu I, Wang X, Johnston P, Lowe JK, Tian Y, Horvath S, Mill J, Cantor RM, Blencowe BJ, Geschwind DH. 2011. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 474:380–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijetunge LS, Angibaud J, Frick A, Kind PC, Nagerl UV. 2014. Stimulated emission depletion (STED) microscopy reveals nanoscale defects in the developmental trajectory of dendritic spine morphogenesis in a mouse model of fragile X syndrome. J Neurosci. 34:6405–6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong HY, Alipanahi B, Lee LJ, Bretschneider H, Merico D, Yuen RK, Hua Y, Gueroussov S, Najafabadi HS, Hughes TR, et al. 2015. RNA splicing. The human splicing code reveals new insights into the genetic determinants of disease. Science. 347:1254806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu LM, Li JR, Huang Y, Zhao M, Tang X, Wei L. 2012. AutismKB: an evidence-based knowledgebase of autism genetics. Nucleic Acids Res. 40:D1016–D1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalfa F, Eleuteri B, Dickson KS, Mercaldo V, De Rubeis S, di Penta A, Tabolacci E, Chiurazzi P, Neri G, Grant SG, et al. 2007. A new function for the fragile X mental retardation protein in regulation of PSD-95 mRNA stability. Nat Neurosci. 10:578–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang JB, Nosyreva ED, Spencer CM, Volk LJ, Musunuru K, Zhong R, Stone EF, Yuva-Paylor LA, Huber KM, Paylor R, et al. 2009. A mouse model of the human fragile X syndrome I304N mutation. PLoS Genet. 5:e1000758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel A, Munoz-Manchado AB, Codeluppi S, Lonnerberg P, La Manno G, Jureus A, Marques S, Munguba H, He L, Betsholtz C, et al. 2015. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 347:1138–1142. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, et al. 2014. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 34:11929–11947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Kang Y, Wang M, Li Y, Xu T, Yang W, Song H, Wu H, Shu Q, Jin P. 2018. Fragile X mental retardation protein modulates the stability of its m6A-marked messenger RNA targets. Hum Mol Genet. 27:3936–3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.