Abstract
Temperature compensation and period determination by casein kinase 1 (CK1) are conserved features of eukaryotic circadian rhythms, whereas the clock gene transcription factors that facilitate daily gene expression rhythms differ between phylogenetic kingdoms. Human red blood cells (RBCs) exhibit temperature-compensated circadian rhythms, which, because RBCs lack nuclei, must occur in the absence of a circadian transcription-translation feedback loop. We tested whether period determination and temperature compensation are dependent on CKs in RBCs. As with nucleated cell types, broad-spectrum kinase inhibition with staurosporine lengthened the period of the RBC clock at 37°C, with more specific inhibition of CK1 and CK2 also eliciting robust changes in circadian period. Strikingly, inhibition of CK1 abolished temperature compensation and increased the Q10 for the period of oscillation in RBCs, similar to observations in nucleated cells. This indicates that CK1 activity is essential for circadian rhythms irrespective of the presence or absence of clock gene expression cycles.
Keywords: casein kinase, temperature compensation, erythrocyte, dielectrophoresis, electrophysiology
Most organisms use endogenous timekeeping mechanisms to coordinate their physiology with the external cycle of day and night. These circadian rhythms have a cell-autonomous basis (Balsalobre et al., 1998; Welsh et al., 2004) orchestrating many cellular functions, such as DNA repair, protein synthesis, and cell division, to an approximately 24-h beat (Dekens et al., 2003; Kang et al., 2009; Lipton et al., 2015; Matsuo et al., 2003). Circadian rhythms bestow a selective advantage upon organisms in rhythmic and arrhythmic settings by synchronising them with predictable daily environmental cycles and facilitating the temporal segregation of mutually antagonistic processes (Beale et al., 2016; Vaze and Sharma, 2013). In most cell types, these temporal programs are thought to be dependent on the rhythmic regulation of gene expression by oscillating “clock gene” activity, in turn facilitated by transcriptional feedback repression of clock gene transcription by their own protein products (Takahashi, 2017).
Transcriptional oscillations are not essential for circadian rhythms; bona fide circadian rhythms occur in the complete absence of transcription in the alga Ostreococcus tauri (O’Neill et al., 2011), in human red blood cells (RBCs) (O’Neill and Reddy, 2011), and in other organisms (Lakin-Thomas, 2006; Tomita et al., 2005; Woolum, 1991). Importantly, these examples of transcription-independent circadian rhythms satisfy the 3 fundamental properties that define circadian clocks (Pittendrigh, 1960): they are entrained by relevant environmental cues, such as light or temperature; they free run with an approximately 24-h period; and they are temperature compensated, meaning that they do not run faster at higher temperatures, i.e., Q10 ≈ 1, unlike most chemical and biochemical reactions, where Q10 ≈ 2.
Although temperature compensation is a defining feature of circadian clocks, it remains poorly understood. Some period-determining processes, and thus candidate mechanisms that facilitate temperature compensation, are known, and these tend to involve the phosphorylation of clock proteins by Ser/Thr kinases (Gallego and Virshup, 2007). One important and evolutionarily conserved post-translational modifier is casein kinase 1 (CK1), which phosphorylates non-orthologous clock protein targets, such as PERIOD (PER) in mammals and FREQUENCY (FRQ) in fungi (He et al., 2006; Meng et al., 2008), and has been shown to regulate period and temperature compensation of circadian rhythms across eukaryotes (Causton et al., 2015; He et al., 2006; Hirota et al., 2010; Meng et al., 2008; van Ooijen et al., 2013).
Whilst mutation of individual PERIOD homologs can alter circadian period by several hours, these perturbations have only modest effects on temperature compensation (Dibner et al., 2009). Recently, it was shown that CK1δ/ε-dependent phosphorylation is intrinsically temperature compensated, with a CK1-specific domain around K224 sufficient to confer temperature compensation on other kinases (Isojima et al., 2009; Shinohara et al., 2017). Since temperature compensation and the circadian function of CK1 exhibit far greater evolutionary conservation than any known clock protein substrate or clock gene transcriptional circuit, we hypothesised that the timekeeping and temperature compensation function of CK1 might extend beyond its known interaction with clock gene transcriptional circuits to include transcription-independent circadian rhythms. Here, we employed a reduced model of non-transcriptional circadian rhythms—isolated human RBCs—to test whether period determination and temperature compensation remain dependent on CK1 activity in the absence of clock protein expression and cycling transcription.
Materials and Methods
Experimental Model and Subject Details
Studies were conducted in accordance with the principles of the Declaration of Helsinki, with approval/favourable opinion from our local Research Ethics Committee. Participants in the study were screened for relevant self-reported health issues, including sleep disorders or excessive daytime sleepiness. All participants provided written, informed consent after having received a detailed explanation of the study procedures. RBCs were isolated from male participants, with RBCs from 4 separate donors for each treatment or experimental condition.
Human RBC Isolation
RBCs were isolated following the protocol of Henslee et al. (2017). RBCs were isolated from anticoagulant-treated whole blood (lithium heparin Vacutainer, BD Biosciences; Franklin Lakes, NJ) by density gradient centrifugation, layering a mixture (1:2.67) of whole blood and Dulbecco’s phosphate-buffered saline (DPBS; Sigma-Aldrich, St Louis, MO) on top of Histopaque-1077 (Sigma-Aldrich) at 50 ×g for 30 min. RBCs were washed once in DPBS (250 ×g for 15 min) and once in modified Krebs-Henseleit buffer (KHB; Sigma-Aldrich) (250 ×g for 10 min). KHB was modified by supplementation with calcium chloride dihydrate (to a final concentration of 2.54 mM), sodium bicarbonate (25 mM), bovine serum albumin (1 g/l), penicillin (100 U/l ) and streptomycin (100 µg/l), and adjusted to pH 7.4 and 290-299 mOsm. After washing, 330 µl of packed RBCs were added to 40 ml of KHB.
Circadian Entrainment
RBCs were subjected to daily temperature cycles (12 h, 37°C; 12 h, 32°C) for 2 days using a programmed thermal cycler before transfer into constant temperature conditions of either 37°C or 32°C (as specified). Samples were taken at 3- or 4-hourly intervals upon transfer into constant conditions, with a single 500-µl PCR tube of RBCs at each time point for each donor. Following convention, the transition to constant conditions was taken as t = 0.
Dielectrophoresis
After the 48-h entrainment, at each timepoint, a single PCR tube was removed from the thermal cycler to assess cell electrophysiology by dielectrophoresis (DEP). The modified KHB medium was removed and the pellet was suspended in iso-osmotic DEP medium (248 mM sucrose, 16.7 mM dextrose, 250 mM MgCl2, 100 mM CaCl2, adjusted to 290-299 mOsm and a conductivity of 0.043 S/m using DPBS). Cells in DEP medium were then centrifuged for 2 min at 500 ×g, and the medium was replaced with fresh DEP medium to reduce ionic carryover from the high ionic strength of the KHB medium. The cells were subsequently adjusted to a final concentration of 106 per ml and pipetted into 3DEP chips (Labtech; Heathfield, East Sussex, UK). Chips were analysed by a 3DEP 3D Dielectrophoresis Cell Analysis System (3DEP reader, Labtech). The wells were energised for 30 s at 10 Vp–p, at 20 frequencies between 10 kHz and 20 MHz. This was repeated 3 to 4 times for each donor at each time point. The raw data were fitted with a single-shell model using 3DEP software (v1.2, Labtech, https://www.labtech.com/3dep-3d-dielectrophoresis-cell-analysis-system) to extract the electrophysiological parameters; models that produced an R value (Pearson correlation coefficient) of 0.95 or lower were excluded.
Bioluminescence Imaging
Human U2OS cells (HTB-96, ATCC; Manassas, VA) were stably transfected with BMAL1:Luc and cultured in Dulbecco’s Modified Essential Medium (DMEM) supplemented with 10% HyClone FetalClone II Serum (Thermo Fisher Scientific; Waltham, MA), 1 mM luciferin (Biosynth AG; Staad, Switzerland), 100 U/mL penicillin, 100 μg/mL streptomycin, and 2 μg/mL puromycin (to maintain selection). Four days before bioluminescence imaging, cells were trypsinized, resuspended in culture medium, seeded into six 96-well white plates at a density of 104 cells per well, and incubated in a humidified incubator for 96 h (37°C, 5% CO2) under daily applied temperature cycles (12 h 32°C; 12 h 37°C). At 4 h before the anticipated transition to the warm phase, cell media was exchanged for recording media (± drug or vehicle) and the plates were placed into an ALLIGATOR for bioluminescence imaging, and were maintained at a constant temperature and 5% CO2 (Crosby et al., 2017). The recording media was identical to the culture media except that the serum concentration was reduced to 1%; B27 supplement (Thermo Fisher) was added to a final concentration of 2%, and puromycin was omitted. For constant culture conditions at 40°C, cells were plated as above but, rather than being subjected to daily temperature cycles, incubation temperature was increased by 1°C per day from 37°C to 40°C to acclimate cells to the higher temperature. Cells were synchronised on the fourth day after plating, with the media changed to media containing B27, as described above. Bioluminescence levels were integrated every 30 min without EM gain.
Gel Electrophoresis and Immunoblotting
RBC pellets were lysed by trituration in 200 µl 1× LDS sample buffer (Thermo Fisher Scientific) supplemented with 10 mM TCEP. Before electrophoresis, samples were heated at 70°C for 10 min on a heat block. Western blotting and SDS-PAGE were performed using the NuPage Novex mini system (Thermo Fisher Scientific) with 4% to 12% bis-tris gradient gels and nitrocellulose transfer stacks, used according to the manufacturer’s instructions. Blots were blocked in 5% (w/v) non-fat dried milk (Marvel) dissolved in Tris-buffered saline containing 0.05% Tween-20 (TBST) for 1 h at room temperature, and then incubated overnight at 4°C on a rotary shaker with primary antibodies diluted in blocking buffer. Primary antibodies were as follows: rabbit anti-CK1α (1:1000, ab108296; Abcam, Cambridge, UK), mouse anti-CK1δ (1:1000, ab85320; Abcam), mouse anti-CK1ε (1:2000, ab82426; Abcam), rabbit anti-CK2α‘ (1:1000, ab10474; Abcam) and rabbit anti-CK2β (1:1000, ab133576; Abcam). Blots were washed for 3 × 5 min in TBST then incubated for 1 h with HRP-conjugated anti-rabbit (1:5000) or anti-mouse (1:10,000) secondary antibodies. Blots were washed again for 3 × 15 min with TBST before chemiluminescence detection using Immobilon Western Chemiluminescent HRP Substrate (Millipore; Billerica, MA), according to manufacturer’s instructions. RIPA extract lysates of U2OS cells transfected with mammalian expression constructs of V5-CK1α (Addgene plasmid #92014, pCAGImC_V5-CSNK1A1, a gift from Joshua Mendell), CK1δ-V5His, HA-CK1ε (Addgene plasmid #13724, V405 4HA-CKIe, a gift from David Virshup), HA-CK2α’ and Myc-CK2β (Addgene plasmids #27087 and #27088, pZW16 [CK2alpha’] and pZW12 [CK2beta] respectively; gifts from David Litchfield) served as positive controls.
Pharmacological Treatments
For each drug, cell viability and morphology were initially screened over a period of 2 to 4 days using a range of concentrations. Concentrations of the drug that did not result in hemolysis or altered viability or morphology under our experimental conditions (following Henslee et al., 2017) were chosen for subsequent time-course experiments. The drugs used in this study—staurosporine, PF-670462, D4476 (all Sigma-Aldrich), and TTP22 (Tocris Biosciences; Bristol, UK)—were prepared in DMSO according to the manufacturer’s instructions. Drugs were diluted in KHB and delivered as a 10X concentration bolus to RBC suspensions to achieve the final concentration. In each case, an equivalent % of DMSO was used as a vehicle control. Drugs were added to RBC suspensions at ZT0 on day 1 (staurosporine) or circadian time (CT)-0 on day 3 (PF-670462, D4476, TTP22), as indicated in the figure legend. For human U2OS cells, PF-670462 was diluted from a 10 mM DMSO stock directly into recording medium, and was delivered to cells through a media change at the beginning of the recording. This was compared with vehicle control (between 0.03% to 0.1% DMSO).
Sequence Comparisons
The following sequences were compared: the catalytic domain (8-282) and C-terminus (283-end) of human CK1 delta isoform 1 (NP_001884), isoform 2 (NP_620693) and isoform 3 (NP_001350678); the catalytic domain (isoform 1: 16-309; isoforms 2 and 3: 16-281) and C-terminus (isoform 1: 309-end; isoforms 2 and 3: 282-end) of human CK1 alpha isoform 1 (NP_001020276), isoform 2 (NP_001883.4) and isoform 3 (NP_001258670.1). Orthologous sequences in Arabidopsis thaliana (NP_193170), Neurospora crassa (ESA44278), Ostreococcus tauri (OUS42226) and Drosophila melanogaster (CK1 alpha: CAA64358; doubletime/discs overgrown: NP_524602) were also compared using NCBI protein-protein Basic Local Alignment Search Tool (BLASTP) using default settings.
Quantification and Statistical Analysis
All data are presented as the mean ± SEM, where n refers to the number of biological replicates. To determine whether a circadian rhythm was present in time-course experiments, and subsequently the circadian parameters period, amplitude and phase, a damped cosine was fit to the data, as described by Hirota et al., (2008), using least squares non-linear regression curve fitting in GraphPad Prism (v7.03, GraphPad; La Jolla, CA). Briefly, a straight line fit () was compared with a damped cosine + baseline fit:
where X = time, using the extra sum-of-squares F comparison test in GraphPad Prism, with the simpler model being preferred unless the p value was < 0.05. For the calculation of Q10 value, the period values were entered into the following equation:
where τ1 and τ2 are the period at temperature T1 and T2, respectively. T1 = 32°C and T2 = 37°C. Rhythm parameters (i.e., period), derived from the best fitcosinor, were compared using the extra sum-of-squares F comparison test and 2-way ANOVA. If ANOVA revealed significant (p < 0.05) effects, Holm-Sidak’s multiple comparison post hoc tests were used to determine p values for all relevant comparisons. All analyses were performed in Graphpad Prism. Statistically significant differences were defined as those with a p value < 0.05. The significance level is indicated as *p < 0.05, **p < 0.01 and ***p < 0.001.
Results
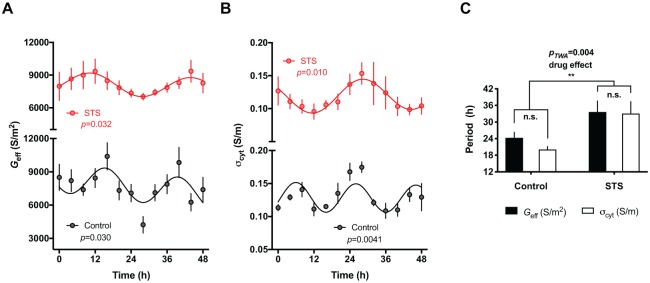
We previously reported the circadian regulation of membrane electrophysiology and potassium transport in isolated human RBCs under constant conditions using an assay based on DEP (Henslee et al., 2017). Kinases fulfill essential timing functions in the circadian clocks of other mammalian cells (i.e., those with nuclei) and we were therefore interested in understanding whether this extended to encompass the transcription-independent circadian clock of RBCs. We used staurosporine, a broad-spectrum kinase inhibitor first shown to dramatically alter the free-running period in the dinoflagellate Gonyaulax polyedra (Comolli and Hastings, 1999), as a proof-of-principle test of the hypothesis that kinases regulate the RBC circadian clock. Staurosporine treatment dramatically increased the period of rhythms in membrane conductance (Geff) and cytoplasmic conductivity (σcyt) (Fig. 1A-C), similar to its activity in nucleated cells (Comolli and Hastings, 1999). RBCs are therefore an amenable platform with which to test kinase function in the RBC clockwork using small-molecule inhibitors.
Figure 1.
RBC circadian rhythms are affected by the inhibition of protein kinases. Isolated RBCs were cultured in the presence of 10 μM staurosporine (STS) or vehicle control at constant 37°C after 2 days of temperature entrainment. DEP measurements of membrane conductance (Geff) (A) and cytoplasmic conductivity (σcyt) (B) are reported (mean ± SEM, n = 4). Damped cosine curves were fitted to the data using non-linear regression comparison of fits (H1 = damped cosine model, H0 = straight line) extra sum-of-squares F test. Cosine fit was preferred over straight line fit if p < 0.05. Geff and σcyt oscillate in antiphase, as previously reported (Henslee et al., 2017). (C) STS treatment significantly increases the period of both Geff and σcyt rhythms relative to vehicle control (period values derived from best fitcosinor ± SEM; n = 4. Two-way ANOVA p value (pTWA) and Holm-Sidak’s post hoc tests reported).
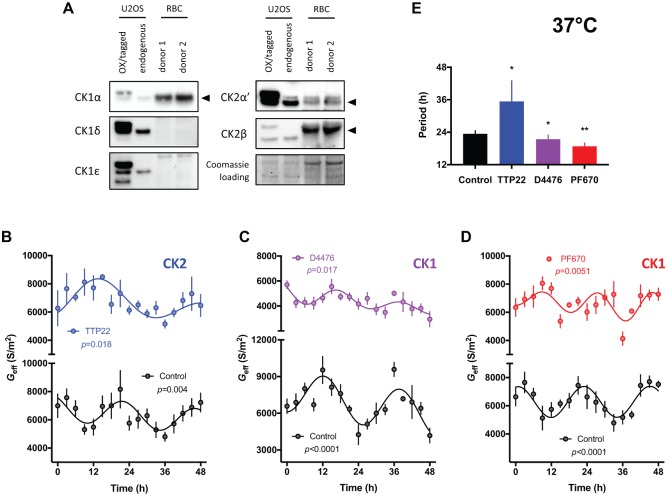
Casein kinases play functionally conserved roles in timekeeping across eukaryotes, with roles attributed to CK1 homologs (CK1α, CK1δ, CK1ε) and CK2 (He et al., 2006; Hirota et al., 2010; Meng et al., 2008; van Ooijen et al., 2013). RBCs express one CK1 paralogue, CK1α, as well as catalytic and regulatory subunits of CK2: CK2α’, and CK2β (Fig. 2A and Supplementary Figure S1) (Bryk and Wiśniewski, 2017). Therefore, we explored whether their timekeeping role extended to non-transcriptional oscillations in RBCs by applying casein kinase inhibitors that are known to affect period determination in nucleated cells (Badura et al., 2007; Meng et al., 2010). We found that TTP22, an inhibitor of CK2 (Golub et al., 2011), and D4476, an inhibitor of CK1 (Bain et al., 2007; Rena et al., 2004), elicited significant effects on the period of the circadian rhythm in RBC membrane electrophysiology, lengthening, and shortening period, respectively (Fig. 2B, C, E). This demonstrates a role for these enzymes in the transcription-independent time-keeping mechanism of RBCs.
Figure 2.
Casein kinase inhibition alters the circadian rhythm of red blood cells (RBCs). (A) Immunoblots showing the expression profiles of human CK1 and CK2 paralogs in human RBCs and in human U2OS cells. Data from 2 donors are shown. U2OS cells transfected with V5-CK1α (Golden et al., 2017), CK1δ-V5, HA-CK1ε (Rivers et al., 1998), HA-CK2α’ and Myc-CK2β (both Turowec et al., 2010) served as positive controls. (B-D) Dielectrophoresis (DEP) measurements of membrane conductance (Geff) in isolated RBCs incubated at constant 37°C over 2 days in the presence of 100 nM TTP22 (CK2 inhibitor) (B), 1 μM D4476 (CK1 inhibitor) (C), or 3 μM PF-670462 (highly potent CK1 inhibitor) (D) versus vehicle control (mean ± SEM, n = 4. Extra sum-of-squares F test p values reported from non-linear regression comparison of fits, H1 = damped cosine model, H0 = straight line). (E) The period of Geff rhythms are significantly affected by inhibition of CK1 and CK2 (period values derived from best fitcosinor ± SEM, n = 4. Extra sum-of-squares F test p values reported following comparison of best fitcosinor. **p < 0.01, *p < 0.05).
A lengthening of the circadian period in response to CK2 inhibition has been observed previously for other mammalian and eukaryotic models (Tsuchiya et al., 2009). We were surprised, however, that CK1 inhibition elicited a significantly shorter circadian period in RBCs compared with controls because, to our knowledge, in all other contexts CK1 inhibition slows down the cellular clock (Badura et al., 2007; Hirota et al., 2010; Isojima et al., 2009; Meng et al., 2010; van Ooijen et al., 2013). We therefore repeated our assays using a pharmacologically distinct and highly potent inhibitor of CK1, PF-670462 (Badura et al., 2007; Bibian et al., 2013). This resulted in a similar reduction in circadian period length as that observed with D4476 (Fig. 2D and E).
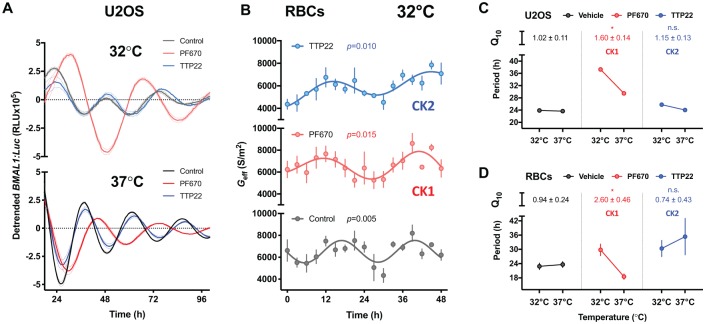
Temperature compensation is an evolutionarily conserved property of the circadian clock and a defining feature of bona fide circadian rhythms. CK1 and CK2 have proposed roles in temperature compensation in organisms as diverse as Neurospora fungi, algae and mammals (Isojima et al., 2009; Mehra et al., 2009; Tosini and Menaker, 1998; Zhou et al., 2015). We examined the role of these cellular kinases in temperature compensation in the absence of known clock protein targets. If temperature compensation is reliant on the activity of either kinase, their inhibition will result in a Q10 for the period of oscillation that lies outside the temperature-compensated range of 0.8 to 1.2; i.e. inhibition of the kinase should have markedly different effects on circadian period at different temperatures. As shown in previous studies in nucleated cells (Isojima et al., 2009), inhibition of CK1 disrupted temperature compensation in (nucleated) human U2OS cells expressing a BMAL1:Luc reporter, with Q10PF670 = 1.60 ± 0.14 (Fig. 3A and C). Consistent with daily rhythms becoming temperature-dependent under CK1 inhibition, in a separate experiment, the magnitude of period lengthening due to CK1 inhibition was further reduced in U2OS cells when cultured at the higher temperature of 40°C (Supplementary Figure S2). In contrast, CK2 inhibition had no effect on temperature compensation, i.e. Q10TTP22 (1.15 ± 0.13) was not significantly different from the vehicle Q10control (1.02 ± 0.11, Fig. 3C). In anucleate RBCs, as with U2OS cells, temperature compensation was observed both for vehicle control cultures (Q10control = 0.94 ± 0.24) and TTP22-treated RBCs (Q10TTP22 = 0.74 ± 0.43) (Fig. 3B and D). Again, as with U2OS cells, CK1 inhibition abolished temperature compensation of circadian rhythms in isolated RBCs, with period lengthening at 32°C as compared with period shortening at 37°C, and a Q10PF670 of 2.60 ± 0.46 (Fig. 3B and D). Therefore, inhibition of CK1, but not CK2, activity abolished temperature compensation of circadian rhythms in the presence and absence of transcription-translation feedback loops (Fig. 3C and D).
Figure 3.
Temperature compensation of RBC circadian rhythms is abolished by inhibition of casein kinase 1 (CK1). (A) Nucleated human U2OS cells (BMAL1:Luc) were cultured at constant 32°C (top, light tone) and 37°C (bottom, full tone) following 4 days of temperature entrainment in the presence of 0.31 µM PF-670462 (CK1 inhibitor, red) or 2.5 µM TTP22 (CK2 inhibitor, blue) vs. vehicle control (black) (mean ± SEM, n = 6), detrended using a moving average of 24-h for vehicle and TTP22 or 30-h for PF-670462. (B) Dielectrophoretic (DEP) measurements of membrane conductance (Geff) in isolated RBCs over 2 days incubated at 32°C in the presence of 3 μM PF-670462 (light red) or 100 nM TTP22 (light blue) vs. vehicle control (mean ± SEM, n = 4). P values for DEP assays report non-linear regression comparison of fits (H1 = damped cosine model, H0 = straight line). (C and D) Inhibition of CK1 abolishes temperature compensation in U2OS (top) and RBCs (bottom). In vehicle control (black) and TTP22-treated (blue) cells, period does not change with increased temperature, reflected by a Q10 of approximately 1. Conversely, treatment with PF-670462 results in a significantly increased Q10 vs. control, reflected by the shorter period at 37°C relative to 32°C. Period values derived from best fitcosinor ± SEM (nU2OS = 6, nRBC = 4). Holm-Sidak’s multiple comparisons p-value reported, following one-way ANOVA (pANOVARBC = 0.015, pANOVAU2OS = 0.017), for Q10 vs. vehicle. *p < 0.05.
Discussion
Both CK1 and CK2 play roles in circadian period determination in many eukaryotic cellular contexts (O’Neill et al., 2013). CK1 and CK2 homologs are present in RBCs (this study and Bryk and Wiśniewski, 2017), whereas the capacity for nascent transcription is not. Here, we have extended the timekeeping function for these ancient and ubiquitously expressed kinases to encompass regulation of circadian period in the non-transcriptional RBC clock. Whilst it is clear that PER (and other clock gene encoded) proteins are functionally relevant substrates for CK1 and CK2 in cells where cycling clock gene transcription is present, our RBC assays suggest clock protein transcription factors (TFs) are unlikely to be their only relevant substrates with respect to cellular timekeeping. Separately, this is also implied by the complete conservation of CK1 and CK2 in period determination across eukaryotes (Causton et al., 2015; He et al., 2006; Lin et al., 2002; Meng et al., 2008; Tsuchiya et al., 2009; van Ooijen et al., 2013; Yang et al., 2002; Zhang et al., 2013), whereas the clock protein TFs, upon which they act, are not. A parsimonious interpretation of this observation is that there exist one or more CK substrates, evolutionarily conserved but as yet unidentified, whose activity determines periodicity.
In the absence of cycling transcription and clock gene activity, we also found that the catalytic activity of CK1, but not CK2, appears to be relevant to the mechanisms that facilitate temperature compensation of RBC circadian rhythms, similar to the results of nucleated mammalian cells. In two very different human cell types, primary RBCs and the U2OS osteosarcoma cell line, inhibition of CK1 abolished temperature compensation and increased the Q10 for the period of oscillation. Inhibition of CK1 with PF-670462 had a greater effect on the Q10 of rhythms in RBCs than in U2OS cells (Q10 of 2.6 compared with 1.6, respectively), thus explaining the period shortening elicited by CK1 inhibition in RBCs at 37°C (Fig. 3 and Supplemental Figure S2). The differential sensitivity of Q10 to CK1 inhibition between the two cell types may be attributable to cell type-dependent variation in the expression of CK1 isoforms; i.e., in U2OS cells, the α, δ, and ε isoforms were all readily detectable, whereas only CK1α was detected in RBCs, both in our study and in an independent mass spectrometry-based investigation (Bryk and Wiśniewski, 2017).
Unlike CK1δ/ε, the timekeeping functions of CK1α are less well established, being reported by two separate studies (Hirota et al., 2010; Lam et al., 2018). Thus, an alternative interpretation of our findings is that temperature-compensated circadian rhythms in RBCs arise by a completely different mechanism from that which operates in other mammalian cell types. In this case, the similar sensitivity to CK inhibition of RBC circadian rhythms compared with other cell types, as well as their common features, such as rhythmic K+ transport and over-oxidised peroxiredoxin levels (Cho et al., 2014; Edgar et al., 2012; Feeney et al., 2016; Henslee et al., 2017; O’Neill and Reddy, 2011), is simply coincidental. We note, however, that the CK1α catalytic domain shares very high sequence identity with human CK1δ/ε (78%) and CK1 orthologs in the metazoan, fungal and green lineages (>71%), whereas the C-terminal domains that distinguish mammalian CK1 paralogs and isoforms from each other show much less conservation across the eukaryotic kingdoms. We also note that, whilst CK1ε is dispensable, CK1δ and CK1α both play essential roles in development and proliferation, possess identical substrate specificities that cannot be distinguished pharmacologically, and have many cellular targets in common (Schittek and Sinnberg, 2014). Therefore, we suggest that there is very little evidence that CK1α is not equally as important as CK1δ for the regulation of circadian gene expression rhythms in mammalian cells.
Unlike most enzymes (including CK2), the activity of CK1δ/ε toward certain polypeptides is insensitive to temperature, with a Q10 of around 1.0 (Isojima et al., 2009; Shinohara et al., 2017; Zhou et al., 2015). This temperature insensitivity is inherent to the catalytic domain of CK1, demonstrated by impaired temperature compensation of catalytic mutants against synthetic substrates compared with native sequences (Shinohara et al., 2017). This “temperature-compensated domain” is conserved in other CK1δ homologs, such as human CK1α (86% similarity), including the critical lysine residue (K224 in CK1δ; K232 in CK1α). This strongly suggests that temperature compensation is a function of CK1 homologs across the eukaryotic kingdom, independent of the presence or absence of the TTFL, and functions similarly between nucleated and non-nucleated cells.
In this, we note similarities with KaiC phosphorylation in cyanobacteria, whose post-translational rhythm persists in the absence of nascent gene expression (Nakajima et al., 2005; Tomita et al., 2005). Here, temperature compensation of circadian period is conferred by the intrinsically temperature-insensitive phosphotransferase activity of KaiC (Egli et al., 2012; Terauchi et al., 2007), with circadian period and temperature compensation also being sensitive to amino acid substitution around the ATP-binding site (Murakami et al., 2008; Terauchi et al., 2007). Our present observations support the possibility that the role of CK1 homologs in eukaryotic circadian rhythms may be functionally analogous to that of KaiC in cyanobacteria (Merrow et al., 2006; Wong and O’Neill, 2018).
The circadian clock regulates biological functions, such as metabolism, protein synthesis, and ion transport, in many cell types across phylogenetic kingdoms, both in the presence and absence of transcriptional feedback cycles (Cho et al., 2014; Feeney et al., 2016; Henslee et al., 2017; Lipton et al., 2015; O’Neill et al., 2011; O’Neill and Reddy, 2011; van Ooijen et al., 2011; Woolum, 1991). Though no definitive adaptive advantage has been ascribed to RBC circadian rhythms, daily variation in CO2-carrying potential, adverse cardiovascular events, and osmotic homeostasis are potential consequences of this reduced and highly specialised cellular clockwork (Henslee et al., 2017). Erythrocytes are the most numerous cell type in the human body. Whereas their circadian clock mechanism is not understood, it does involve the same conserved kinases that regulate timekeeping fidelity in other mammalian cell types. Future studies should identify which RBC proteins might be subject to circadian post-translational modification by CK1 and CK2, and whether these also exhibit phosphorylation rhythms in other cell types.
Supplemental Material
Supplemental material, Supplementary_data for Casein Kinase 1 Underlies Temperature Compensation of Circadian Rhythms in Human Red Blood Cells by Andrew D. Beale, Emily Kruchek, Stephen J. Kitcatt, Erin A. Henslee, Jack S.W. Parry, Gabriella Braun, Rita Jabr, Malcolm von Schantz, John S. O’Neill and Fatima H. Labeed in Journal of Biological Rhythms
Acknowledgments
We are grateful to the donors and the Clinical Research Centre at the University of Surrey for assistance with the blood samples, and Michael P. Hughes and members of the O’Neill lab for useful discussions. This research was supported by a Biotechnology and Biological Research Council (BBSRC) grant (BB/M021556/1) and the Medical Research Council (MC_UP_1201/4).
Supplemental material is available for this article online.
Conflict of Interest Statement: The author(s) have no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
ORCID iD: Andrew D. Beale  https://orcid.org/0000-0002-2051-0919
https://orcid.org/0000-0002-2051-0919
References
- Badura L, Swanson T, Adamowicz W, Adams J, Cianfrogna J, Fisher K, Holland J, Kleiman R, Nelson F, Reynolds L, et al. (2007) An inhibitor of casein kinase I epsilon induces phase delays in circadian rhythms under free-running and entrained conditions. J Pharmacol Exp Therap 322:730-738. [DOI] [PubMed] [Google Scholar]
- Bain J, Plater L, Elliott M, Shpiro N, Hastie CJ, Mclauchlan H, Klevernic I, Arthur JSC, Alessi DR, Cohen P. (2007) The selectivity of protein kinase inhibitors: a further update. Biochem J 408:297-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsalobre A, Damiola F, Schibler U. (1998) A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell 93:929-937. [DOI] [PubMed] [Google Scholar]
- Beale AD, Whitmore D, Moran D. (2016) Life in a dark biosphere: a review of circadian physiology in “arrhythmic” environments. J Comp Physiol B 186:947-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibian M, Rahaim RJ, Choi JY, Noguchi Y, Schürer S, Chen W, Nakanishi S, Licht K, Rosenberg LH, Li L, et al. (2013) Development of highly selective casein kinase 1δ/1ε (CK1δ/ε) inhibitors with potent antiproliferative properties. Bioorg Med Chem Lett 23:4374-4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryk AH, Wiśniewski JR. (2017) Quantitative analysis of human red blood cell proteome. J Proteome Res 16:2752-2761. [DOI] [PubMed] [Google Scholar]
- Causton HC, Feeney KA, Ziegler CA, O’Neill JS. (2015) Metabolic cycles in yeast share features conserved among circadian rhythms. Curr Biol 25:1056-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho C-S, Yoon HJ, Kim JY, Woo HA, Rhee SG. (2014) Circadian rhythm of hyperoxidized peroxiredoxin II is determined by hemoglobin autoxidation and the 20S proteasome in red blood cells. Proc Natl Acad Sci U S A 111:12043-12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comolli JC, Hastings JW. (1999) Novel effects on the Gonyaulax circadian system produced by the protein kinase inhibitor staurosporine. J Biol Rhythms 14:11-19. [DOI] [PubMed] [Google Scholar]
- Crosby P, Hoyle NP, O’Neill JS. (2017) Flexible measurement of bioluminescent reporters using an automated longitudinal luciferase imaging gas- and temperature-optimized recorder (ALLIGATOR). J Visualized Exp (130): 1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekens MPS, Santoriello C, Vallone D, Grassi G, Whitmore D, Foulkes NS. (2003) Light regulates the cell cycle in zebrafish. Curr Biol 13:2051-2057. [DOI] [PubMed] [Google Scholar]
- Dibner C, Sage D, Unser M, Bauer C, D’Eysmond T, Naef F, Schibler U. (2009) Circadian gene expression is resilient to large fluctuations in overall transcription rates. EMBO J 28:123-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RS, Green EW, Zhao Y, van Ooijen G, Olmedo M, Qin X, Xu Y, Pan M, Valekunja UK, Feeney KA, et al. (2012) Peroxiredoxins are conserved markers of circadian rhythms. Nature 485:459-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egli M, Mori T, Pattanayek R, Xu Y, Qin X, Johnson CH. (2012) Dephosphorylation of the core clock protein KaiC in the cyanobacterial KaiABC circadian oscillator proceeds via an ATP synthase mechanism. Biochemistry 51:1547-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feeney KA, Hansen LL, Putker M, Olivares-Yañez C, Day J, Eades LJ, Larrondo LF, Hoyle NP, O’Neill JS, van Ooijen G, et al. (2016) Daily magnesium fluxes regulate cellular timekeeping and energy balance. Nature 532:375-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego M, Virshup DM. (2007) Post-translational modifications regulate the ticking of the circadian clock. Nat Rev Mol Cell Biol 8:139-148. [DOI] [PubMed] [Google Scholar]
- Golden RJ, Chen B, Li T, Braun J, Manjunath H, Chen X, Wu J, Schmid V, Chang T-C, Kopp F, et al. (2017) An argonaute phosphorylation cycle promotes microRNA-mediated silencing. Nature 542:197-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golub AG, Bdzhola VG, Briukhovetska N V., Balanda AO, Kukharenko OP, Kotey IM, Ostrynska O V., Yarmoluk SM. (2011) Synthesis and biological evaluation of substituted (thieno[2,3-d]pyrimidin-4-ylthio)carboxylic acids as inhibitors of human protein kinase CK2. Eur J Med Chem 46:870-876. [DOI] [PubMed] [Google Scholar]
- He Q, Cha J, He Q, Lee HC, Yang Y, Liu Y. (2006) CKI and CKII mediate the FREQUENCY-dependent phosphorylation of the WHITE COLLAR complex to close the Neurospora circadian negative feedback loop. Genes Dev 20:2552-2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henslee EA, Crosby P, Kitcatt SJ, Parry JSW, Bernardini A, Abdallat RG, Braun G, Fatoyinbo HO, Harrison EJ, Edgar RS, et al. (2017) Rhythmic potassium transport regulates the circadian clock in human red blood cells. Nat Commun 8:1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota T, Lewis WG, Liu AC, Lee JW, Schultz PG, Kay SA. (2008) A chemical biology approach reveals period shortening of the mammalian circadian clock by specific inhibition of GSK-3. Proc Natl Acad Sci U S A 105:20746-20751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota T, Lee JW, Lewis WG, Zhang EE, Breton G, Liu X, Garcia M, Peters EC, Etchegaray J-P, Traver D, et al. (2010) High-throughput chemical screen identifies a novel potent modulator of cellular circadian rhythms and reveals CKIα as a clock regulatory kinase. PLoS Biol 8:e1000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isojima Y, Nakajima M, Ukai H, Fujishima H, Yamada RG, Masumoto K -h., Kiuchi R, Ishida M, Ukai-Tadenuma M, Minami Y, et al. (2009) CKI epsilon/delta-dependent phosphorylation is a temperature-insensitive, period-determining process in the mammalian circadian clock. Proc Natl Acad Sci U S A 106:15744-15749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang T-H, Reardon JT, Kemp M, Sancar A. (2009) Circadian oscillation of nucleotide excision repair in mammalian brain. Proc Natl Acad Sci U S A 106:2864-2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakin-Thomas PL. (2006) Transcriptional feedback oscillators: Maybe, maybe not... J Biol Rhythms 21:83-92. [DOI] [PubMed] [Google Scholar]
- Lam VH, Li YH, Liu X, Murphy KA, Diehl JS, Kwok RS, Chiu JC. (2018) CK1α collaborates with DOUBLETIME to regulate PERIOD function in the Drosophila circadian clock. J Neurosci 38:10631-10643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J-M, Kilman VL, Keegan K, Paddock B, Emery-Le M, Rosbash M, Allada R. (2002) A role for casein kinase 2α in the Drosophila circadian clock. Nature 420:816-820. [DOI] [PubMed] [Google Scholar]
- Lipton JO, Yuan ED, Boyle LM, Ebrahimi-Fakhari D, Kwiatkowski E, Nathan A, Güttler T, Davis F, Asara JM, Sahin M. (2015) The circadian protein BMAL1 regulates translation in response to S6K1-mediated phosphorylation. Cell 161:1138-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo T, Yamaguchi S, Mitsui S, Emi A, Shimoda F, Okamura H. (2003) Control mechanism of the circadian clock for timing of cell division in vivo. Science 302:255-259. [DOI] [PubMed] [Google Scholar]
- Mehra A, Shi M, Baker CL, Colot H V., Loros JJ, Dunlap JC. (2009) A role for casein kinase 2 in the mechanism underlying circadian temperature compensation. Cell 137:749-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng QJ, Logunova L, Maywood ES, Gallego M, Lebiecki J, Brown TM, Sládek M, Semikhodskii AS, Glossop NRJ, Piggins HD, et al. (2008) Setting clock speed in mammals: The CK1ε tau mutation in mice accelerates circadian pacemakers by selectively destabilizing PERIOD proteins. Neuron 58:78-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng QJ, Maywood ES, Bechtold DA, Lu W-Q, Li J, Gibbs JE, Dupre SM, Chesham JE, Rajamohan F, Knafels J, et al. (2010) Entrainment of disrupted circadian behavior through inhibition of casein kinase 1 (CK1) enzymes. Proc Natl Acad Sci U S A 107:15240-15245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrow M, Mazzotta G, Chen Z, Roenneberg T. (2006) The right place at the right time: regulation of daily timing by phosphorylation. Genes Dev 20:2629-2633. [DOI] [PubMed] [Google Scholar]
- Murakami R, Miyake A, Iwase R, Hayashi F, Uzumaki T, Ishiura M. (2008) ATPase activity and its temperature compensation of the cyanobacterial clock protein KaiC. Genes Cells 13:387-395. [DOI] [PubMed] [Google Scholar]
- Nakajima M, Imai K, Ito H, Nishiwaki T, Murayama Y, Iwasaki H, Oyama T, Kondo T. (2005) Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science 308:414-415. [DOI] [PubMed] [Google Scholar]
- O’Neill JS, Reddy AB. (2011) Circadian clocks in human red blood cells. Nature 469:498-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill JS, Van Ooijen G, Dixon LE, Troein C, Corellou F, Bouget FY, Reddy AB, Millar AJ. (2011) Circadian rhythms persist without transcription in a eukaryote. Nature 469:554-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill JS, Maywood ES, Hastings MH. (2013) Cellular mechanisms of circadian pacemaking: Beyond transcriptional loops. In: Kramer A, Merrow M. (eds), Circadian Clocks. Handbook of Experimental Pharmacology. Berlin, Heidelberg: Springer, pp 63-103. [DOI] [PubMed] [Google Scholar]
- Pittendrigh CS. (1960) Circadian Rhythms and the Circadian Organization of Living Systems. Cold Spring Harbor Symp Quant Biol 25:159-184. [DOI] [PubMed] [Google Scholar]
- Rena G, Bain J, Elliott M, Cohen P. (2004) D4476, a cell-permeant inhibitor of CK1, suppresses the site-specific phosphorylation and nuclear exclusion of FOXO1a. EMBO Rep 5:60-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivers A, Gietzen KF, Vielhaber E, Virshup DM. (1998) Regulation of casein kinase I ε and casein kinase I δ by an in vivo futile phosphorylation cycle. J Biol Chem 273:15980-15984. [DOI] [PubMed] [Google Scholar]
- Schittek B, Sinnberg T. (2014) Biological functions of casein kinase 1 isoforms and putative roles in tumorigenesis. Mol Cancer 13:231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara Y, Koyama YM, Ukai-Tadenuma M, Hirokawa T, Kikuchi M, Yamada RG, Ukai H, Fujishima H, Umehara T, Tainaka K, et al. (2017) Temperature-sensitive substrate and product binding underlie temperature-compensated phosphorylation in the clock. Mol Cell 67:783-798.e20. [DOI] [PubMed] [Google Scholar]
- Takahashi JS. (2017) Transcriptional architecture of the mammalian circadian clock. Nat Rev Genet 18:164-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terauchi K, Kitayama Y, Nishiwaki T, Miwa K, Murayama Y, Oyama T, Kondo T. (2007) ATPase activity of KaiC determines the basic timing for circadian clock of cyanobacteria. Proc Natl Acad Sci U S A 104:16377-16381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita J, Nakajima M, Kondo T, Iwasaki H. (2005) No transcription-translation feedback in circadian rhythm of KaiC phosphorylation. Science 307:251-254. [DOI] [PubMed] [Google Scholar]
- Tosini G, Menaker M. (1998) The tau mutation affects temperature compensation of hamster retinal circadian oscillators. NeuroReport 9:1001-1005. [DOI] [PubMed] [Google Scholar]
- Tsuchiya Y, Akashi M, Matsuda M, Goto K, Miyata Y, Node K, Nishida E. (2009) Involvement of the protein kinase CK2 in the regulation of mammalian circadian rhythms. Sci Signal 2:ra26-ra26. [DOI] [PubMed] [Google Scholar]
- Turowec JP, Duncan JS, French AC, Gyenis L, St. Denis NA, Vilk G, Litchfield DW. (2010) Protein kinase CK2 is a constitutively active enzyme that promotes cell survival: Strategies to identify CK2 substrates and manipulate its activity in mammalian cells. In Methods in Enzymology, pp 471-493. [DOI] [PubMed] [Google Scholar]
- van Ooijen G, Dixon LE, Troein C, Millar AJ. (2011) Proteasome function is required for biological timing throughout the twenty-four hour cycle. Curr Biol 21:869-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ooijen G, Hindle M, Martin SF, Barrios-Llerena M, Sanchez F, Bouget FY, O’Neill JS, Le Bihan T, Millar AJ, O’Neill JS, et al. (2013) Functional analysis of casein kinase 1 in a minimal circadian system. PLoS ONE 8:e70021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaze KM, Sharma VK. (2013) On the adaptive significance of circadian clocks for their owners. Chronobiol Int 30:413-433. [DOI] [PubMed] [Google Scholar]
- Welsh DK, Yoo S-H, Liu AC, Takahashi JS, Kay SA. (2004) Bioluminescence imaging of individual fibroblasts reveals persistent, independently phased circadian rhythms of clock gene expression. Curr Biol 14:2289-2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong DC, O’Neill JS. (2018) Non-transcriptional processes in circadian rhythm generation. Curr Opin Physiol 5:117-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolum JC. (1991) A re-examination of the role of the nucleus in generating the circadian rhythm in acetabularia. J Biol Rhythms 6:129-136. [DOI] [PubMed] [Google Scholar]
- Yang Y, Cheng P, Liu Y. (2002) Regulation of the Neurospora circadian clock by casein kinase II. Genes Dev 16:994-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Hastings MH, Green EW, Tauber E, Sladek M, Webster SG, Kyriacou CP, Wilcockson DC. (2013) Dissociation of circadian and circatidal timekeeping in the marine crustacean Eurydice pulchra. Curr Biol 23:1863-1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Kim JK, Eng GWL, Forger DB, Virshup DM. (2015) A period2 phosphoswitch regulates and temperature compensates circadian period. Mol Cell 60:77-88. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, Supplementary_data for Casein Kinase 1 Underlies Temperature Compensation of Circadian Rhythms in Human Red Blood Cells by Andrew D. Beale, Emily Kruchek, Stephen J. Kitcatt, Erin A. Henslee, Jack S.W. Parry, Gabriella Braun, Rita Jabr, Malcolm von Schantz, John S. O’Neill and Fatima H. Labeed in Journal of Biological Rhythms