Key Points
Question
Does an association exist between systemic antibiotic treatment and its withdrawal and changes in skin microbiota in individuals with acne?
Findings
In this study of 4 women with acne, there was a significant decrease in the relative abundance of Cutibacterium acnes (formerly Propionibacterium acnes) concurrent with a significant increase in the relative abundance of Pseudomonas species across participants following 4 weeks of treatment with oral minocycline. After 8 weeks of antibiotic treatment withdrawal, C acnes levels recovered, while Streptococcus species significantly increased and Lactobacillus species significantly decreased from baseline.
Meaning
Systemic antibiotic treatment of acne may be associated with transient and persistent changes in the skin microbiota that may underlie skin comorbidities related to microbial dysbiosis.
Abstract
Importance
Given the widespread use of systemic antibiotics for treatment of moderate to severe acne, it is important to understand the associations of such antibiotic use with changes not only in Cutibacterium acnes (formerly Propionibacterium acnes) but also in the complete bacterial community of the skin.
Objective
To examine the composition, diversity, and resilience of skin microbiota associated with systemic antibiotic perturbation in individuals with acne.
Design, Setting, and Participants
This longitudinal cohort study conducted at an academic referral center in Maryland from February 11 to September 23, 2014, included 4 female participants who had received a recent diagnosis of acne vulgaris, showed comedonal and inflammatory acne on the face, were at least 18 years old, and had no recent use of systemic or topical treatments for acne, including antibiotics and retinoids. Data analysis was performed between July 5, 2017, and November 7, 2018.
Interventions
Participants were prescribed oral minocycline, 100 mg, twice daily for 4 weeks. Skin areas on the forehead, cheek, and chin were sampled for 16S ribosomal RNA gene sequencing at baseline, 4 weeks after starting minocycline treatment, and then 1 week and 8 weeks after discontinuation of treatment.
Main Outcomes and Measures
Skin microbiota examined with respect to relative abundance of bacterial taxa, α diversity (represents within-sample microbial diversity), and β diversity (represents between-sample microbial diversity). Acne status evaluated with photography and lesion count.
Results
Of the 4 patients included in this study, 2 were 25 years old, 1 was 29 years old, and 1 was 35 years old; 2 were white women, 1 was an African American woman, and 1 was an Asian woman. Across all 4 patients, antibiotic treatment was associated with a 1.4-fold reduction in the level of C acnes (difference, −10.3%; 95% CI, −19.9% to −0.7%; P = .04) with recovery following cessation of treatment. Distinct patterns of change were identified in multiple bacterial genera, including a transient 5.6-fold increase in the relative abundance of Pseudomonas species (difference, 2.2%; 95% CI, 0.9%-3.4%; P < .001) immediately following antibiotic treatment, as well as a persistent 1.7-fold increase in the relative abundance of Streptococcus species (difference, 5.4%; 95% CI, 0.3%-10.6%; P = .04) and a 4.7-fold decrease in the relative abundance of Lactobacillus species (difference, −0.8%; 95% CI, −1.4% to −0.2%; P = .02) 8 weeks following antibiotic treatment withdrawal. In general, antibiotic administration was associated with an initial decrease from baseline of bacterial diversity followed by recovery. Principal coordinates analysis results showed moderate clustering of samples by patient (analysis of similarity, R = 0.424; P = .001) and significant clustering of samples by time in one participant (analysis of similarity, R = 0.733; P = .001).
Conclusions and Relevance
In this study, systemic antibiotic treatment of acne was associated with changes in the composition and diversity of skin microbiota, with variable rates of recovery across individual patients and parallel changes in specific bacterial populations. Understanding the association between systemic antibiotic use and skin microbiota may help clinicians decrease the likelihood of skin comorbidities related to microbial dysbiosis.
This pilot study of 4 patients assesses the composition, diversity, and resilience of skin microbiota following systemic antibiotic administration to individuals with facial acne.
Introduction
Acne vulgaris is a common disease of the pilosebaceous unit characterized by release of inflammatory mediators, hyperkeratinization, increased production of sebum, and colonization by Propionibacterium acnes,1,2,3 recently reclassified as Cutibacterium acnes.4 This bacterium promotes inflammation through toll-like receptors, neutrophil chemotactic factors, and complement pathways.5,6 Cutibacterium acnes also secretes lipases, proteases, and hyaluronidases that damage the pilosebaceous unit.6 Tetracycline-class antibiotics, such as minocycline and doxycycline, are commonly used as first-line treatment of moderate to severe acne because of their antimicrobial and anti-inflammatory actions.2,3 Although recent guidelines recommend limiting systemic antibiotic treatment to 3 to 4 months,3 retrospective studies reveal that the duration of antibiotic treatment often exceeds recommendations,7,8,9,10,11 with a mean treatment duration of 6.5 months among patients with acne who were treated by dermatologists in the United States between 2004 and 2013.11 Given the widespread use of systemic antibiotics for acne, it is important to understand their effects not only on C acnes but also on the complete bacterial community of the skin.
Amplification and sequencing of the 16S ribosomal RNA (rRNA) gene has been used to identify bacterial communities in a variety of body sites, including the skin.12,13 Using this culture-independent method, many studies have investigated the effects of antibiotics on gut microbiota, often noting shifts in bacterial diversity following antibiotic treatment and incomplete recovery in subsets of bacterial taxa following cessation of antibiotic treatments.14,15,16,17 Given these findings in gut microbiota, the aim of the present study was to examine the composition, diversity, and resilience of skin microbiota following antibiotic perturbation in individuals with acne.
Methods
Study Participants
Four women who had received a recent diagnosis of acne by a dermatologist were enrolled from February 11 through September 23, 2014, at the Johns Hopkins Department of Dermatology, Baltimore, Maryland. Inclusion criteria included the presence of comedonal and inflammatory acne on the face, age of 18 years or older, and willingness to use nonantibacterial bar soap for facial cleansing for the duration of the study. Exclusion criteria included history of systemic or topical antibiotic use within 1 month of the baseline study visit, hypersensitivity to tetracycline-class antibiotics, dermatologic procedural treatment within 6 months, systemic acne treatment within 4 weeks, topical acne treatment within 2 weeks, significant facial hair, pregnancy or breastfeeding status, and inability to provide informed consent. No participant had a history of topical or systemic retinoid use within 2 years of the baseline study visit, and no concomitant medication use was reported. The study was approved by the Johns Hopkins University Institutional Review Board, and participants provided written informed consent prior to participation.
Antibiotic Treatment and Sample Collection
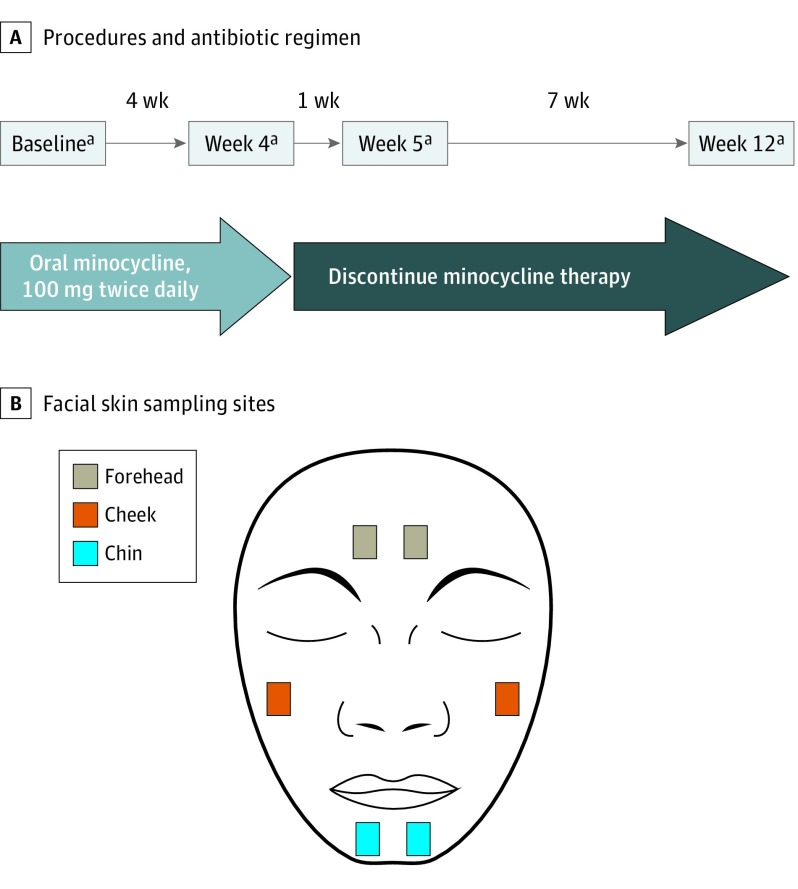
Participants were prescribed minocycline, 100 mg, to be taken orally twice daily for 4 weeks. Skin samples were collected at 4 visits across 12 weeks (Figure 1A), including a baseline visit before antibiotic treatment and visits 4 weeks after starting minocycline therapy (week 4), 1 week after discontinuation of minocycline therapy (week 5), and 8 weeks after discontinuation of minocycline therapy (week 12). Participants’ adherence to antibiotic therapy was followed with a self-reported diary; all participants took minocycline as directed, with participants 2 and 4 each missing 2 doses on nonconsecutive days at least 2 weeks apart. At each visit, facial skin was sampled bilaterally from the forehead, cheek, and chin using sterile cotton swabs (Figure 1B), yielding a total of 96 skin samples from 4 participants across 4 time points. Clinical evaluation of acne status at each visit included photography and lesion counts, which were performed by a trained postdoctoral fellow.
Figure 1. Schematic Diagram of Study Design.
A, Time course of antibiotic therapy and procedures performed at each study visit. B, Bilateral sites of the face sampled by sterile cotton swabs at each visit for 16S ribosomal RNA (rRNA) gene amplification and sequencing.
aBilateral skin sampling (forehead, cheek, and chin), 16S rRNA gene sequencing, photography, and lesion count.
DNA Extraction and 16S rRNA Gene Polymerase Chain Reaction Amplification and Sequencing
Extraction of DNA from skin samples involved an enzymatic lysis and bead-based tissue homogenization protocol; samples were briefly incubated in a lytic enzyme mixture of lysozyme, mutanolysin, proteinase K, and lysostaphin, followed by mechanical lysis with silica beads (0.1 mm), as previously published.18 The DNA cleanup was then performed with a fecal DNA extraction kit (ZR Fecal DNA MiniPrep; Zymo Research). Following DNA extraction, the V3-V4 hypervariable region of the 16S rRNA gene was amplified by polymerase chain reaction and sequenced using the Illumina MiSeq platform (300 base pairs, paired-end reads) as previously described.18,19
Statistical Analysis
After screening raw sequences for low-quality bases and short read lengths, read pairs were assembled using PANDAseq, demultiplexed, trimmed of barcodes and primers, and assessed for chimeras, using UCHIME in de novo mode implemented in the bioinformatic platform QIIME, version 1.9.1.20 Quality-trimmed sequences were then clustered de novo into operational taxonomic units (97% similarity threshold), after which taxonomic assignments were performed using the Ribosomal Database Project Classifier21,22 and the Greengenes database (version 13.8).23 For all downstream analyses, samples were normalized to 8746 sequences. Microbiota α diversity, which represents microbial diversity within an individual sample, was computed in QIIME through the whole tree phylogenetic diversity metric.24,25,26 Microbiota β diversity, which indicates intervariability of microbial diversity between samples, was examined through principal coordinates analysis of weighted UniFrac distances27 in QIIME and hierarchical clustering based on the unweighted pair group method with arithmetic mean algorithm in the R statistical software (R Core Team). Comparisons of α diversity and relative abundance of bacterial taxa between samples were performed with t tests with Monte Carlo permutations in QIIME and Metastats,28 respectively. Comparison of β diversity among samples was performed with QIIME using the analysis of similarity (ANOSIM).29 All other analyses and visualizations were performed with R and the ggplot2 package.30 For all statistical analyses, a 2-sided P < .05 was accepted as statistically significant.
Results
Demographic and relevant clinical characteristics of the 4 patients included in this study are shown in the Table. Two patients were 25 years old, 1 was 29 years old, and 1 was 35 years old; 2 were white women, 1 was an African American woman, and 1 was an Asian woman.
Table. Demographic and Clinical Characteristics of the 4 Patients.
| Patient No. | Age, y | Race/Ethnicity | Fitzpatrick Skin Type | Duration of Acne, y | Baseline Inflamed Lesion Count |
|---|---|---|---|---|---|
| 1 | 35 | White | 2 | 22 | 7 |
| 2 | 25 | Asian | 3 | 14 | 4 |
| 3 | 29 | African American | 5 | 13 | 7 |
| 4 | 25 | White | 2 | 11 | 22 |
Taxonomic Assignment
Of 96 skin samples obtained bilaterally from 3 sites (forehead, cheek, and chin) of 4 patients, 8 samples were excluded from the analysis because of low concentration of extracted DNA, and 4 additional samples were excluded because of low read numbers from 16S rRNA gene sequencing (eTable in the Supplement). The final data set included 84 samples across 4 patients sequenced to a mean (SD) read count of 66 414 (24 141). We identified 30 phyla, 79 classes, 131 orders, 217 families, 447 genera, and 232 species that were unique and present in at least 1 sample. There was predominance of Actinobacteria (41.9%), Firmicutes (31.7%), and Proteobacteria (19.5%) at the phylum level across all samples (eFigure 1A in the Supplement).
Relative Abundance of Individual Bacterial Taxa
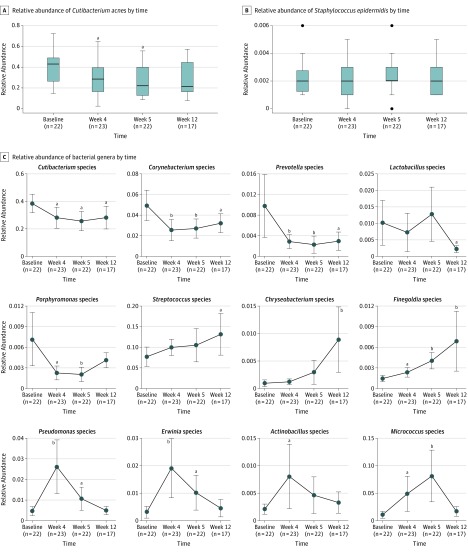
We focused on the genus and species levels when examining changes in abundance of individual bacterial taxa relative to the entire bacterial community in samples. Although the majority of operational taxonomic units in this study could not be speciated beyond the genus level, we did characterize levels of Cutibacterium acnes and Staphylococcus epidermidis over time (Figure 2A and B). Across all patients and sites, the mean relative abundance of C acnes decreased 1.4-fold from baseline to week 4 (difference, −10.3%; 95% CI, −19.9% to −0.7%; P = .04) following treatment with minocycline and decreased further by week 5 (difference, −12.8%; 95% CI, −21.9% to −3.6%; P = .02). At week 12, or 8 weeks after discontinuation of minocycline treatment, the relative abundance of C acnes returned to a level that was quantitatively lower but not statistically different from baseline (difference, −10.4%; 95% CI, −20.3% to −0.4%; P = .06). Staphylococcus epidermidis showed a consistently low relative abundance across all time points, at a level approximately 100-fold lower than that of C acnes.
Figure 2. Dynamics of Bacterial Populations in the Presence and After Withdrawal of Antibiotic Treatment.
Relative abundance of Cutibacterium acnes (formerly Propionibacterium acnes) (A) and Staphylococcus epidermidis (B) at various times during and after antibiotic treatment. Horizontal bar within box plots represents the median; bottom and top of each box, first and third quartiles; lower error bar extends to the lowest data point within 1.5 times the interquartile range from the first quartile; upper error bar extends to the highest data point within 1.5 times the interquartile range from the third quartile. Data points beyond 1.5 times the interquartile range above the third quartile or below the first quartile are plotted individually as outliers. Relative abundance of selected bacterial genera at various times during and after antibiotic treatment (C). Error bars indicate 95% CIs. P values calculated using t tests with 1000 Monte Carlo permutations and 5% false-discovery rate adjustment. Sample sizes shown on x-axes indicate distinct skin samples obtained from all 4 participants.
aP < .05 compared with baseline.
bP < .01 compared with baseline.
We identified 12 genera with relative abundance greater than 0.1% across all samples and with statistically significant changes over time (Figure 2C). Among these 12 genera, we noted 4 distinct patterns of change, namely, decrease in relative abundance following antibiotic treatment with or without recovery at week 12, as well as increase in relative abundance following antibiotic treatment with or without recovery at week 12. Five genera showed decreases in relative abundance on treatment with minocycline: Cutibacterium (baseline [BL]-week [W]4 difference, −10.3%; 95% CI, −19.9% to −0.7%; P = .04), Corynebacterium (BL-W4 difference, −2.4%; 95% CI, −4.1% to −0.7%; P = .005), Prevotella (BL-W4 difference, −0.7%; 95% CI, −1.3% to −0.1%; P = .008), Lactobacillus (BL-W12 difference, −0.8%; 95% CI, −1.4% to −0.2%; P = .02), and Porphyromonas (BL-W4 difference, −0.5%; 95% CI, −0.9% to −0.1%; P = .01). Of these 5 genera, only Porphyromonas recovered at week 12 to a level that was not statistically different from baseline (BL-W12 difference, −0.3%; 95% CI, −0.7% to 0.1%; P = .13). By contrast, 7 genera showed increases in relative abundance on treatment with minocycline: Streptococcus (BL-W12 difference, 5.4%; 95% CI, 0.3%-10.6%; P = .04), Chryseobacterium (BL-W12 difference, 0.8%; 95% CI, 0.2%-1.3%; P = .01), Finegoldia (BL-W4 difference, 0.1%; 95% CI, 0.01%-0.2%; P = .03), Pseudomonas (BL-W4 difference, 2.2%; 95% CI, 0.9%-3.4%; P < .001), Erwinia (BL-W4 difference, 1.6%; 95% CI, 0.6%-2.6%; P < .001), Actinobacillus (BL-W4 difference, 0.6%; 95% CI, 0.04%-1.2%; P = .04), and Micrococcus (BL-W4 difference, 3.8%; 95% CI, 0.7%-6.9%; P = .03). Only the latter 4 genera returned at week 12 to a level that was not statistically different from baseline: Pseudomonas (BL-W12 difference, 0.02%; 95% CI, −0.3% to 0.3%; P = .90), Erwinia (BL-W12 difference, 0.1%; 95% CI, −0.2% to 0.5%; P = .45), Actinobacillus (BL-W12 difference, 0.1%; 95% CI, −0.1% to 0.3%; P = .27), and Micrococcus (BL-W12 difference, 0.6%; 95% CI, −0.4% to 1.6%; P = .29).
Acne Severity and α Diversity
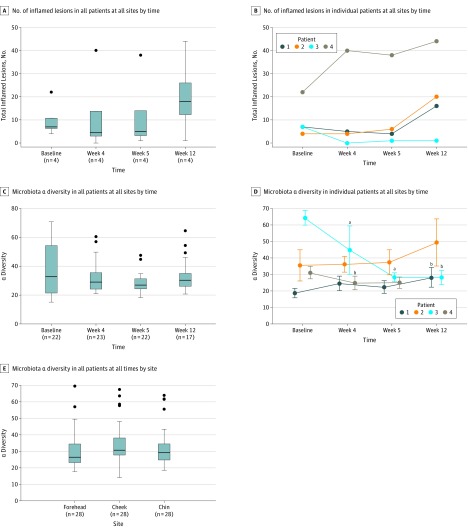
When aggregated across all 4 patients and sampled sites, the median number of inflamed lesions decreased following treatment with minocycline (median at baseline, 7.00; interquartile range [IQR], 6.20-10.75; median at W4, 4.50; IQR, 3.00-13.75) and increased after discontinuation of treatment (median at W12, 18.00; IQR, 12.25-26.00) (Figure 3A). Median α diversity, representative of within-sample microbial diversity, showed a similar temporal trend, that is, decrease following antibiotic treatment (median at baseline, 32.73; IQR, 21.43-54.25; median at W4, 28.99; IQR, 24.18-35.49) and increase to near original baseline levels after 8 weeks of treatment withdrawal (median at W12, 30.29; IQR, 26.08-34.98) (Figure 3C). Observed changes in inflamed lesion count and α diversity over time did not reach statistical significance. However, we noted distinct and statistically significant changes in α diversity in individual patients (Figure 3D), highlighting the intrapersonal responses of the skin microbiota to antibiotics. Both patients 3 and 4 had significant decreases in α diversity at week 4 compared with baseline (patient 3 difference, −19.16; 95% CI, −31.76 to −6.56; P = .006; patient 4 difference, −6.41; 95% CI, −10.53 to −2.29; P = .048), while patient 3 showed a continued loss of α diversity at week 5 (difference, −36.26; 95% CI, −40.39 to −32.12; P = .006). Patients 1 and 3 exhibited opposite changes in α diversity at week 12 compared with baseline; α diversity for patient 1 at week 12 was significantly higher than baseline (difference, 9.02; 95% CI, 3.96-14.08; P = .01), whereas α diversity for patient 3 remained significantly lower than baseline (difference, −36.33; 95% CI, −41.07 to −31.60; P = .01). When aggregated across all patients and time points, α diversity showed no difference among the forehead, cheek, and chin (Figure 3E).
Figure 3. Trends in Acne Severity and Microbiota α Diversity.
Numbers of inflamed lesions in all (A) or individual (B) patients across all sites at different times. Microbiota α diversity (represents within-sample microbial diversity), based on the whole tree phylogenetic diversity metric, of all (C) or individual (D) patients across all sites at various times and across all patients and times at different sites (E). Horizontal bar within box plots represents the median; bottom and top of each box, first and third quartiles; lower error bar extends to the lowest data point within 1.5 times the interquartile range from the first quartile; upper error bar extends to the highest data point within 1.5 times the interquartile range from the third quartile. Data points beyond 1.5 times the interquartile range above the third quartile or below the first quartile are plotted individually as outliers. P values were calculated using t tests with 999 Monte Carlo permutations and Bonferroni corrections. Sample sizes shown on x-axes of (C) and (E) indicate distinct skin samples obtained from all 4 patients.
aP < .01 compared with baseline.
bP < .05 compared with baseline.
β Diversity
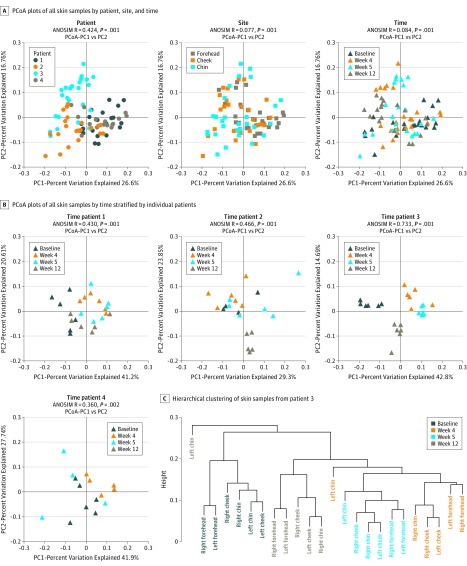
We also examined intersample diversity, or β diversity, based on principal coordinates analyses of weighted UniFrac distances. Figure 4 displays principal coordinates analysis plots with intersample distances represented by 2 principal coordinates (PC1 and PC2), with closely positioned samples being more similar in composition. The ANOSIM, which generates an R test statistic ranging from −1 to 1, was used to examine clustering of samples by patient, site, and time (Figure 4A). A positive R value suggests greater within-group similarity than between-group similarity, with greater magnitudes of the R value suggesting more pronounced clustering of samples. An R value of 0 indicates no clustering of samples, whereas a negative R value suggests greater between-group similarity than within-group similarity. Although there was moderate clustering of samples by patient (ANOSIM, R = 0.424; P = .001), showing the unique microbial signature for each person, clustering by site (ANOSIM, R = 0.077; P = .001) or by time (ANOSIM, R = 0.084; P = .001) was much less apparent. Within individual patients, however, there was clustering of samples by time (Figure 4B), especially in patient 3 (ANOSIM, R = 0.733; P = .001). We performed hierarchical clustering of samples obtained from patient 3, and the resulting cluster dendrogram (Figure 4C) also showed grouping of samples by time, with samples from weeks 4 and 5 most similar to each other, a finding supported by the proximity of samples from weeks 4 and 5 on the principal coordinates analysis plot for patient 3 (Figure 4B).
Figure 4. Microbiota β Diversity.
Microbiota β diversity (between-sample microbial diversity) based on principal coordinates analysis (PCoA) of weighted UniFrac distances. A and B, PCoA plots show intersample distances by 2 principal coordinates (PC1 and PC2), with labeling of individual samples by patient, site, and time across all samples and by time for patients 1 to 4. Principal coordinates, calculated from a distance matrix of weighted Unifrac distances, and have no units. C, Hierarchical clustering of skin samples from patient 3 with the unweighted pair group method with arithmetic mean algorithm. Analysis of similarity (ANOSIM) test statistic R ranges from −1 to 1; values closer to 1 indicate more pronounced clustering of samples by the examined categorical variable (patient, site, or time).
Discussion
This pilot longitudinal cohort study examined the association of systemic antibiotic treatment with acne severity and skin microbiota. Through 16S rRNA gene sequencing, we confirmed the predominance of Cutibacterium species and C acnes as well as bacterial taxa belonging to phyla Actinobacteria, Firmicutes, and Proteobacteria in skin samples, in accordance with previous studies.12,31,32,33 Treatment of acne with minocycline was associated with a 1.4-fold reduction of C acnes across all patients, with recovery toward baseline C acnes levels 8 weeks after discontinuation of minocycline therapy. Two patients (1 and 3) displayed clinical improvements in acne severity concurrent with the reduction in C acnes, although our small sample size and short duration of antibiotic treatment may have limited the power of our study to detect changes in acne severity.
Our finding of significant changes in relative abundance of multiple bacterial genera across all 4 patients (Figure 2C) may have important clinical implications. In addition to reductions in C acnes, we also observed a 5.6-fold increase in Pseudomonas species immediately following 4 weeks of antibiotic treatment. As C acnes levels recovered after 8 weeks of antibiotic treatment withdrawal, Pseudomonas species levels also returned to baseline. These observations parallel previous findings by Hall et al,34 who noted a negative correlation between the abundance of C acnes and Pseudomonas species in skin samples of individuals with acne. Our study shows that this negative association between C acnes and Pseudomonas species persists on disruption of skin microbiota with minocycline and suggests that C acnes and Pseudomonas species may compete for the same microenvironments within skin microbiota. The transient growth of bacterial populations, such as Pseudomonas species, immediately following antibiotic treatment points toward the possibility of opportunistic skin infections, such as gram-negative folliculitis,35,36 with prolonged antibiotic therapy for acne.
Our observation of a 1.7-fold increase in Streptococcus species relative to baseline 8 weeks following antibiotic treatment withdrawal adds to existing discussions on the association between antibiotic therapy and Streptococcus species in individuals with acne. In a cross-sectional study, Levy et al37 previously found a 3-fold increase in colonization of the oropharynx by Streptococcus pyogenes among patients with acne receiving antibiotic therapy. Subsequent studies by Margolis et al38,39 also suggested an association between antibiotic treatment of acne and upper respiratory tract infections, particularly pharyngitis. Although we did not sample the oropharynx in the present study, our observation of increased Streptococcus species on facial skin following systemic antibiotic treatment raises the question of whether a similar growth of Streptococcus species occurs in the oropharynx—a process that may mediate previously discovered associations between antibiotic therapy for acne and oral pharyngitis.
Systemic antibiotic treatment of acne may also reduce bacterial populations beneficial to skin health. Several members of the Lactobacillus genus, for instance, have been associated with protective effects against Staphylococcus aureus infections,40 atopic dermatitis,41,42 and acne.43 In the present study, we observed a sustained, 4.7-fold decrease in Lactobacillus species relative to baseline 8 weeks following discontinuation of minocycline treatment, but whether this presents increased risk for skin infections remains to be determined. Understanding the timing and significance of shifts in microbial flora associated with antibiotic treatment, both during and after therapy, may help clinicians decrease the likelihood of skin comorbidities related to dysbiosis. This may involve concurrent monitoring of the skin microbiota, a more targeted antibiotic approach, or additional therapies to sustain “good” flora during treatment.
Limitations
We did not notice definite trends in acne severity and α diversity across patients in this study. At the individual level, we observed opposite changes in α diversity in 2 patients (Figure 3D) that may be associated with the recurrence status of inflammatory acne following withdrawal of minocycline therapy (Figure 3B). Patient 1, who showed excessive recovery of microbial diversity 8 weeks after discontinuation of minocycline treatment, had an increased number of inflamed lesions at week 12 relative to baseline. By contrast, patient 3, who showed lack of total recovery of microbial diversity even 8 weeks after discontinuation of minocycline treatment, showed little recurrence of inflammatory acne. These findings, however, were confounded by both patients 3 and 4 showing decreases in α diversity following 4 weeks of antibiotic treatment but having opposite changes in inflamed lesion counts, with patient 3 showing improvement in acne severity and patient 4 displaying exacerbation. Although our results indicated variable rates of microbiota recovery to antibiotic therapy across individuals, we could not determine whether a relationship existed between acne severity and microbial diversity of the skin.
The small participant size of our study meant that statistically significant changes in bacterial taxa over time may have been driven by individual patients. We compared relative abundance of bacteria across time in individual patients (eFigure 2 in the Supplement) and found variations in the bacterial taxa change associated with antibiotic treatment among the patients. Although multiple bacterial taxa—particularly C acnes, Pseudomonas species, Corynebacterium species, and Erwinia species—showed similar temporal patterns of change in more than 1 patient, the small participant size of our study ultimately limits the generalizability of our findings.
Although we set the duration of antibiotic treatment to 4 weeks to maximize patient compliance and retention, the short course of treatment may have also limited improvements in acne severity, particularly in patients with greater acne severity at baseline. Of the 4 patients in this study, patients 1 through 3 had fewer inflamed lesions than patient 4 at baseline and showed either improvement or no change in acne severity following 4 weeks of antibiotic treatment. Patient 4, by contrast, had significantly more inflamed lesions at baseline and showed worsening of acne after 4 weeks of treatment with minocycline. The use of a larger sample size and longer duration of antibiotic treatment would enable better characterization of changes in inflamed lesion count and α diversity, which may respond differently to antibiotic treatment depending on acne severity at baseline. Longitudinal data of negative control participants receiving no antibiotic treatment, which we did not collect in the present study, would also enable more accurate characterization of the changes in skin microbiota associated with antibiotic treatment.
Another potential limitation of our study was the use of swabs for skin sampling, which has been characterized as unsuitable for capturing the bacterial community of the pilosebaceous follicle.44 Although a recent study by Hall et al34 showed no difference in C acnes–associated factors between surface and follicular sampling methods, concurrent examination of the follicular microbiota may have provided additional insights on the clinical significance of shifts in the skin microbiota observed in the present study. Specifically, whether changes in bacteria, such as C acnes and Pseudomonas species, on superficial skin layers contribute to or reflect changes in follicular microbiota may be investigated. Future iterations of our study may also use mock community controls,45 which we did not use, to account for foreign DNA that may have been introduced into 16S rRNA gene analysis through use of cotton swabs.
Our use of primers targeting the V3-V4 hypervariable region of the 16S rRNA gene identified significant levels of C acnes and Cutibacterium species, thereby avoiding one of the major limitations of skin microbiome studies targeting the V4 region in isolation.46,47 However, the relative abundance of Cutibacterium species that we observed (mean of 38.4% at baseline, 29.8% across all time points; see eFigure 1B in the Supplement) was still lower than that reported in previous studies targeting the V1-V3 region.34,46 Finally, we were unable to obtain strain-level resolution of C acnes. Considering the emerging association of type IA1 strains of C acnes with inflammatory skin disease,48,49,50 it would be valuable to examine the association of changes in specific strains of C acnes with antibiotic treatment.
Conclusions
Antibiotic treatment was associated with changes in the composition and diversity of skin microbiota with variable rates of recovery across individuals. Reductions in the relative abundance of C acnes associated with systemic antibiotic treatment was accompanied by concurrent growth and suppression of various bacterial populations, with possible clinical implications. Understanding the associations between antibiotics and skin microbiota may help clinicians decrease the likelihood of skin comorbidities related to microbial dysbiosis.
eTable. Read Count and Operational Taxonomic Unit (OTU) Assignment of Individual Skin Samples
eFigure 1. Microbial Distributions of All Samples at the Phylum and Genus Levels
eFigure 2. Dynamics of Bacterial Populations in the Presence and After Withdrawal of Antibiotic Treatment Stratified by Individual Patients
eReference
References
- 1.Thiboutot D, Gollnick H, Bettoli V, et al. ; Global Alliance to Improve Outcomes in Acne . New insights into the management of acne: an update from the Global Alliance to Improve Outcomes in Acne group. J Am Acad Dermatol. 2009;60(5)(suppl):S1-S50. doi: 10.1016/j.jaad.2009.01.019 [DOI] [PubMed] [Google Scholar]
- 2.Williams HC, Dellavalle RP, Garner S. Acne vulgaris. Lancet. 2012;379(9813):361-372. doi: 10.1016/S0140-6736(11)60321-8 [DOI] [PubMed] [Google Scholar]
- 3.Zaenglein AL, Pathy AL, Schlosser BJ, et al. . Guidelines of care for the management of acne vulgaris. J Am Acad Dermatol. 2016;74(5):945-73.e33. doi: 10.1016/j.jaad.2015.12.037 [DOI] [PubMed] [Google Scholar]
- 4.Scholz CF, Kilian M. The natural history of cutaneous propionibacteria, and reclassification of selected species within the genus Propionibacterium to the proposed novel genera Acidipropionibacterium gen. nov., Cutibacterium gen. nov. and Pseudopropionibacterium gen. nov. Int J Syst Evol Microbiol. 2016;66(11):4422-4432. doi: 10.1099/ijsem.0.001367 [DOI] [PubMed] [Google Scholar]
- 5.Heymann WR. Toll-like receptors in acne vulgaris. J Am Acad Dermatol. 2006;55(4):691-692. doi: 10.1016/j.jaad.2006.05.049 [DOI] [PubMed] [Google Scholar]
- 6.Dessinioti C, Katsambas AD. The role of Propionibacterium acnes in acne pathogenesis: facts and controversies. Clin Dermatol. 2010;28(1):2-7. doi: 10.1016/j.clindermatol.2009.03.012 [DOI] [PubMed] [Google Scholar]
- 7.Lee YH, Liu G, Thiboutot DM, Leslie DL, Kirby JS. A retrospective analysis of the duration of oral antibiotic therapy for the treatment of acne among adolescents: investigating practice gaps and potential cost-savings. J Am Acad Dermatol. 2014;71(1):70-76. doi: 10.1016/j.jaad.2014.02.031 [DOI] [PubMed] [Google Scholar]
- 8.Straight CE, Lee YH, Liu G, Kirby JS. Duration of oral antibiotic therapy for the treatment of adult acne: a retrospective analysis investigating adherence to guideline recommendations and opportunities for cost-savings. J Am Acad Dermatol. 2015;72(5):822-827. doi: 10.1016/j.jaad.2015.01.048 [DOI] [PubMed] [Google Scholar]
- 9.Nagler AR, Milam EC, Orlow SJ. The use of oral antibiotics before isotretinoin therapy in patients with acne. J Am Acad Dermatol. 2016;74(2):273-279. doi: 10.1016/j.jaad.2015.09.046 [DOI] [PubMed] [Google Scholar]
- 10.Barbieri JS, Hoffstad O, Margolis DJ. Duration of oral tetracycline-class antibiotic therapy and use of topical retinoids for the treatment of acne among general practitioners (GP): a retrospective cohort study. J Am Acad Dermatol. 2016;75(6):1142-1150. doi: 10.1016/j.jaad.2016.06.057 [DOI] [PubMed] [Google Scholar]
- 11.Barbieri JS, James WD, Margolis DJ. Trends in prescribing behavior of systemic agents used in the treatment of acne among dermatologists and nondermatologists: a retrospective analysis, 2004-2013. J Am Acad Dermatol. 2017;77(3):456-463. doi: 10.1016/j.jaad.2017.04.016 [DOI] [PubMed] [Google Scholar]
- 12.Grice EA, Kong HH, Conlan S, et al. ; NISC Comparative Sequencing Program . Topographical and temporal diversity of the human skin microbiome. Science. 2009;324(5931):1190-1192. doi: 10.1126/science.1171700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326(5960):1694-1697. doi: 10.1126/science.1177486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Antonopoulos DA, Huse SM, Morrison HG, Schmidt TM, Sogin ML, Young VB. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect Immun. 2009;77(6):2367-2375. doi: 10.1128/IAI.01520-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A. 2011;108(suppl 1):4554-4561. doi: 10.1073/pnas.1000087107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fouhy F, Guinane CM, Hussey S, et al. . High-throughput sequencing reveals the incomplete, short-term recovery of infant gut microbiota following parenteral antibiotic treatment with ampicillin and gentamicin. Antimicrob Agents Chemother. 2012;56(11):5811-5820. doi: 10.1128/AAC.00789-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Panda S, El khader I, Casellas F, et al. . Short-term effect of antibiotics on human gut microbiota. PLoS One. 2014;9(4):e95476. doi: 10.1371/journal.pone.0095476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roghmann MC, Lydecker AD, Hittle L, et al. . Comparison of the microbiota of older adults living in nursing homes and the community. mSphere. 2017;2(5):e00210-17. doi: 10.1128/mSphere.00210-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fadrosh DW, Ma B, Gajer P, et al. . An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome. 2014;2(1):6. doi: 10.1186/2049-2618-2-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caporaso JG, Kuczynski J, Stombaugh J, et al. . QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335-336. doi: 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73(16):5261-5267. doi: 10.1128/AEM.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cole JR, Wang Q, Cardenas E, et al. . The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37(Database issue):D141-D145. doi: 10.1093/nar/gkn879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DeSantis TZ, Hugenholtz P, Larsen N, et al. . Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72(7):5069-5072. doi: 10.1128/AEM.03006-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Faith DP. Conservation Evaluation and Phylogenetic Diversity. Biol Conserv. 1992;61(1):1-10. doi: 10.1016/0006-3207(92)91201-3 [DOI] [Google Scholar]
- 25.Faith DP, Baker AM. Phylogenetic diversity (PD) and biodiversity conservation: some bioinformatics challenges. Evol Bioinform Online. 2007;2:121-128. [PMC free article] [PubMed] [Google Scholar]
- 26.Faith DP. The role of the phylogenetic diversity measure, PD, in bio-informatics: getting the definition right. Evol Bioinform Online. 2007;2:277-283. [PMC free article] [PubMed] [Google Scholar]
- 27.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71(12):8228-8235. doi: 10.1128/AEM.71.12.8228-8235.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.White JR, Nagarajan N, Pop M. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput Biol. 2009;5(4):e1000352. doi: 10.1371/journal.pcbi.1000352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clarke KR. Non-parametric multivariate analyses of changes in community structure. Aust J Ecol. 1993;18(1):117-143. doi: 10.1111/j.1442-9993.1993.tb00438.x [DOI] [Google Scholar]
- 30.Wickham H. ggplot2: Elegant Graphics For Data Analysis. New York: Springer; 2009. doi: 10.1007/978-0-387-98141-3 [DOI] [Google Scholar]
- 31.Byrd AL, Belkaid Y, Segre JA. The human skin microbiome. Nat Rev Microbiol. 2018;16(3):143-155. doi: 10.1038/nrmicro.2017.157 [DOI] [PubMed] [Google Scholar]
- 32.Schommer NN, Gallo RL. Structure and function of the human skin microbiome. Trends Microbiol. 2013;21(12):660-668. doi: 10.1016/j.tim.2013.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grice EA, Segre JA. The skin microbiome. Nat Rev Microbiol. 2011;9(4):244-253. doi: 10.1038/nrmicro2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hall JB, Cong Z, Imamura-Kawasawa Y, et al. . Isolation and identification of the follicular microbiome: implications for acne research. J Invest Dermatol. 2018;138(9):2033-2040. doi: 10.1016/j.jid.2018.02.038 [DOI] [PubMed] [Google Scholar]
- 35.Böni R, Nehrhoff B. Treatment of gram-negative folliculitis in patients with acne. Am J Clin Dermatol. 2003;4(4):273-276. doi: 10.2165/00128071-200304040-00005 [DOI] [PubMed] [Google Scholar]
- 36.Leyden JJ, Marples RR, Mills OH Jr, Kligman AM. Gram-negative folliculitis—a complication of antibiotic therapy in acne vulgaris. Br J Dermatol. 1973;88(6):533-538. doi: 10.1111/j.1365-2133.1973.tb08015.x [DOI] [PubMed] [Google Scholar]
- 37.Levy RM, Huang EY, Roling D, Leyden JJ, Margolis DJ. Effect of antibiotics on the oropharyngeal flora in patients with acne. Arch Dermatol. 2003;139(4):467-471. doi: 10.1001/archderm.139.4.467 [DOI] [PubMed] [Google Scholar]
- 38.Margolis DJ, Bowe WP, Hoffstad O, Berlin JA. Antibiotic treatment of acne may be associated with upper respiratory tract infections. Arch Dermatol. 2005;141(9):1132-1136. doi: 10.1001/archderm.141.9.1132 [DOI] [PubMed] [Google Scholar]
- 39.Margolis DJ, Fanelli M, Kupperman E, et al. . Association of pharyngitis with oral antibiotic use for the treatment of acne: a cross-sectional and prospective cohort study. Arch Dermatol. 2012;148(3):326-332. doi: 10.1001/archdermatol.2011.355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prince T, McBain AJ, O’Neill CA. Lactobacillus reuteri protects epidermal keratinocytes from Staphylococcus aureus-induced cell death by competitive exclusion. Appl Environ Microbiol. 2012;78(15):5119-5126. doi: 10.1128/AEM.00595-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kalliomäki M, Salminen S, Poussa T, Arvilommi H, Isolauri E. Probiotics and prevention of atopic disease: 4-year follow-up of a randomised placebo-controlled trial. Lancet. 2003;361(9372):1869-1871. doi: 10.1016/S0140-6736(03)13490-3 [DOI] [PubMed] [Google Scholar]
- 42.Kalliomäki M, Salminen S, Arvilommi H, Kero P, Koskinen P, Isolauri E. Probiotics in primary prevention of atopic disease: a randomised placebo-controlled trial. Lancet. 2001;357(9262):1076-1079. doi: 10.1016/S0140-6736(00)04259-8 [DOI] [PubMed] [Google Scholar]
- 43.Baquerizo Nole KL, Yim E, Keri JE. Probiotics and prebiotics in dermatology. J Am Acad Dermatol. 2014;71(4):814-821. doi: 10.1016/j.jaad.2014.04.050 [DOI] [PubMed] [Google Scholar]
- 44.Omer H, McDowell A, Alexeyev OA. Understanding the role of Propionibacterium acnes in acne vulgaris: the critical importance of skin sampling methodologies. Clin Dermatol. 2017;35(2):118-129. doi: 10.1016/j.clindermatol.2016.10.003 [DOI] [PubMed] [Google Scholar]
- 45.Kim D, Hofstaedter CE, Zhao C, et al. . Optimizing methods and dodging pitfalls in microbiome research. Microbiome. 2017;5(1):52. doi: 10.1186/s40168-017-0267-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meisel JS, Hannigan GD, Tyldsley AS, et al. . Skin microbiome surveys are strongly influenced by experimental design. J Invest Dermatol. 2016;136(5):947-956. doi: 10.1016/j.jid.2016.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zeeuwen PLJM, Boekhorst J, Ederveen THA, et al. . Reply to Meisel et al. J Invest Dermatol. 2017;137(4):961-962. doi: 10.1016/j.jid.2016.11.013 [DOI] [PubMed] [Google Scholar]
- 48.Dréno B, Pécastaings S, Corvec S, Veraldi S, Khammari A, Roques C. Cutibacterium acnes (Propionibacterium acnes) and acne vulgaris: a brief look at the latest updates. J Eur Acad Dermatol Venereol. 2018;32(suppl 2):5-14. doi: 10.1111/jdv.15043 [DOI] [PubMed] [Google Scholar]
- 49.Fitz-Gibbon S, Tomida S, Chiu BH, et al. . Propionibacterium acnes strain populations in the human skin microbiome associated with acne. J Invest Dermatol. 2013;133(9):2152-2160. doi: 10.1038/jid.2013.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lomholt HB, Kilian M. Population genetic analysis of Propionibacterium acnes identifies a subpopulation and epidemic clones associated with acne. PLoS One. 2010;5(8):e12277. doi: 10.1371/journal.pone.0012277 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eTable. Read Count and Operational Taxonomic Unit (OTU) Assignment of Individual Skin Samples
eFigure 1. Microbial Distributions of All Samples at the Phylum and Genus Levels
eFigure 2. Dynamics of Bacterial Populations in the Presence and After Withdrawal of Antibiotic Treatment Stratified by Individual Patients
eReference