Key Points
Question
What is the germline mutational landscape in unselected patients with prostate cancer, and do guidelines for genetic testing adequately identify patients at risk for aggressive disease?
Findings
This cross-sectional study of 3607 men with a personal history of prostate cancer found that 620 (17.2%) had a pathogenic germline variant, of which 229 (37%) did not qualify (at time of testing) for genetic testing per National Comprehensive Cancer Network recommendations.
Meaning
National Comprehensive Cancer Network guidelines and Gleason scores are not reliable for stratifying patients with prostate cancer for the presence or absence of pathogenic germline variants; simplification and expansion of testing guidelines will improve medical management of these patients.
This cross-sectional study assesses the frequency and distribution of positive germline variants in patients with prostate cancer and evaluates the usefulness of current practice guidelines in recognizing individuals with prostate cancer who would benefit from diagnostic genetic testing.
Abstract
Importance
Prostate cancer is the third leading cause of cancer-related death in men in the United States. Although serious, most of these diagnoses are not terminal. Inherited risk for prostate cancer is associated with aggressive disease and poorer outcomes, indicating a critical need for increased genetic screening to identify disease-causing variants that can pinpoint individuals at increased risk for metastatic castration-resistant prostate cancer.
Objective
To identify positive (pathogenic, likely pathogenic, and increased risk) germline variants in a large prostate cancer cohort and to evaluate the usefulness of current practice guidelines in recognizing individuals at increased risk for prostate cancer who would benefit from diagnostic genetic testing.
Design, Setting, and Participants
Cross-sectional study of data from 3607 men with a personal history of prostate cancer who underwent germline genetic testing between 2013 and 2018 and were unselected for family history, stage of disease, or age at diagnosis. Referral-based testing was performed at a Clinical Laboratory Improvement Amendments/College of American Pathologists–certified diagnostic laboratory. All analysis took place between February 2017 and August 2018.
Main Outcomes and Measures
The frequency and distribution of positive germline variants, and the percentage of individuals with prostate cancer who met National Comprehensive Cancer Network (NCCN) guidelines for germline genetic testing.
Results
Of 3607 men (mean [SD] age at testing, 67 [9.51] years; mean age at diagnosis, 60 [9.05] years) with a personal diagnosis of prostate cancer who were referred for genetic testing, 620 (17.2%) had positive germline variants, of which only 30.7% were variants in BRCA1/2. Positive variants in HOXB13, a gene associated only with prostate cancer risk, were identified in 30 patients (4.5%). DNA mismatch repair variants with substantial known therapeutic implications were detected in 1.74% of variants in the total population tested. Examination of self-reported family histories indicated that 229 individuals (37%) with positive variants in this cohort would not have been approved for genetic testing using the NCCN genetic/familial breast and ovarian guidelines for patients with prostate cancer.
Conclusions and Relevance
Current NCCN guidelines and Gleason scores cannot reliably stratify patients with prostate cancer for the presence or absence of pathogenic germline variants. Most positive genetic test results identified in this study have important management implications for patients and their families, which underscores the need to revisit current guidelines.
Introduction
Prostate cancer is a leading cause of death in men, but despite its prevalence, its cause remains unclear in most men. Inherited mutations have considerable implications at presentation for staging,1 screening,2 treatment,3 genetic counseling, and cascade testing of family members.4 However, many questions remain given the limited data available for various ethnic populations, geographic heterogeneity in published reports, the presence of rare variants, and incomplete family histories.3 Overcoming these limitations is particularly critical because inherited risk of prostate cancer is associated with more aggressive disease and poorer outcomes. Increased genetic screening is needed to identify individuals at high risk for metastatic prostate cancer and will help guide treatment.
Findings reported by Pritchard and colleagues5 clearly delineate the prevalence of selected inherited DNA repair mutations in men with metastatic prostate cancer. Smaller studies support these findings, but there is considerable geographic heterogeneity in the results.6 For example, in the study by Pritchard et al,5 8.8% of patients with metastatic disease treated at the University of Washington had germline mutations compared with 18.5% of those treated at Memorial Sloan Kettering. The prevalence of rare variants in prostate cancer is also overlooked. Several studies report the frequency of germline DNA repair mutations but have predominantly focused on a limited number of genes (BRCA1/2 and ATM).7,8,9 Although these genes are clearly important, other potentially important genes such as CHEK2 and those involved in DNA mismatch repair mechanisms (MLH1, MSH2, MSH6, PMS2) are evaluated less frequently. The underlying cause of these limitations is unknown but may represent a lack of access to genetic testing or may be a result of the complicated guidelines that indicate when genetic testing should be conducted.
Guidelines specifically addressing genetic testing for patients with prostate cancer were sparse until recently. However, the National Comprehensive Cancer Network (NCCN) guidelines for prostate cancer (version 3.2018),10 the guidelines for genetic/familial high-risk assessment for breast and ovarian cancers (version 1.2019)11 as they relate to prostate cancer, and the genetic/familial high-risk assessment for colorectal cancer (version 1.2018)12 are inconsistent with one another, and consensus documents add additional inconsistencies.2 Current NCCN prostate cancer guidelines state that clinicians should consider genetic testing for all patients with metastatic, regional, very high-risk disease, or high-risk disease regardless of family history. Testing is suggested for the following genes: BRCA1, BRCA2, ATM, PALB2, and FANCA. For those with lower-risk disease, germline genetic testing should be considered when there is a strong family history, which is defined as a brother or father or multiple family members diagnosed with prostate cancer before age 60 years; known germline abnormalities (particularly in BRCA2 or genes associated with Lynch syndrome), and/or more than 1 family member with breast, ovarian, or pancreatic cancer (suggestive of BRCA2 mutations); or more than 1 family member with a cancer associated with Lynch syndrome (colorectal, endometrial, gastric, ovarian, endometrial, pancreatic, urothelial, kidney, or bile duct cancer). Adding to the complexity, current NCCN guidelines for colorectal cancer state that “there is insufficient evidence to support prostate cancer screening among high-risk men with Lynch syndrome.”12
Perhaps most complicated are the familial breast and ovarian guideline recommendations for BRCA1/2 testing of men, which introduce a complex decision tree for determining which individuals qualify for testing. One branch of this tree requires a personal history of metastatic prostate cancer or a Gleason score of 7 or higher, in addition to 1 or more close relatives with ovarian cancer at any age, or a family history of female breast cancer (diagnosed before age 50 years). If these requirements are not met, genetic testing may still be recommended for patients with a personal history of prostate cancer (Gleason score ≥7) with 2 or more close relatives with any combination of breast, prostate (Gleason score ≥7), or pancreatic cancer. Patients with or without a history of prostate cancer qualify for genetic testing if they have a family history of BRCA1/2 mutation or if BRCA1/2 mutations were detected in the tumor. Additionally, testing would be warranted in men with a first- or second-degree relative who meets BRCA1/2 guidelines or a third-degree relative with breast and/or ovarian cancer who also has 2 or more close relatives with breast and/or ovarian cancer. It is unclear whether these guidelines are followed by practitioners, but the discordance and complexity of the recommendations are problematic. Simplifying and expanding current testing guidelines would provide several benefits for the medical management of men with prostate cancer and their family members.
The purpose of this study was to evaluate the prevalence of germline mutations in a large, diverse cohort with prostate cancer with respect to current genetic testing guidelines and to bridge gaps in knowledge given the heterogeneity in both prostate cancer and germline testing recommendations. Herein, we report a study of 3607 patients with prostate cancer at various stages of disease. Family history, age at testing, ancestry/ethnicity, and Gleason scores were analyzed. This study provides a broader ascertainment of patients than previous reports. To our knowledge, this series of multigene germline testing is the largest in a population with prostate cancer.
Methods
Patient Cohort
Personal and family history information from submitted requisition forms and medical records (when available) were reviewed for 3607 individuals with a personal history of prostate cancer who underwent germline genetic testing between 2013 and 2018. All patient data were deidentified before analysis under Western Institutional Review Board protocol number 1167406, which waived the requirement to obtain written patient informed consent. Ethnicity was provided by all patients at the time of test ordering and was grouped for analysis based on categories used in population databases.
Panel Composition
Genes analyzed for each patient were chosen at the discretion of the ordering clinician and ranged from 2 to 80 genes. The 14 genes included on the Invitae curated prostate cancer panel (ATM, BRCA1, BRCA2, CHEK2, EPCAM, HOXB13, MLH1, MSH2, MSH6, NBN, PMS2, TP53, RAD51D, and PALB2) were requisitioned in 2250 (62%) of 3608 orders. Analysis of other genes was ordered from larger hereditary cancer panels or through customized requests. The price of a panel was the same regardless of the genes tested, making test selection dependent only on clinician preferences.
Sequencing and Variant Interpretation
Sample types for this cohort included blood and saliva. Once extracted, DNA was processed and subjected to paired-end sequencing on an Illumina next-generation sequencing platform, as described previously.13 Pathogenic (P), likely pathogenic (LP), and increased-risk allele variants were confirmed using orthogonal technology in accordance with Invitae standard operating practices.13 Variants were subjected to clinical interpretation using a refined American College of Medical Genetics and Genomics criteria (Sherloc).14 Variants interpreted as P, LP, or increased-risk allele were considered positive.
Statistical Analysis
The frequency of variants identified in the cohort were evaluated relative to the expected frequency in the normal population using the genome Aggregation Database (gnomAD). The analysis was similar to that used by Pritchard et al.5 An “N−1” χ2 test was used to compare proportions among cohorts, and CIs were calculated using SAS Studio 3.71 (SAS Institute). P values less than .05 were considered significant. To compare proportions between cohorts when the number of occurrences in a cell were fewer than 5, the Fisher exact test was used.
Results
Positive variants were identified in 620 (17.2%) of 3607 patients (mean [SD] age at testing, 67 [9.51] years; mean age at diagnosis, 60 [9.05] years). The general age distribution, ancestry/ethnicity, family history, and Gleason scores of the patients at the time of testing are described in eTable 1 in the Supplement. Age at testing was distinct from age at diagnosis (eFigure 1 in the Supplement), and a considerable lag between the initial diagnosis of prostate cancer and referral for germline genetic testing was observed across almost all age groups. Age at testing was not associated with the risk of finding positive variants in this cohort. Positive results by ancestry/ethnicity varied, with the highest rates found among those identified as Ashkenazi Jewish (n = 234; 22.7%) and white (n = 2594; 17.8%). Compared with the other ethnic groups, African Americans (n = 227 vs 3385; 10.1%; odds ratio, 0.527; P = .006) and Hispanics (n = 78 vs 3534; 6.4%; odds ratio, 0.325; P = .02) had lower rates of positive variants. The positive variants detected in the various ethnic groups are listed in eTable 2 in the Supplement. DNA repair defects were less frequently found in nonwhite, non-Jewish subsets of patients, but statistical testing was not applied because individual testing frequencies were unavailable. Corresponding family histories were available for 558 (90%) of 620 patients with positive variants. There was an inconsistent depth of information provided for primary and secondary relatives, and deidentification of patients prevented follow-up inquiries. Family history of breast, prostate, ovarian, colon, pancreatic, or other cancers did not correlate with positive variant detection. Metastatic disease was reported in 542 (15%) patients, and Gleason scores were available for 1539 (43%) patients. Gleason scores were not associated with the presence of positive mutations. In patients with Gleason scores of 6 or lower for a primary tumor (n = 148), positive germline variants were found in 15.1% compared with 16.3% in those with Gleason scores of 7 or higher (n = 1391).
As reported in the Table, among the entire cohort, the top 10 genes with positive variants as a percentage of men tested were as follows: BRCA2, 4.74%; CHEK2, 2.88%; ATM, 2.03%; MUTYH, 2.37%; APC, 1.28%; BRCA1, 1.25%; HOXB13, 1.12%; MSH2, 0.69%; TP53, 0.66%; and PALB2, 0.56%. Notably, positive variants in mismatch repair genes (PMS2, MLH1, MSH2, MSH6) accounted for 1.74% of the variants in the total population tested. A complete breakdown of the variants detected in this cohort is shown in eFigure 2 in the Supplement. The breadth of genes with positive findings indicates the long tail of abnormalities observed in prostate cancer and offers insights into the value of extended genetic testing.
Table. Most Frequently Detected Variants in Patients With a Personal History of Prostate Cancer.
| Gene | No. of Requisitions | Variants of Uncertain Significance Detected | Positive Variants Detected, n = 674, (%) | Positive Variants per Requisition, %a |
|---|---|---|---|---|
| BRCA2 | 3459 | 75 | 164 (24.3) | 4.74 |
| CHEK2 | 3300 | 71 | 95 (14.1) | 2.88 |
| ATM | 3207 | 160 | 65 (9.6) | 2.03 |
| MUTYH | 2322 | 27 | 55 (8.2) | 2.37 |
| BRCA1 | 3436 | 38 | 43 (6.4) | 1.25 |
| HOXB13 | 2667 | 0 | 30 (4.5) | 1.12 |
| APC | 2345 | 76 | 30 (4.5) | 1.28 |
| MSH2 | 3350 | 48 | 23 (3.4) | 0.69 |
| TP53 | 3329 | 30 | 22 (3.3) | 0.66 |
| PALB2 | 3014 | 42 | 17 (2.5) | 0.56 |
| PMS2 | 3345 | 50 | 18 (2.7) | 0.54 |
| MSH6 | 3346 | 75 | 15 (2.2) | 0.45 |
| NBN | 3145 | 41 | 10 (1.5) | 0.32 |
| RAD50 | 2173 | 40 | 7 (1.0) | 0.32 |
| BRIP1 | 2461 | 36 | 7 (1.0) | 0.28 |
| RAD51C | 2438 | 21 | 5 (0.7) | 0.21 |
| RAD51D | 2689 | 12 | 4 (0.6) | 0.15 |
| CDKN2A | 2277 | 6 | 3 (0.4) | 0.13 |
| CDH1 | 2504 | 28 | 3 (0.4) | 0.12 |
| NF1 | 2347 | 35 | 2 (0.3) | 0.09 |
| MLH1 | 3343 | 25 | 2 (0.3) | 0.06 |
Percentages of positive variants per total gene requisitions are calculated as the number of positive variants in a gene divided by the total number of requisitions for that gene.
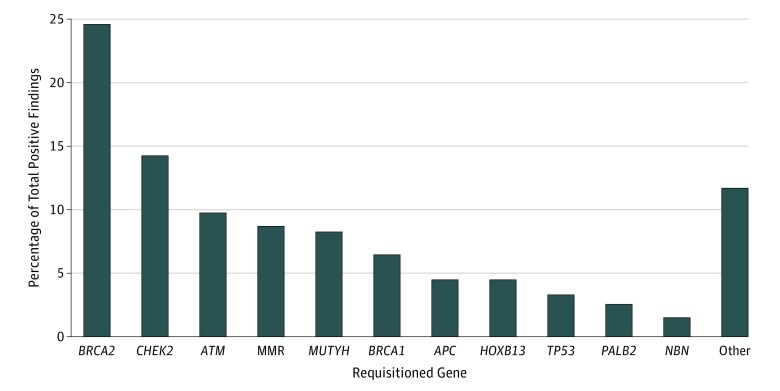
Among patients with positive results, as reported in the Table, the top 10 genes were as follows: BRCA2, 24.3%; CHEK2, 14.1%; ATM, 9.6%; MUTYH, 8.2%; BRCA1, 6.4%; HOXB13, 4.5%; APC, 4.5%; MSH2, 3.4%; TP53, 3.3%; and PMS2, 2.7% (Figure). A number of rare but important pathogenic variants implicated in hereditary cancer risk were identified through extended genetic testing. Only 207 of 674 positive variants (30.7%) occurred in the commonly tested BRCA1/2 genes, and only 291 of 674 positive variants (43.8%) were detected in genes indicated for testing by the 2018 prostate cancer guidelines (BRCA1/2, ATM, PALB2, FANCA).5 More than half of the positive variants detected (386 of 674; 57.2%) were identified in genes not currently recommended for genetic testing. Of note, a variant in the HOXB13 homeobox gene (p.Gly84Glu), implicated in prostate cancer risk, was detected in 1.1% of men (n = 30). Variants of uncertain significance were also identified (eTable 3 in the Supplement).
Figure. Frequency by Gene of Pathogenic, Likely Pathogenic, and Increased-Risk Allele Variants Detected in This Study.
All positive variants detected in a gene were combined and divided by the total number of pathogenic variants detected (n = 674). Mismatch repair (MMR) genes included MLH1, MSH2, MSH6, and PMS2 (no MSH1 variants were detected in this cohort).
The occurrence of 2 FANCA alterations among 194 patients with prostate cancer is noteworthy, given that FANCA may have a role in prostate cancer.10 By comparison, no pathogenic variants in FANCA were detected in a sample of 3679 patients with no known cancer indication referred to Invitae for cardiovascular disease testing (data available from the authors). This finding supports a role for germline FANCA mutations in prostate cancer.
To compare our findings with a well-known data set, we evaluated the top genes identified in our cohort against the metastatic prostate cancer series published by Pritchard et al8 (eTable 4 in the Supplement). The overall findings are consistent between cohorts even though Pritchard et al8 examined patients with metastatic prostate cancer and we evaluated an unselected patient population.
Discussion
To our knowledge, this study reports the largest series of patients with prostate cancer who were tested for pathogenic germline mutations. Comparison of these data with those from another study5 yielded important commonalities. Among the top annotated genes with P and/or LP alterations, BRCA2 is consistently the most frequently mutated gene, followed by ATM, CHEK2, and BRCA1. The percentage of BRCA1/2 mutations in this study (5.99%) is very similar to the 6.22% noted by Pritchard et al,5 who focused on metastatic disease (eTable 4 in the Supplement). Despite the differences between the populations of these 2 studies, the overall findings among commonly tested genes are more similar than distinct. The inclusion of germline genetic testing in the medical management of these patients may help identify deleterious variants and potentially guide treatment decision making. Similar to the management of breast and ovarian cancers, early identification of positive variants may encourage more frequent assessment of tumor progression, screening of at-risk family members, and surgery before metastasis, which can improve prognosis and quality of life for patients with prostate cancer. Furthermore, several of the variants identified in our study are treatable with specific US Food and Drug Administration–approved therapies.
Examination of family histories provided by the ordering clinician indicated that 229 patients (37%) with the positive variants detected in this study would not have been identified had they been tested using only the NCCN genetic/familial breast and ovarian guidelines.11 Guidelines that rely heavily on family history have limitations, including the fact that family history, as shown in this cohort, is only partially informative for many individuals. After the collection and analysis of this data set, NCCN revised and expanded its recommendations for prostate cancer testing. These new guidelines rely heavily on Gleason scores. Direct comparison of our findings related to these new guidelines is infeasible given the absence of staging data for a large subset of the patients in our data set. However, we found that Gleason scores do not correlate with the detection of positive variants, and staging data are not always readily available to practitioners or family members requesting germline testing, both of which limit the value of these new prostate cancer guidelines.
The implications of genetic testing are becoming stronger. Patients with prostate cancer and a BRCA2 germline mutation are more likely to respond to a carboplatin-based regimen.12 It is also known that 59% of those with a germline mutation will have a second allele mutated in the tumor, which adds therapeutic complexity.3 In our population, we found positive variants in a large number of genes that are less well known but are likely relevant from a patient and familial medical management perspective. Specifically, men with prostate cancer owing to a hereditary cancer syndrome may have an increased risk of certain second primary cancers such as male breast, pancreatic, and colon cancers. Information about the presence of pathogenic variants can be used to tailor screening and preventive measures, possibly improving patient outcomes. These same principles could then be applied to family members who may benefit from early detection and prevention measures. Furthermore, the identification of family members who do not harbor a familial risk factor allows individuals the opportunity to forego unnecessary and often costly testing.
The importance of BRCA1/2 and other DNA repair genes outlined in the NCCN guidelines represents a small subset of the findings that would benefit patients with prostate cancer. Therapeutic implications of non-BRCA germline alterations remain unclear, but identification of positive variants in genes not included in the current guidelines have serious implications for treatment options, including clinical trials, particularly basket trials that examine specific genetic biomarkers regardless of tumor type. Clinical trials for poly (ADP-ribose) polymerase (PARP) inhibitor therapy, are available to patients with prostate cancer who carry specific germline variants in DNA repair genes (NCT03413995, NCT02955082). Increasing the availability of genetic testing for prostate cancer may have a substantial influence on opportunities for targeted therapies that may improve patient outcomes. Genetic testing is a critical tool in the optimal management of patients.
Our data show a diagnostic yield (17.2%) across patients with prostate cancer who were referred for clinical genetic testing. Furthermore, the data indicate that factors frequently used to identify patients who qualify for germline testing, including age, race, and family history, did not correlate with positive test results. Based on these findings, we propose that genetic testing guidelines should be simplified and expanded to include genetic testing of all men diagnosed with prostate cancer similar to guidelines for pancreatic and colorectal cancer.15,16 Simplification of testing guidelines would facilitate informed decision making for patients and their family members and provide the foundation for cascade testing of at-risk relatives before they develop cancer, initiating both surveillance and risk-reduction options.
Limitations
This study has limitations. First, a number of ethnic groups, particularly African Americans, Asians, and Hispanics, were underrepresented in this cohort. This is a major limitation of all germline studies to date. Second, Gleason data and family history are not directly derived from medical records. Instead, they are derived from information provided by clinicians, a method prone to errors and omission. Third, this study is not population based; additional information, such as known familial DNA repair mutations, may have influenced who in our cohort was referred for testing. These data also include results for genes that have not been clearly linked to prostate cancer risk (MUTYH, APC). In terms of prostate cancer risk, the importance of the alterations annotated herein cannot be accurately determined, and we make no conclusions regarding the risk attributable to the positive variants identified. Additional studies are required to better discern cancer risks at various ages. Such studies will ideally consider polygenic risk scores in addition to studies of single-allele changes.17
Despite the limitations of this data set, certain findings of this study are noteworthy. We identified a substantial number of germline variants in individuals who do not meet current NCCN guidelines for genetic testing. We also found that BRCA1/2 mutations represent a minority of the detected abnormalities and that there is a long tail of genes with important variants.
Conclusions
Any conversation about expanding germline genetic testing needs to take into consideration the burden such an expansion would create. A persistent shortage of genetics professionals and coordination of follow-up when indicated are issues being addressed by multiple national organizations. For example, various models have been proposed that improve access to genetic counseling, including novel genetic counselor extender models.18,19,20 The capacity and number of accredited genetic counseling training programs has also grown considerably. The economic implications of genetic testing must also be considered. However, as costs continue to decrease, the benefits of a single germline genetic test become apparent when compared with the potentially dramatic cost of managing advanced prostate cancer. Finally, the benefit of increased genetic testing also needs to be considered in the context of preventive management opportunities. Comparing the 1-time cost of genetic testing to the high cost of treating catastrophic late-stage cancer in patients with a genetic risk that was not otherwise identified, as well as the benefit to family members from early screening, provides substantial justification for the simplification and expansion of current guidelines.
eTable 1. Demographic Information for 3607 Patients with a Personal History of Prostate Cancer Who Underwent Germline Genetic Testing Between 2013 and 2018
eTable 2. Distribution by Ethnicity of Pathogenic/Likely Pathogenic/Risk Allele Variants Detected in Men with Prostate Cancer
eTable 3. Pathogenic/Likely Pathogenic/Risk Allele (Positive Variants) and Variants of Unknown Significance Detected in This Study
eTable 4. Comparison of Frequency of Mutations among Tested Patients From This Data Set Compared With Those in the Pritchard et al5 Metastatic Series.
eFigure 1. Age at Diagnosis Compared With Age at Testing in Men with a Personal
History of Prostate Cancer.
eFigure 2. Distribution of Genes in Which Pathogenic, Likely Pathogenic, and Increased Risk Allele Variants Were Detected in This Study.
References
- 1.Castro E, Goh C, Olmos D, et al. Germline BRCA mutations are associated with higher risk of nodal involvement, distant metastasis, and poor survival outcomes in prostate cancer. J Clin Oncol. 2013;31(14):1748-1757. doi: 10.1200/JCO.2012.43.1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bancroft EK, Page EC, Castro E, et al. ; IMPACT Collaborators . Targeted prostate cancer screening in BRCA1 and BRCA2 mutation carriers: results from the initial screening round of the IMPACT study. Eur Urol. 2014;66(3):489-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pomerantz MM, Spisák S, Jia L, et al. The association between germline BRCA2 variants and sensitivity to platinum-based chemotherapy among men with metastatic prostate cancer. Cancer. 2017;123(18):3532-3539. doi: 10.1002/cncr.30808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giri VN, Knudsen KE, Kelly WK, et al. Role of genetic testing for inherited prostate cancer risk: Philadelphia prostate cancer consensus conference 2017. J Clin Oncol. 2018;36(4):414-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pritchard CC, Mateo J, Walsh MF, et al. Inherited DNA-repair gene mutations in men with metastatic prostate cancer. N Engl J Med. 2016;375(5):443-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robinson D, Van Allen EM, Wu YM, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161(5):1215-1228. doi: 10.1016/j.cell.2015.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leongamornlert D, Saunders E, Dadaev T, et al. ; UKGPCS Collaborators . Frequent germline deleterious mutations in DNA repair genes in familial prostate cancer cases are associated with advanced disease. Br J Cancer. 2014;110(6):1663-1672. doi: 10.1038/bjc.2014.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Na R, Zheng SL, Han M, et al. Germline mutations in ATM and BRCA1/2 distinguish risk for lethal and indolent prostate cancer and are associated with early age at death. Eur Urol. 2017;71(5):740-747. doi: 10.1016/j.eururo.2016.11.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castro E, Goh C, Leongamornlert D, et al. Effect of BRCA mutations on metastatic relapse and cause-specific survival after radical treatment for localised prostate cancer. Eur Urol. 2015;68(2):186-193. doi: 10.1016/j.eururo.2014.10.022 [DOI] [PubMed] [Google Scholar]
- 10.National Comprehensive Cancer Network Prostate cancer (version 3.2018). https://www.nccn.org/professionals/physician_gls/pdf/prostate.pdf Accessed July 27, 2018.
- 11.National Comprehensive Cancer Network Genetic/familial high-risk assessment: breast and ovarian (version 2.2019). https://www.nccn.org/professionals/physician_gls/pdf/genetics_screening.pdf Accessed July 30, 2018.
- 12.National Comprehensive Cancer Network Genetic/familial high-risk assessment: colorectal (version 1.2018). https://www.nccn.org/professionals/physician_gls/pdf/genetics_colon.pdf Accessed July 27, 2018.
- 13.Lincoln SE, Kobayashi Y, Anderson MJ, et al. A systematic comparison of traditional and multigene panel testing for hereditary breast and ovarian cancer genes in more than 1000 patients. J Mol Diagn. 2015;17(5):533-544. [DOI] [PubMed] [Google Scholar]
- 14.Nykamp K, Anderson M, Powers M, et al. ; Invitae Clinical Genomics Group . Sherloc: a comprehensive refinement of the ACMG-AMP variant classification criteria. Genet Med. 2017;19(10):1105-1117. doi: 10.1038/gim.2017.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hampel H, Bennett RL, Buchanan A, Pearlman R, Wiesner GL; Guideline Development Group, American College of Medical Genetics and Genomics Professional Practice and Guidelines Committee and National Society of Genetic Counselors Practice Guidelines Committee . A practice guideline from the American College of Medical Genetics and Genomics and the National Society of Genetic Counselors: referral indications for cancer predisposition assessment. Genet Med. 2015;17(1):70-87. doi: 10.1038/gim.2014.147 [DOI] [PubMed] [Google Scholar]
- 16.Shindo K, Yu J, Suenaga M, et al. Deleterious germline mutations in patients with apparently sporadic pancreatic adenocarcinoma. J Clin Oncol. 2017;35(30):3382-3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lecarpentier J, Silvestri V, Kuchenbaecker KB, et al. ; EMBRACE; GEMO Study Collaborators; HEBON; KConFab Investigators . Prediction of breast and prostate cancer risks in male BRCA1 and BRCA2 mutation carriers using polygenic risk scores. J Clin Oncol. 2017;35(20):2240-2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stoll K, Kubendran S, Cohen SA. The past, present and future of service delivery in genetic counseling: keeping up in the era of precision medicine. Am J Med Genet C Semin Med Genet. 2018;178(1):24-37. doi: 10.1002/ajmg.c.31602 [DOI] [PubMed] [Google Scholar]
- 19.Cohen SA, Nixon DM. A collaborative approach to cancer risk assessment services using genetic counselor extenders in a multi-system community hospital. Breast Cancer Res Treat. 2016;159(3):527-534. doi: 10.1007/s10549-016-3964-z [DOI] [PubMed] [Google Scholar]
- 20.Buchanan AH, Rahm AK, Williams JL. Alternate service delivery models in cancer genetic counseling: a mini-review. Front Oncol. 2016;6:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eTable 1. Demographic Information for 3607 Patients with a Personal History of Prostate Cancer Who Underwent Germline Genetic Testing Between 2013 and 2018
eTable 2. Distribution by Ethnicity of Pathogenic/Likely Pathogenic/Risk Allele Variants Detected in Men with Prostate Cancer
eTable 3. Pathogenic/Likely Pathogenic/Risk Allele (Positive Variants) and Variants of Unknown Significance Detected in This Study
eTable 4. Comparison of Frequency of Mutations among Tested Patients From This Data Set Compared With Those in the Pritchard et al5 Metastatic Series.
eFigure 1. Age at Diagnosis Compared With Age at Testing in Men with a Personal
History of Prostate Cancer.
eFigure 2. Distribution of Genes in Which Pathogenic, Likely Pathogenic, and Increased Risk Allele Variants Were Detected in This Study.