Key Points
Question
Is history of hypertension and apolipoprotein E (APOE) associated with intracerebral hemorrhage risk in participants stratified by self-reported race/ethnicity?
Findings
In this case-control study of 13 124 adults, having a copy of APOE ε4 alleles increased the risk for lobar intracerebral hemorrhage only in white individuals, but after propensity score matching for hypertension burden, Hispanic individuals showed the same risk of APOE ε4.
Meaning
APOE ε4 appears to be confirmed as a risk factor for lobar intracerebral hemorrhage in nonwhite populations but is masked by differential hypertension burden in Hispanic individuals; further studies are needed to explore the interactions between APOE alleles and environmental exposures.
Abstract
Importance
Genetic studies of intracerebral hemorrhage (ICH) have focused mainly on white participants, but genetic risk may vary or could be concealed by differing nongenetic coexposures in nonwhite populations. Transethnic analysis of risk may clarify the role of genetics in ICH risk across populations.
Objective
To evaluate associations between established differences in ICH risk by race/ethnicity and the variability in the risks of apolipoprotein E (APOE) ε4 alleles, the most potent genetic risk factor for ICH.
Design, Setting, and Participants
This case-control study of primary ICH meta-analyzed the association of APOE allele status on ICH risk, applying a 2-stage clustering approach based on race/ethnicity and stratified by a contributing study. A propensity score analysis was used to model the association of APOE with the burden of hypertension across race/ethnic groups. Primary ICH cases and controls were collected from 3 hospital- and population-based studies in the United States and 8 in European sites in the International Stroke Genetic Consortium. Participants were enrolled from January 1, 1999, to December 31, 2017. Participants with secondary causes of ICH were excluded from enrollment. Controls were regionally matched within each participating study.
Main Outcomes and Measures
Clinical variables were systematically obtained from structured interviews within each site. APOE genotype was centrally determined for all studies.
Results
In total, 13 124 participants (7153 [54.5%] male with a median [interquartile range] age of 66 [56-76] years) were included. In white participants, APOE ε2 (odds ratio [OR], 1.49; 95% CI, 1.24-1.80; P < .001) and APOE ε4 (OR, 1.51; 95% CI, 1.23-1.85; P < .001) were associated with lobar ICH risk; however, within self-identified Hispanic and black participants, no associations were found. After propensity score matching for hypertension burden, APOE ε4 was associated with lobar ICH risk among Hispanic (OR, 1.14; 95% CI, 1.03-1.28; P = .01) but not in black (OR, 1.02; 95% CI, 0.98-1.07; P = .25) participants. APOE ε2 and ε4 did not show an association with nonlobar ICH risk in any race/ethnicity.
Conclusions and Relevance
APOE ε4 and ε2 alleles appear to affect lobar ICH risk variably by race/ethnicity, associations that are confirmed in white individuals but can be shown in Hispanic individuals only when the excess burden of hypertension is propensity score–matched; further studies are needed to explore the interactions between APOE alleles and environmental exposures that vary by race/ethnicity in representative populations at risk for ICH.
This case-control study examines whether the risk for intracerebral hemorrhage presented by apolipoprotein E ε4 and ε2 alleles varies by race/ethnicity.
Introduction
Spontaneous intracerebral hemorrhage (ICH) is the most severe form of stroke. In the United States, 160 000 people experience an ICH each year with a case fatality rate of 54% at 1 year.1 The prevalence of ICH has increased 47% between 1990 and 2010,2 and ICH risk appears to vary among white, black and Hispanic populations.3,4,5,6 Compared with white individuals, young and middle-aged black individuals have an almost 2-fold increased risk for ICH.3,4 Similarly, Hispanic individuals have a relative risk increase that ranges from 1.4 for lobar ICH to 3.7 for nonlobar ICH.5 Moreover, not only is hypertension prevalence among older adults lower among non-Hispanic white groups (76.3%) than among non-Hispanic black (82.5%) and Hispanic (79.2%) groups, but the risk of ICH in the presence of hypertension increases more than 50% from white to Hispanic groups.7,8,9 The associations of genetic and acquired ICH risk factors with these observed risk differences are poorly understood.
Previous studies conducted in predominantly European-ancestry populations have demonstrated that apolipoprotein E (APOE [OMIM 107741]) ε2 and ε4 alleles potently increase risk of lobar ICH.10 In Alzheimer disease, another disorder associated with APOE ε allele status, the degree of risk contributed by APOE genotype varies substantially by the ancestry of the population studied. Among non-Hispanic white people, homozygous carriers of APOE ε4 exhibit up to a 12-fold higher risk of Alzheimer disease, but this same haplotype exerts little or no risk for black or Hispanic people.11,12,13
Understanding how genetic risk factors vary across race/ethnicity may highlight novel underlying disease mechanisms and identify populations who may be particularly responsive to specific prevention strategies, as has previously been shown in treatment response for heart failure by race/ethnicity.14 Unfortunately, with individuals of African American and Hispanic ancestry representing less than 4% of all samples in genome-wide association studies, only recently has it become possible to study genetic risk of common disease across representative US populations.15
We tested the associations of APOE ε alleles with risk of lobar and nonlobar ICH among white, black, and Hispanic individuals, using direct genotyping data supplemented by genome-wide genotyping as available in cases and controls from the International Stroke Genetics Consortium. Because these analyses revealed substantial heterogeneity by race/ethnicity, we further explored the degree to which the differential burden of hypertension across populations is associated with the variability in observed APOE risks.
Methods
Participating Studies and Data Collection
Case and control participants included in the study were gathered from 3 multicenter studies in the United States and from 8 European sites participating in the International Stroke Genetics Consortium, according to availability of directly ascertained APOE ε genotypes and a harmonized local acute case recruitment scheme. These participants were enrolled in the aforementioned studies from January 1, 1999, to December 31, 2017. Institutional review board approval was obtained from all participating centers, and informed consent was obtained from all participants or their legally authorized representative.
The ICH cases from population-based cohorts were not included because of potential imbalances in lethal case recruitment between the 2 sampling approaches.16 Studies included the Genetics of Cerebral Hemorrhage with Anticoagulation (GOCHA) study,17 the Genetic and Environmental Risk Factors for Hemorrhagic Stroke (GERFHS) study,18 the Ethnic/Racial Variations of Intracerebral Hemorrhage (ERICH) study,19 the Hospital del Mar and Vall d’Hebron Hospital ICH studies,20,21 the Jagiellonian University Hemorrhagic Stroke Study,22 the Lund Stroke Register study,23 the Edinburgh Stroke Study and LINCHPIN,24 the UMC Utrecht ICH study, and the Brescia Stroke Registry.25 Because of variable sample sizes from contributing centers, data from European studies were analyzed together for association testing in a meta-analysis (International Stroke Genetics Consortium Europe), as done previously.26,27
Participants with secondary causes of ICH were excluded from enrollment. More specific inclusion and exclusion criteria for each of the included studies are reported in eTable 1 in the Supplement. Demographic variables, including self-identified race/ethnicity,8 were systematically obtained from structured patient and family member interviews within each site,19,28 along with additional covariates.29 Computed tomographic images on admission were analyzed at each participating site for classification as lobar (involving predominantly the cortex and underlying white matter) and nonlobar (involving predominately the basal ganglia, periventricular white matter, or internal capsule) according to prespecified criteria.26,27 The APOE genotype was centrally determined according to standard procedures.30 Genomewide data were available for a subgroup of participants. Genetic and bioinformatic analysis followed standardized prespecified quality-control procedures31 (eMethods in the Supplement).
Population Stratification
Fifteen ancestry informative markers were selected from participants with direct or genomewide genotyping and subjected to principal component (PC) analysis in accordance with previously published methods.32,33,34,35 The first 4 PCs were included in regression models to adjust for population stratification in this subgroup. This PC analysis was not used to reclassify participants, as self-identified race/ethnicity may capture exposures that transcend genetic ancestry and could help explain the stratification among different populations. A complete description of the genetic analysis, the participants genotyped, and the markers selected is available in eTable 2 in the Supplement.
Statistical Analysis
Categorical variables were expressed as count (%), and continuous variables were expressed as median (interquartile range [IQR]) or mean (SD), as appropriate. Categorical variables were compared using the 2-tailed χ2 test, whereas continuous variables were compared with unpaired Mann-Whitney tests. The threshold for statistical significance was set at P = .05.
We tested APOE allele association with ICH risk using 3 logistic regression models. Model 1 was adjusted for age, sex, and history of hypertension. Model 2 included variables from model 1 in addition to history of hypercholesterolemia; history of ischemic stroke; warfarin, statin and antiplatelet use; smoking; and alcohol use. Model 3 also included variables from model 1 with the addition of the first 4 principal components derived from ancestry-informative genotypes. APOE risk allele status was modeled as 2 variables, ε2 and ε4, coded for allele counts (0, 1, or 2 for each) in an additive model referent to the wildtype ε3 allele.17 Analyses were performed in lobar and nonlobar ICH, given the known differences in underlying biology between the 2 ICH locations.36 All statistical analyses were performed using Stata, version 13.0 (Stata Corp LLC), and R statistical software (R Foundation for Statistical Computing).
Transethnic Meta-analysis
We applied a 2-stage clustering approach for meta-analysis, based on race/ethnicity and stratified by study.37 Cases and controls in each study were divided into black, white, and Hispanic groups, based on self-identified race/ethnicity. Each race/ethnicity group within each study was allocated to the same cluster and tested using the regression models described above. Individual cluster results were presented graphically by plotting OR estimates on a forest plot to visually assess heterogeneity. The effect sizes obtained were then used for a DerSimonian-Laird random-effects, inverse-weighted nonparametric meta-analysis.38 Cochran Q and I2 tests were used to quantify heterogeneity.
Propensity Score Modeling of APOE and Hypertension
To address imbalances in the burden of hypertension across ICH populations, and associated imbalances of baseline characteristics among participants with and without hypertension, 2 propensity score (PS) analyses were performed using the nearest neighbor matching method to compare participants of similar underlying hypertensive pathophysiologic burden.39,40 The first PS analysis was constructed using history of hypertension and included variables of age, sex, and self-identified race/ethnicity. The second PS analysis, leveraging data only available in the ERICH study, contained the same variables as the first PS analysis, in addition to the number of medications prescribed to treat hypertension as well as systolic and diastolic blood pressure readings at ICH presentation. Propensity score results were used in a logistic regression model for ICH risk identical to model 1, as described. In a sensitivity analysis, the same PS procedure was tested against age (older or younger than 65 years), sex, and hypercholesterolemia to increase the confidence that the PS findings were specific for hypertension.
Power Calculation
Using empirical data from our analyses, we performed a post hoc calculation of the statistical power to detect an association of APOE ε alleles with lobar ICH risk in black and Hispanic participants, commensurate with the effect size detected in white participants. Type I error rate of 0.05, log additive inheritance mode, and 0.01 of population risk were assumed, with analyses performed using Quanto software, version 1.2.4 (University of Southern California).41
Results
In total, 13 124 individuals (47.2% cases) were included from the participating studies. Among this sample, 7153 (54.5%) were male, with a median (IQR) age of 66 (56-76) years and with 8334 white (63.5%), 2272 black (17.3%), 1781 Hispanic (13.6%), and 736 other (5.6%) race/ethnicity (Table). The latter group was excluded from the primary analyses given its low statistical power. Rates of APOE ε4 homozygosity in cases were 128 (3.6%) in white, 62 (5.3%) in black, and 22 (1.8%) in Hispanic participants, and the rates of APOE ε2 homozygosity in cases were 36 (1.0%) in white, 14 (1.2%) in black, and 5 (0.4%) in Hispanic participants. Among participants, 56.8% (4069 cases and 3379 controls of 13 124 participants) had genomewide or direct genotyping data on ancestry informative markers for PC analysis (eTable 3 in the Supplement). Self-identified race/ethnicity showed overall strong concordance with PC-based ancestry (eFigure in the Supplement). Additional clinical covariates were available for a subset of participants (eTable 4 in the Supplement).
Table. Demographic Characteristics and Clinical and APOE Allele Frequencies Across Participating Studies.
| Source | No. (%) | |||
|---|---|---|---|---|
| ERICH (n = 5017) | GOCHA (n = 2297) | ISGC Europe (n = 3471) | GERFHS (n = 2339) | |
| Male sex | 2866 (57.1) | 1266 (55.1) | 1891 (54.6) | 1130 (48.3) |
| Age, median (IQR) | 61 (52-72) | 73 (65-80) | 70 (61-77) | 65 (51-75) |
| Cases | 2880 (57.4) | 1322 (57.6) | 1281 (36.9) | 811 (34.7) |
| Lobar ICH | 882 (30.6) | 613 (47.8) | 493 (40.2) | 316 (39.0) |
| Nonlobar ICH | 1998 (69.4) | 670 (52.2) | 734 (59.8) | 495 (61.0) |
| Hypertension | 3364/4976 (67.6) | 1667/2275 (73.3) | 1673/2893 (57.8) | 1264/2337 (54.1) |
| Self-reported race/ethnicity | ||||
| White | 1739 (34.7) | 2024 (88.1) | 2622 (75.5) | 1949 (83.3) |
| Black | 1751 (34.9) | 131 (5.7) | NA | 390 (16.7) |
| Hispanic | 1527 (30.4) | 60 (2.6) | 194 (5.6) | NA |
| Other/missing | NA | 82 (3.6) | 654 (18.8) | NA |
| APOE ε4 allele count | ||||
| 0 | 3553 (70.8) | 1664 (71.8) | 2789 (80.4) | 1664 (71.1) |
| 1 | 1298 (25.9) | 570 (24.8) | 637 (18.4) | 601 (25.7) |
| 2 | 166 (3.3) | 77 (3.4) | 45 (1.3) | 74 (3.2) |
| APOE ε2 allele count | ||||
| 0 | 4262 (85.0) | 1916 (83.4) | 3034 (87.4) | 1881 (80.4) |
| 1 | 710 (14.2) | 363 (15.8) | 413 (11.9) | 431(18.4) |
| 2 | 45 (0.9) | 18 (0.8) | 24 (0.7) | 27 (1.2) |
Abbreviations: APOE, apolipoprotein E; ERICH, Ethnic/Racial Variations of Intracerebral Hemorrhage; GERFHS, Genetic and Environmental Risk Factors for Hemorrhagic Stroke; GOCHA, Genetics of Cerebral Hemorrhage with Anticoagulation; ICH, intracerebral hemorrhage; IQR, interquartile range; ISGC, International Stroke Genetics Consortium; NA, not applicable.
Lobar ICH
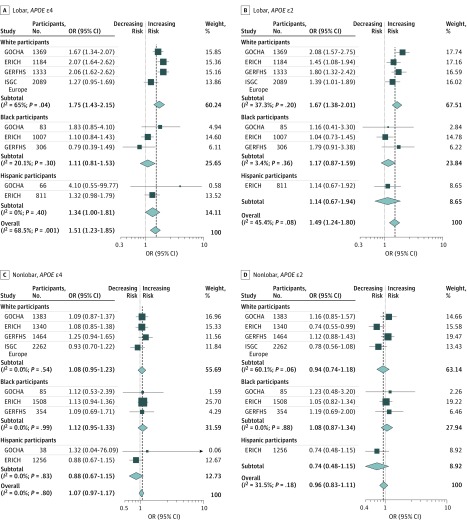
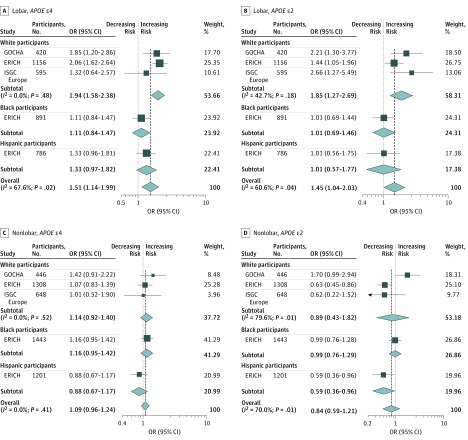
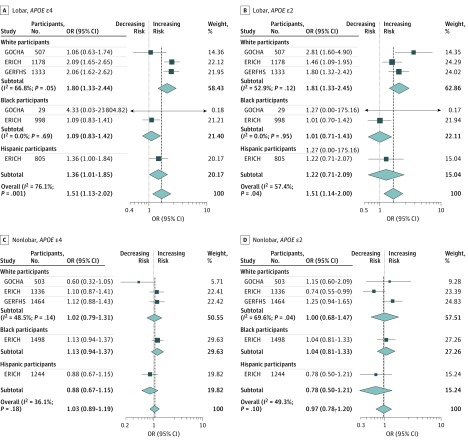
We analyzed 2305 lobar ICH cases from all studies. Model 1 confirmed the previously reported association of APOE ε2 (pooled OR, 1.49; 95% CI, 1.24-1.80; P < .001) and APOE ε4 (pooled OR, 1.51; 95% CI, 1.23-1.85; P < .001) with ICH risk; however, within self-identified Hispanic and black groups, no associations were found (Figure 1). Model 2 was used to interrogate the independent association of APOE alleles with ICH, controlled for established ICH factors (Figure 2). Here, APOE ε2 and ε4 alleles retained an association with lobar ICH. As with model 1, this association was observed in the white group, but not in black or Hispanic groups (for APOE ε2 OR, 1.45 [95% CI, 1.04-2.03; P = .03]; for APOE ε4 OR, 1.51 [95% CI, 1.14-1.99; P = .004]). Model 3 considered population stratification (Figure 3). In the white group, both APOE ε2 (OR, 1.81; 95% CI, 1.33-2.45; P < .001) and APOE ε4 (OR, 1.80; 95% CI, 1.33-2.44; P < .001) conferred higher risk for lobar ICH. For APOE ε4 alone, we found a similar association in the Hispanic group, suggesting that population stratification may have played some role in the lack of ε4 association found in models 1 and 2, particularly for the large and ethnically diverse Hispanic population recruited through the ERICH study. In contrast, for the black population, neither APOE ε2 nor APOE ε4 conferred a statistically significant risk for lobar ICH after controlling for population structure.
Figure 1. Forest Plots of Meta-analysis of Apolipoprotein E (APOE) in Lobar and Nonlobar Intracerebral Hemorrhage Cases and Controls in Model 1, Stratified Across Participating Studies and Race/Ethnicity.
ERICH indicates Ethnic/Racial Variations of Intracerebral Hemorrhage; GERFHS, Genetic and Environmental Risk Factors for Hemorrhagic Stroke; GOCHA, Genetics of Cerebral Hemorrhage with Anticoagulation; ISGC, International Stroke Genetics Consortium; OR, odds ratio. Weights are from random-effects analysis.
Figure 2. Forest Plots of Meta-analysis of Apolipoprotein E (APOE) in Lobar and Nonlobar Intracerebral Hemorrhage Cases and Controls in Model 2, Stratified Across Participating Studies and Race/Ethnicity.
ERICH indicates Ethnic/Racial Variations of Intracerebral Hemorrhage; GERFHS, Genetic and Environmental Risk Factors for Hemorrhagic Stroke; GOCHA, Genetics of Cerebral Hemorrhage with Anticoagulation; ISGC, International Stroke Genetics Consortium; OR, odds ratio. Weights are from random-effects analysis.
Figure 3. Forest Plots of Meta-analysis of Apolipoprotein E (APOE) in Lobar and Nonlobar Intracerebral Hemorrhage Cases and Controls in Model 3, Stratified Across Participating Studies and Race/Ethnicity.
ERICH indicates Ethnic/Racial Variations of Intracerebral Hemorrhage; GERFHS, Genetic and Environmental Risk Factors for Hemorrhagic Stroke; GOCHA, Genetics of Cerebral Hemorrhage with Anticoagulation; ISGC, International Stroke Genetics Consortium; OR, odds ratio. Weights are from random-effects analysis.
Nonlobar ICH
We analyzed 3897 nonlobar ICH cases (Figure 1). In model 1, APOE ε2 and ε4 did not show an association with nonlobar ICH risk, across any of the self-identified race/ethnicity groups. When comparing nonlobar ICH cases with controls, we found that the APOE ε4 P values were all P > .10. For model 2 and model 3 in nonlobar ICH, again neither APOE ε2 nor APOE ε4 showed an association with disease risk across all the studies and racial/ethnic groups (Figure 2 and Figure 3).
Power Calculation (Lobar ICH)
Given the differences in sample sizes between white, black, and Hispanic groups, we performed post hoc power calculations to determine whether the study was powered to detect a comparable APOE association in the smaller populations of blacks and Hispanics. Given the frequency of APOE ε4 in black participants (ε4 frequency 37.7%), the sample size (assuming an unmatched case-control ratio of 1:1) would provide 99% power to detect an ε4 association similar to the lower bound of the 95% CI seen in white participants (OR, 1.43). Our analyses of APOE ε4 association in Hispanic participants were similarly powered at 90%. Furthermore, the APOE ε2 frequency in black participants (19.9%) at the reported sample sizes would provide 93.8% power to detect the lower bound of the association seen in white participants (OR, 1.38). For Hispanic participants, given the lower APOE ε2 frequency (0.8%), 80% power would be achieved at a slightly higher effect size (OR, 1.60) but still would be below that found in white participants.
Propensity Score Modeling for Hypertension
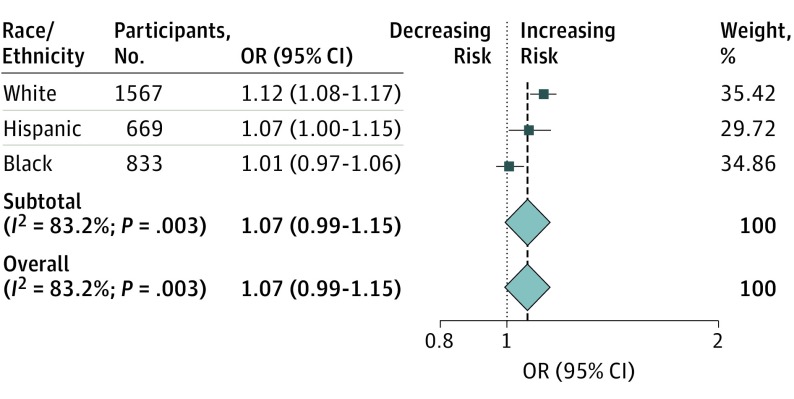
We used a PS analysis to attempt to isolate the association of APOE against the imbalanced burden of hypertension across race/ethnicity. In the first PS, we selected case and control participants with a balanced hypertension burden to form a group comprising individuals of white, black, or Hispanic ancestry. In this matched and homogeneous group, we were able to detect an association of APOE ε4 with lobar ICH risk among Hispanic participants (OR, 1.14; 95% CI, 1.03-1.28; P = .01) but not in black participants (OR, 1.02; 95% CI, 0.98-1.07; P = .25). Results were confirmed in the second PS analysis performed only in the ERICH data set, which included hypertension diagnosis as well as additional hypertension severity variables, including number of medications used to treat hypertension and systolic and diastolic blood pressure readings (Figure 4).
Figure 4. Risk of Apolipoprotein E (APOE) ε4 Allele for Lobar Intracerebral Hemorrhage Across Race/Ethnicity After Propensity Score Matching Based on Hypertension Burden.
Weights are from random-effects analysis.
Discussion
Although APOE associations with ICH risk have been characterized in multiple previous studies and meta-analyses for populations with European, and more recently Asian, ancestries, there have been fewer opportunities for examination of US minority populations at disproportionate risk for ICH. Supplemented by data from the ERICH study, we are now able to confirm variability in associations between APOE ε genotypes and lobar ICH risk across white, black, and Hispanic groups and to explore the degree to which differences in genetic risk are attributable to comorbid exposures. Our results demonstrate an association of APOE ε4 and ε2 alleles with lobar ICH led primarily by white individuals and confirmed by additional models adjusting for known covariates.29 When the association of hypertension is propensity matched across race/ethnicity, APOE ε4 emerges as a risk factor for lobar ICH among self-identified Hispanic individuals.
These results highlight the challenges of generalizing genetic risk factors across ancestries, where nongenetic exposures are known to vary by race/ethnicity. In Alzheimer disease, the relative risks for Hispanic or black individuals associated with an APOE ε 4 allele become progressively weaker or disappear entirely in comparison with white individuals.42,43,44,45,46 In ICH, APOE ε alleles have already been shown to exert higher risks in East Asian individuals when compared with those of European ancestry.47 Although recent analyses by Sawyer et al48 demonstrate the association of hypertension and APOE ε allele status with ICH risk across race/ethnicity specific to the ERICH study, the present analysis has the advantage of a larger sample size via formal transethnic meta-analysis as well as a PS matching approach that helps to illustrate the potential mechanisms underlying the observed variability of APOE ε alleles on lobar ICH risk across populations.
The observed differences in association between APOE ε alleles and lobar ICH risk do not provide direct evidence that the biological risks of the APOE gene or associations with underlying cerebral amyloid angiopathy (CAA, a major cause of lobar ICH) are necessarily different across racial/ethnic boundaries. It would seem more likely that genetic and/or environmental risk exposures covarying with race/ethnicity exert a role in modifying or mitigating the underlying APOE genetic risks. The PS analysis supports this conjecture, demonstrating that hypertension, the most important known risk factor for ICH, may simply obscure the APOE risks that may be common across ancestries. Aside from variation in environmental risk exposures, variants in a modifier gene (or genes) that differ across populations may alter the biological risk of APOE and consequently vary ICH risk, as has been hypothesized for Alzheimer disease.49,50 Furthermore, genetic variants that are racially stratified and not associated with APOE may directly modify the risk of ICH. This hypothesis represents an alternative explanation for why the PS matching for hypertension only partially remediated the association of APOE ε4 with ICH risk in Hispanic participants and had little to no association in black participants. APOE interaction studies and transethnic genome-wide association studies for ICH will likely provide insights on these hypotheses. Similarly, analyses of CAA in nonwhite populations will also clarify the association of race/ethnicity with this pathologic pathway.
In this study, we have not attempted to stratify subsets of participants by probable CAA status using magnetic resonance imaging data as has been done in previous meta-analyses. Most studies linking lobar hemorrhage locations to the pathologic diagnosis of CAA were performed in largely white populations.51 As such, widely accepted criteria for classifying probable and possible CAA using hemorrhage location and microbleed counts have not been validated in nonwhite populations.52 Validating CAA burdens across multiethnic populations will require concomitant neuroimaging and/or tissue pathologic data in genotyped individuals of many races/ethnicities to ensure patients are not misassigned.
Previously demonstrated associations between APOE ε4 and nonlobar ICH risk, also seen in nonlobar ICH recurrence,17,53 were not replicated in this study. Potential explanations include a higher rate of participants affected by hypertension and an overall younger age of participants in this study. These factors may reflect the associations of environmental or non-APOE genetic exposures in younger populations with nonlobar ICH in particular. Demographic heterogeneity is also higher in this study, and the reduced availability of covariates such as steady state lipid levels53 for risk modeling may have affected this finding. In addition, a previous meta-analysis of APOE risks in ICH also did not show the association between APOE ε4 and nonlobar ICH in black individuals, a finding supported by the present analyses.17 Future studies in larger data sets with well-phenotyped cases are needed to further elucidate the potential role of APOE ε4 in nonlobar ICH.
Strengths and Limitations
The targeted enrollment of Hispanic and black individuals through the ERICH study, lacking in previous reports,17,54 is an important strength of the present study. The high number of nonwhite individuals enrolled permits well-powered analyses in these populations and promotes confidence that the lack of observed associations is not a false acceptance of the null hypothesis, as supported by our post hoc power calculations.
Some limitations of the study should be acknowledged. Diagnoses of comorbidities were based on self-identified attestation and therefore affected by patient or caregiver awareness. This concern is present in both cases and controls, however, and internal consistency between diagnoses and prescribed medications helps to limit this potential source of bias. Furthermore, the propensity score was based on variables that only partially captured the complex phenotype represented by hypertension. However, this lack of information content is likely to bias our score results toward the null; we expect that a more precise index of hypertension burden would have increased our ability to normalize this phenotype across race/ethnicity and to demonstrate even more homogeneous APOE ε4 risks. Finally, genomewide genotypes for the ERICH study participants are not currently available, preventing us from determining whether additional genetic exposures modify the association of APOE with ICH risk across race/ethnicity.
Conclusions
In this meta-analysis, APOE ε2 and ε4 alleles remain genetic risk factors for lobar ICH, but these results are largely driven by the associations in white individuals. However, the results support a biological risk of APOE ε4 alleles that seems to transcend ancestral backgrounds,55 albeit with varying effect because of the presence of racial/ethnic disparities across associated risk factors. As availability of genetic data on US minority populations continues to increase, it is hoped that improved modeling of covarying genetic and nongenetic exposures in these populations will provide new insights into treatment and prevention strategies in ICH that maximize the potential advantages for all.
eMethods.
eTable 1. ICH Case Inclusion and Exclusion Criteria by Recruitment Site
eTable 2. Ancestry Informative Markers Selected
eTable 3. Comparison Between With Subjects With and Without Ancestry Informative Markers Available for Population Stratification Analysis
eTable 4. Frequencies of Additional Diseases and Risk Factors Among Cases and Controls of the Available Data
eFigure. Distribution of Subjects in Principal Component (PA) Space and the Relation with Self-identified Race and Ethnicity (Ellipses Cluster 95% of PC Analysis Data)
References
- 1.An SJ, Kim TJ, Yoon B-W. Epidemiology, Risk Factors, and Clinical Features of Intracerebral Hemorrhage: An Update. J Stroke. 2017;19(1):3-10. doi: 10.5853/jos.2016.00864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krishnamurthi RV, Moran AE, Forouzanfar MH, et al. ; Global Burden of Diseases, Injuries, and Risk Factors 2010 Study Stroke Expert Group . The global burden of hemorrhagic stroke: a summary of findings from the GBD 2010 study. Glob Heart. 2014;9(1):101-106. doi: 10.1016/j.gheart.2014.01.003 [DOI] [PubMed] [Google Scholar]
- 3.Qureshi AI, Giles WH, Croft JB. Racial differences in the incidence of intracerebral hemorrhage: effects of blood pressure and education. Neurology. 1999;52(8):1617-1621. doi: 10.1212/WNL.52.8.1617 [DOI] [PubMed] [Google Scholar]
- 4.Broderick JP, Brott T, Tomsick T, Huster G, Miller R. The risk of subarachnoid and intracerebral hemorrhages in blacks as compared with whites. N Engl J Med. 1992;326(11):733-736. doi: 10.1056/NEJM199203123261103 [DOI] [PubMed] [Google Scholar]
- 5.Labovitz DL, Halim A, Boden-Albala B, Hauser WA, Sacco RL. The incidence of deep and lobar intracerebral hemorrhage in whites, blacks, and Hispanics. Neurology. 2005;65(4):518-522. doi: 10.1212/01.wnl.0000172915.71933.00 [DOI] [PubMed] [Google Scholar]
- 6.van Asch CJJ, Luitse MJA, Rinkel GJ, van der Tweel I, Algra A, Klijn CJM. Incidence, case fatality, and functional outcome of intracerebral haemorrhage over time, according to age, sex, and ethnic origin: a systematic review and meta-analysis. Lancet Neurol. 2010;9(2):167-176. doi: 10.1016/S1474-4422(09)70340-0 [DOI] [PubMed] [Google Scholar]
- 7.Giles T, Aranda JM Jr, Suh DC, et al. . Ethnic/racial variations in blood pressure awareness, treatment, and control. J Clin Hypertens (Greenwich). 2007;9(5):345-354. doi: 10.1111/j.1524-6175.2007.06432.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walsh KB, Woo D, Sekar P, et al. . Untreated Hypertension: A Powerful Risk Factor for Lobar and Nonlobar Intracerebral Hemorrhage in Whites, Blacks, and Hispanics. Circulation. 2016;134(19):1444-1452. doi: 10.1161/CIRCULATIONAHA.116.024073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sturgeon JD, Folsom AR, Longstreth WT Jr, Shahar E, Rosamond WD, Cushman M. Risk factors for intracerebral hemorrhage in a pooled prospective study. Stroke. 2007;38(10):2718-2725. doi: 10.1161/STROKEAHA.107.487090 [DOI] [PubMed] [Google Scholar]
- 10.Carpenter AM, Singh IP, Gandhi CD, Prestigiacomo CJ. Genetic risk factors for spontaneous intracerebral haemorrhage. Nat Rev Neurol. 2016;12(1):40-49. doi: 10.1038/nrneurol.2015.226 [DOI] [PubMed] [Google Scholar]
- 11.Reitz C, Mayeux R. Genetics of Alzheimer’s disease in Caribbean Hispanic and African American populations. Biol Psychiatry. 2014;75(7):534-541. doi: 10.1016/j.biopsych.2013.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murrell JR, Price B, Lane KA, et al. . Association of apolipoprotein E genotype and Alzheimer disease in African Americans. Arch Neurol. 2006;63(3):431-434. doi: 10.1001/archneur.63.3.431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Michaelson DM. APOE ε4: the most prevalent yet understudied risk factor for Alzheimer’s disease. Alzheimers Dement. 2014;10(6):861-868. doi: 10.1016/j.jalz.2014.06.015 [DOI] [PubMed] [Google Scholar]
- 14.Taylor MR, Sun AY, Davis G, Fiuzat M, Liggett SB, Bristow MR. Race, common genetic variation, and therapeutic response disparities in heart failure. JACC Heart Fail. 2014;2(6):561-572. doi: 10.1016/j.jchf.2014.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Popejoy AB, Fullerton SM. Genomics is failing on diversity. Nature. 2016;538(7624):161-164. doi: 10.1038/538161a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anderson CD, Nalls MA, Biffi A, et al. . The effect of survival bias on case-control genetic association studies of highly lethal diseases. Circ Cardiovasc Genet. 2011;4(2):188-196. doi: 10.1161/CIRCGENETICS.110.957928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Biffi A, Sonni A, Anderson CD, et al. ; International Stroke Genetics Consortium . Variants at APOE influence risk of deep and lobar intracerebral hemorrhage. Ann Neurol. 2010;68(6):934-943. doi: 10.1002/ana.22134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woo D, Sauerbeck LR, Kissela BM, et al. . Genetic and environmental risk factors for intracerebral hemorrhage: preliminary results of a population-based study. Stroke. 2002;33(5):1190-1195. doi: 10.1161/01.STR.0000014774.88027.22 [DOI] [PubMed] [Google Scholar]
- 19.Woo D, Rosand J, Kidwell C, et al. . The ethnic/racial variations of intracerebral hemorrhage (ERICH) study protocol. Stroke. 2013;44(10):e120-e125. doi: 10.1161/STROKEAHA.113.002332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gomis M, Ois A, Rodríguez-Campello A, et al. . Outcome of intracerebral haemorrhage patients pre-treated with statins. Eur J Neurol. 2010;17(3):443-448. doi: 10.1111/j.1468-1331.2009.02838.x [DOI] [PubMed] [Google Scholar]
- 21.Domingues-Montanari S, Hernandez-Guillamon M, Fernandez-Cadenas I, et al. ; Stroke Project Cerebrovascular Diseases Study Group, Spanish Society of Neurology . ACE variants and risk of intracerebral hemorrhage recurrence in amyloid angiopathy. Neurobiol Aging. 2011;32(3):551.e13-551.e22. doi: 10.1016/j.neurobiolaging.2010.01.019 [DOI] [PubMed] [Google Scholar]
- 22.Pera J, Slowik A, Dziedzic T, Pulyk R, Wloch D, Szczudlik A. Glutathione peroxidase 1 C593T polymorphism is associated with lobar intracerebral hemorrhage. Cerebrovasc Dis. 2008;25(5):445-449. doi: 10.1159/000126918 [DOI] [PubMed] [Google Scholar]
- 23.Hallström B, Jönsson AC, Nerbrand C, Norrving B, Lindgren A. Stroke incidence and survival in the beginning of the 21st century in southern Sweden: comparisons with the late 20th century and projections into the future. Stroke. 2008;39(1):10-15. doi: 10.1161/STROKEAHA.107.491779 [DOI] [PubMed] [Google Scholar]
- 24.Deary IJ, Gow AJ, Pattie A, Starr JM. Cohort profile: the lothian birth cohorts of 1921 and 1936. Int J Epidemiol. 2012;41(6):1576-1584. doi: 10.1093/ije/dyr197 [DOI] [PubMed] [Google Scholar]
- 25.Pezzini A, Grassi M, Iacoviello L, et al. ; Multicenter Study on Cerebral Haemorrhage in Italy (MUCH-Italy) Investigators . Serum cholesterol levels, HMG-CoA reductase inhibitors and the risk of intracerebral haemorrhage. J Neurol Neurosurg Psychiatry. 2016;87(9):924-929. doi: 10.1136/jnnp-2015-312736 [DOI] [PubMed] [Google Scholar]
- 26.Woo D, Falcone GJ, Devan WJ, et al. ; International Stroke Genetics Consortium . Meta-analysis of genome-wide association studies identifies 1q22 as a susceptibility locus for intracerebral hemorrhage. Am J Hum Genet. 2014;94(4):511-521. doi: 10.1016/j.ajhg.2014.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anderson CD, Falcone GJ, Phuah CL, et al. ; Global Lipids Genetics Consortium and International Stroke Genetics Consortium . Genetic variants in CETP increase risk of intracerebral hemorrhage. Ann Neurol. 2016;80(5):730-740. doi: 10.1002/ana.24780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marini S, Morotti A, Ayres AM, et al. . Sex differences in intracerebral hemorrhage expansion and mortality. J Neurol Sci. 2017;379:112-116. doi: 10.1016/j.jns.2017.05.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woo D, Kissela BM, Khoury JC, et al. . Hypercholesterolemia, HMG-CoA reductase inhibitors, and risk of intracerebral hemorrhage: a case-control study. Stroke. 2004;35(6):1360-1364. doi: 10.1161/01.STR.0000127786.16612.A4 [DOI] [PubMed] [Google Scholar]
- 30.Koch W, Ehrenhaft A, Griesser K, et al. . TaqMan systems for genotyping of disease-related polymorphisms present in the gene encoding apolipoprotein E. Clin Chem Lab Med. 2002;40(11):1123-1131. doi: 10.1515/cclm.2002.197 [DOI] [PubMed] [Google Scholar]
- 31.Anderson CA, Pettersson FH, Clarke GM, Cardon LR, Morris AP, Zondervan KT. Data quality control in genetic case-control association studies. Nat Protoc. 2010;5(9):1564-1573. doi: 10.1038/nprot.2010.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Purcell S, Neale B, Todd-Brown K, et al. . PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559-575. doi: 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Biffi A, Anderson CD, Desikan RS, et al. ; Alzheimer’s Disease Neuroimaging Initiative (ADNI) . Genetic variation and neuroimaging measures in Alzheimer disease. Arch Neurol. 2010;67(6):677-685. doi: 10.1001/archneurol.2010.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904-909. doi: 10.1038/ng1847 [DOI] [PubMed] [Google Scholar]
- 35.Marini S, Lena UK, Crawford KM, et al. . Comparison of Genetic and Self-Identified Ancestry in Modeling Intracerebral Hemorrhage Risk. Front Neurol. 2018;9:514. https://www.frontiersin.org/article/10.3389/fneur.2018.00514. doi: 10.3389/fneur.2018.00514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Devan WJ, Falcone GJ, Anderson CD, et al. ; International Stroke Genetics Consortium . Heritability estimates identify a substantial genetic contribution to risk and outcome of intracerebral hemorrhage. Stroke. 2013;44(6):1578-1583. doi: 10.1161/STROKEAHA.111.000089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hong J, Lunetta KL, Cupples LA, Dupuis J, Liu CT. Evaluation of a Two-Stage Approach in Trans-Ethnic Meta-Analysis in Genome-Wide Association Studies. Genet Epidemiol. 2016;40(4):284-292. doi: 10.1002/gepi.21963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.DerSimonian R, Laird N. Meta-analysis in clinical trials revisited. Contemp Clin Trials. 2015;45(Pt A):139-145. doi: 10.1016/j.cct.2015.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Austin PC. An introduction to propensity score methods for reducing the effects of confounding in observational studies. Multivariate Behav Res. 2011;46(3):399-424. doi: 10.1080/00273171.2011.568786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ho D, Imai K, King G, Stuart E. Matching as nonparametric preprocessing for reducing model dependence in parametric causal inference. Polit Anal. 2007;15(3):199-236. [Google Scholar]
- 41.Evans DM, Purcell S. Power calculations in genetic studies. Cold Spring Harb Protoc. 2012;2012(6):664-674. doi: 10.1101/pdb.top069559 [DOI] [PubMed] [Google Scholar]
- 42.Tang MX, Stern Y, Marder K, et al. . The APOE-epsilon4 allele and the risk of Alzheimer disease among African Americans, whites, and Hispanics. JAMA. 1998;279(10):751-755. doi: 10.1001/jama.279.10.751 [DOI] [PubMed] [Google Scholar]
- 43.O’Bryant SE, Johnson L, Reisch J, et al. . Risk factors for mild cognitive impairment among Mexican Americans. Alzheimers Dement. 2013;9(6):622-631.e1. doi: 10.1016/j.jalz.2012.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Farrer LA, Cupples LA, Haines JL, et al. ; APOE and Alzheimer Disease Meta Analysis Consortium . Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. JAMA. 1997;278(16):1349-1356. doi: 10.1001/jama.1997.03550160069041 [DOI] [PubMed] [Google Scholar]
- 45.Evans DA, Bennett DA, Wilson RS, et al. . Incidence of Alzheimer disease in a biracial urban community: relation to apolipoprotein E allele status. Arch Neurol. 2003;60(2):185-189. doi: 10.1001/archneur.60.2.185 [DOI] [PubMed] [Google Scholar]
- 46.Graff-Radford NR, Green RC, Go RCP, et al. . Association between apolipoprotein E genotype and Alzheimer disease in African American subjects. Arch Neurol. 2002;59(4):594-600. doi: 10.1001/archneur.59.4.594 [DOI] [PubMed] [Google Scholar]
- 47.Tzourio C, Arima H, Harrap S, et al. . APOE genotype, ethnicity, and the risk of cerebral hemorrhage. Neurology. 2008;70(16):1322-1328. doi: 10.1212/01.wnl.0000308819.43401.87 [DOI] [PubMed] [Google Scholar]
- 48.Sawyer RP, Sekar P, Osborne J, et al. . Racial/ethnic variation of APOE alleles for lobar intracerebral hemorrhage. Neurology. 2018;91(5):e410-e420. doi: 10.1212/WNL.0000000000005908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee JH, Cheng R, Vardarajan B, et al. . Genetic modifiers of age at onset in carriers of the G206A mutation in PSEN1 with familial Alzheimer disease among Caribbean hispanics. JAMA Neurol. 2015;72(9):1043-1051. doi: 10.1001/jamaneurol.2015.1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maestre G, Ottman R, Stern Y, et al. . Apolipoprotein E and Alzheimer’s disease: ethnic variation in genotypic risks. Ann Neurol. 1995;37(2):254-259. doi: 10.1002/ana.410370217 [DOI] [PubMed] [Google Scholar]
- 51.Charidimou A, Martinez-Ramirez S, Reijmer YD, et al. . Total magnetic resonance imaging burden of small vessel disease in cerebral amyloid angiopathy: an imaging-pathologic study of concept validation. JAMA Neurol. 2016;73(8):994-1001. doi: 10.1001/jamaneurol.2016.0832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith EE, Greenberg SM. Clinical diagnosis of cerebral amyloid angiopathy: validation of the Boston criteria. Curr Atheroscler Rep. 2003;5(4):260-266. doi: 10.1007/s11883-003-0048-4 [DOI] [PubMed] [Google Scholar]
- 53.Raffeld MR, Biffi A, Battey TW, et al. . APOE ε4 and lipid levels affect risk of recurrent nonlobar intracerebral hemorrhage. Neurology. 2015;85(4):349-356. doi: 10.1212/WNL.0000000000001790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Woo D, Kaushal R, Chakraborty R, et al. . Association of apolipoprotein E4 and haplotypes of the apolipoprotein E gene with lobar intracerebral hemorrhage. Stroke. 2005;36(9):1874-1879. doi: 10.1161/01.STR.0000177891.15082.b9 [DOI] [PubMed] [Google Scholar]
- 55.Tai LM, Thomas R, Marottoli FM, et al. . The role of APOE in cerebrovascular dysfunction. Acta Neuropathol. 2016;131(5):709-723. doi: 10.1007/s00401-016-1547-z [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eMethods.
eTable 1. ICH Case Inclusion and Exclusion Criteria by Recruitment Site
eTable 2. Ancestry Informative Markers Selected
eTable 3. Comparison Between With Subjects With and Without Ancestry Informative Markers Available for Population Stratification Analysis
eTable 4. Frequencies of Additional Diseases and Risk Factors Among Cases and Controls of the Available Data
eFigure. Distribution of Subjects in Principal Component (PA) Space and the Relation with Self-identified Race and Ethnicity (Ellipses Cluster 95% of PC Analysis Data)