Abstract
Background
The deep-seated infections caused by the Candida genus are associated with a high mortality rate, and Candida albicans is the most frequent species associated with these diseases. The fungal wall is composed of macromolecules not synthesized by the host, and therefore is a source of ligands recognized by innate immune cells.
Methods
We performed a comparative study analyzing the cell wall composition and organization of Candida tropicalis, Candida guilliermondii, Candida krusei, and Candida auris, along with their ability to stimulate cytokine production and phagocytosis by human innate immune cells.
Results
We found that the wall of these species had the basic components already described in C. albicans, with most of the chitin and b1,3-glucan located underneath the mannan layer. However, the walls of C. krusei and C. auris were rich in chitin and the former had a lower content of mannans. C. guilliermondii contained changes in the mannan and the b1,3-glucan levels. These species were differentially phagocytosed by human macrophages and stimulated cytokine production in a dectin-1-dependent pathway. C. krusei showed the most significant changes in the tested parameters, whereas C. auris behaved like C. albicans.
Conclusion
Our results suggest that the cell wall and innate immune recognition of C. tropicalis, C. guilliermondii, C. krusei, and Candida auris is different from that reported for C. albicans.
Keywords: cell wall, protein glycosylation, host–fungus interplay, phagocytosis, cytokine production
Introduction
The species of the Candida genus are responsible for superficial and systemic can-didiasis, and the latter represents a significant burden to health care systems and patients because of the high rate of morbidity and mortality associated with these infections, especially in immunocompromised patients.1,2 Although Candida albicans is the most frequently isolated species from patients with diagnosed candidiasis, other Candida species are responsible for about 35%–65% of candidemia cases.3,4 Among them, Candida tropicalis, Candida parapsilosis, Candida guilliermondii, Candida glabrata, and Candida krusei top the epidemiological lists of causative agents of systemic candidiasis.5,6
C. tropicalis is considered as an important causative agent of invasive candidiasis and colonizes 60%–80% of immunocompromised patients.6 This species has been associated with similar or higher mortality rates than those reported for C. albicans.7–9 C. krusei and C. guilliermondii are also part of the emergent group of Candida species related to systemic candidiasis, being the causative agents in 2%–5% of the reported cases.6,7 A significant difference between infections associated with C. guilliermondii and C. krusei is the mortality rate: in C. guilliermondii this is similar to that recorded for C. albicans (27%–49%),1,10 while in C. krusei the mortality rate is higher (20%–67%), likely related to their poor response to standard antifungal therapies.6,8,9,11 Candida auris is an emergent fungal species, firstly identified in 2009 as an etiological agent of candidiasis, and has a natural resistance to several of the antifungal drugs used to treat these infections.12 Despite not being related to high numbers of cases of nosocomial candidiasis, it is distributed worldwide, and most of the clinical isolates have shown high minimal inhibitory concentrations for azoles, polyenes, and echinocandins.12
The fungal immune sensing is a key step in the establishment of a protective anti-fungal immune response, and among the first events that trigger this interaction is the recognition of the fungal cell wall.13,14 The wall is an essential cell component that provides protection from extracellular insults, controls communication with the extracellular environment, and is the molecular scaffold that displays virulence factors in fungal pathogens.15 As it is composed of polysaccharides which are absent in the human cells, the cell wall is the main source of pathogen-associated molecular patterns that the immune system recognizes through pattern recognition receptors (PRRs), mainly localized at the cell surface of immune cells.13,15
The C. albicans cell wall has been thoroughly studied, and nowadays, there is enough information to propose general models regarding its composition, organization, and relevance for viability, virulence, and immune sensing.13,15–17 Despite this significant advance, little is known about the cell wall organization and composition of other species of the Candida genus, such as C. guilliermondii, C. tropicalis, C. krusei, and C. auris. A possible reason is that it is assumed that the C. albicans cell wall model should apply to all the members of this genus. In C. albicans, the cell wall is composed of two well-defined layers,13,16 visible under electronic microscopy inspection: an inner layer composed of chitin, β1,3- and β1,6-glucans, and an outer layer rich in proteins modified with N- or O-linked mannans, named mannoproteins.18 These components are not synthesized by the host cells, which in turn are recognized by a variety of PRRs that will ultimately contribute to the establishment of a protective antifungal response by the host immunity.13 The b1,3-glucans are sensed by a heterodimer composed of toll-like receptor (TLR) 2 and TLR6 and by dectin-1, the latter being a specific receptor for this polysaccharide;13,19 while TLR4 participates in the recognition of O-linked mannans20 and mannose receptor,20 dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin,21 dectin-2,22 mincle,23 and dectin-324 recognize the N-linked mannans. The chitin–receptor interaction can block the fungal sensing,25 or promote an anti-inflammatory state in a pathway dependent on the mannose receptor, nucleotide-binding oligomerization domain-containing protein 2, and TLR9.26
So far, little is known about the immune sensing of C. tropicalis, C. krusei, C. guilliermondii, and C. auris. It has been shown that neutrophils and murine phagocytic cells are capable of discriminating among Candida species, the former displaying reduced uptake against C. krusei,27 and the latter an increased ability to kill C. guilliermondii and C. krusei.28 In addition, C. tropicalis is more susceptible to damage by neutrophils than C. albicans.29 Human monocytes differentially recognize Candida species, such as C. albicans, C. parapsilosis, C. tropicalis, and C. krusei, but not C. glabrata and C. guilliermondii, which are good stimuli for C3 and granulocyte-macrophage colony-stimulating factor.30 On the contrary, there are studies indicating that different Candida species have similar abilities to interact with innate immune cells, triggering similar killing rates and phagocytosis as C. albicans.31–33 Although the C. auris-immune effector interplay is an area that is poorly explored, it has been demonstrated that neutrophils are not capable of forming extracellular traps when interacting with C. auris.34
Here, in order to assess insights about the relevance of the cell wall in the immune sensing of C. tropicalis, C. krusei, C. guilliermondii, and C. auris by human peripheral blood mononuclear cells (PBMCs) and human monocyte-derived macrophages, we determined the basic composition of the cell wall of these fungal species and their ability to stimulate cytokine production and phagocytic uptake.
Materials and methods
Strains and culture media
C. albicans SC5314,35 C. guilliermondii ATCC 6260,36 C. krusei ATCC 6258, C. tropicalis ATCC MYA-3404,37 and C. auris VPCI 479/P/13,38 a genome-sequenced strain, were used in this study. Cell cultures were propagated at 30°C in Sabouraud broth (1% [w/v] mycological peptone, 4% [w/v] glucose). Unless otherwise indicated, aliquots containing 500 µL of overnight-grown cells were inoculated in 100 mL of fresh medium and further incubated at 30°C with constant shaking (200 rpm) until reaching the mid-log growth phase (typically 5–6 hours). For heat inactivation, yeast cells were incubated at 56°C for 1 hour.39 In all cases, loss of cell viability was confirmed by incubating cells in Sabouraud medium for 72 hours at 30°C. To remove O-linked mannans, cells were β-eliminated overnight with 100 mM NaOH as described.40 Following this protocol, >96% of the cells retained their viability, as tested by cell growth before and after treatment with the alkali.41
Cell wall analysis
Cell homogenates were obtained in a Braun homogenizer, as previously described,41 and walls were separated from cellular debris by centrifuging at 21,000× g for 10 minutes at 4°C. The cell walls were washed six times with deionized water; adsorbed intracellular components were removed with hot 2% (v/v) SDS, 0.3 M β-mercaptoethanol, and 1 M NaCl; freeze-dried; and hydrolyzed with hot 2 M trifluoroacetic acid for 3 hours, as described previously.39 The content of N-acetylglucosamine, glucose, and mannose in the wall hydrolysates was measured by high-performance anion-exchange chromatography coupled to pulsed amperometric detection (HPAEC-PAD) in a Dionex system (Thermo Fisher Scientific, Waltham, MA, USA) using similar separation conditions to those described earlier.41 Protein content was determined in alkali-hydrolyzed samples from the cell wall,39 using the Bradford protein assay.
Alcian blue binding assays
Yeast cells grown at the exponential phase were pelleted, washed three times with deionized water, and the cell concentration adjusted at an OD600 of 0.2 in deionized water. Aliquots of 1 mL were pelleted, and cells were suspended in 1 mL of Alcian blue (30 µg·mL−1 in 0.02 M HCl; Sigma-Aldrich Co., St Louis, MO, USA) and assayed as described.42
Cell wall porosity assay
The relative cell wall porosity to polycations was estimated as described previously.43 Cells were grown until reaching the exponential phase, washed twice with PBS, and cell concentration was adjusted at 1×108 cells mL−1. Aliquots containing 1 mL were centrifuged, the pellet was saved and resuspended in either 10 mM Tris-HCl, pH 7.4 (buffer A), buffer A plus 30 µg·mL−1 poly-L-lysine (Mw 30–70 kDa, Sigma-Aldrich Co.), or buffer A plus 30 µg·mL−1 diethylaminoethyl (DEAE)-dextran (Mw 500 kDa, Sigma-Aldrich Co.). Cells were incubated for 30 minutes at 30°C with constant shaking (200 rpm), centrifuged, and the supernatants were saved, centrifuged again, and used to measure the absorbance at 260 nm. The relative cell wall porosity to DEAE-dextran was quantified as reported elsewhere.43
Analysis of the exposure of the cell wall polysaccharide at the fungal wall surface
Chitin was labeled by incubating cells with 1 mg·mL−1 fluorescein-5-isothiocyanate conjugated lectin from Triticum vulgaris (WGA-FITC; Sigma-Aldrich Co.) for 60 minutes at room temperature, as reported previously,25 while b1,3-glucan was labeled with 5 µg·mL−1 IgG Fc-Dectin-1 chimera44 for 40 minutes at room temperature. The binding of the lectin to the fungal wall was revealed by incubating cells with 1 µg·mL−1 donkey anti-Fc IgG-FITC (Sigma-Aldrich Co.) for 40 minutes at room temperature.45 Samples were examined by fluorescence microscopy using a Zeiss Axioscope-40 microscope and an Axiocam MRc camera. From the pictures acquired, the fluorescence associated to 300 cells was collected using the software Adobe Photoshop™ CS6, Adobe Systems Incorporated (San Jose, CA, USA) with the formula: ([total of green pixels – background green pixels] × 100)/total pixels.46
Quantification of N- and O-linked mannans
Mannan trimming was performed as reported.47 The N-linked mannans were removed from the cell wall by incubating yeast cells with 25 U endoglycosidase H (New England Biolabs, Ipswich, MA, USA) for 20 hours at 37°C; while removal of O-linked mannans was performed by β-elimination with 1N NaOH and gently shaking for 18 hours at room temperature. Mannan release and quantification was performed by HPAEC-PAD, as previously described.48
Ethics statement
Healthy adult volunteers were enrolled in the study and venous blood samples were withdrawn after information about the study was disclosed and written informed consent was obtained. This procedure was conducted in accordance with the Declaration of Helsinki. The use of human cells was approved by the Ethics Committee of Universidad de Guanajuato (permission code 17082011).
Isolation of human PBMCs and cytokine stimulation
The venous blood was mixed with Histopaque-1077 (Sigma-Aldrich Co.) and density centrifugation was performed as described.49 The PBMC–Candida interactions were carried out using 5 × 105 PBMCs in 100 µL of RPMI 1640 Dutch modification (added with 2 mM glutamine, 0.1 mM pyruvate, and 0.05 mg·mL−1 gentamycin; all reagents from Sigma-Aldrich Co.) and 100 µL containing 1×105 yeast cells freshly harvested or treated. The interactions were placed in round-bottom 96-well microplates and incubated for 24 hours at 37°C with 5% (v/v) CO2. Then, the plates were centrifuged for 10 minutes at 3,000× g at 4°C before the supernatants were saved and kept at –20°C until further use. The concentrations of tumor necrosis factor alpha (TNFα), IL-6, and IL-10 were quantified by ELISA using the kit ABTS ELISA Development from Peprotech, while the IL-1β levels were measured using a DuoSet ELISA Development kit (R&D systems). In all plates, mock interactions containing only PBMCs were included as controls and produced threshold levels that were subtracted to the quantifications of all the cytokines analyzed.
When indicated, human PBMCs were preincubated at 37°C for 60 minutes with 200 µg·mL−1 laminarin (Sigma-Aldrich Co.) before adding the fungal stimuli.
Phagocytosis assays
Human monocyte-derived macrophages were obtained by incubating the PBMCs with recombinant human granulocyte-macrophage colony-stimulating factor (Sigma-Aldrich Co.) as previously reported.46 Yeast cells were washed twice with PBS, stained with 1 mg·mL−1 acridine orange (Sigma-Aldrich Co) as described,50 washed again with PBS, and the cell concentration was adjusted at 3×107 yeast cells mL−1. The macrophage–fungus interactions were performed in aliquots of 800 µL of DMEM medium (Sigma-Aldrich Co), in six-well plates with a macrophage-to-yeast ratio of 1:6. Plates were incubated for 2 hours at 37°C and 5% (v/v) CO2, and then macrophages were washed twice with cold PBS and suspended in 1.25 mg·mL−1 trypan blue as an external fluorescence quencher, as described.37,51 A MoFlo XDP (Beckman Coulter) fuorescence-activated cell sorting system was used to analyze samples by flow cytometry, collecting 50,000 events gated for macrophage cells. Signals were obtained using the FL1 (green fluorescence) and FL3 (red fluorescence) channels previously compensated with macrophage cells without any labeling. The phagocytosis of fungal cells was analyzed from counted events in the green channel (early stage of the phagocytic event) and red channel (cells within acidified phagolysosomes, ie, in the late stage of the phagocytic event).37
Statistical analysis
Statistical analysis was performed using GraphPad Prism 6 software. Stimulation of cytokine production and phagocytosis by human cells were carried out in duplicate with samples from six healthy donors, while the other experiments were performed at least three times in duplicate. Data represent cumulative results of all experiments performed and are shown as the mean and the SD. The Mann–Whitney U test was used to analyze data with a significance level set at P<0.05.
Results
Cell wall composition and organization of the Candida species under study
To quantify the basic polysaccharides and oligosaccharides of the cell wall of C. guilliermondii, C. krusei, C. tropicalis, and C. auris, the walls were extracted, depleted of cell components, acid-hydrolyzed, and analyzed by HPAEC-PAD. This methodology has been applied before in the analysis of other Candida species, including C. albicans, and allowed the quantification of chitin, glucan, and mannan, by assessment of the content of N-acetylglucosamine, glucose, and mannose, respectively.37,39,41,42,46–48,52–54 Although it has been extensively characterized previously, here we included the analysis of the C. albicans cell wall for comparison purposes. The cell walls of the species analyzed showed similar levels of chitin, glucan, and mannan (Table 1), with significant differences in some cases: C. krusei and C. auris showed higher content of chitin when compared to the other species, and they were also significantly different when compared between each other (Table 1). Mannan was significantly higher in C. guilliermon-dii cell wall, but lower in C. krusei wall, and the former also showed lower levels of glucan (Table 1). All strains tested showed similar levels of cell wall protein content, with exception of C. krusei that showed low levels of cell wall protein (Table 1). To confirm the changes in the cell wall mannan content in C. guilliermondii and C. krusei, we measured the content of O-linked and N-linked mannans decorating the cell wall glycoproteins, by trimming the oligosaccharides with b-elimination and endoglycosidase H digestion, respectively.47 Results shown in Figure 1 indicated that upon β-elimination, the O-linked mannan content in the five species analyzed was lower when compared to the N-linked mannans, and C. guil-liermondii and C. krusei were the only two strains that showed changes in the mannan content: the former had increased levels of both mannans, whereas C. krusei showed low levels of both N-linked and O-linked mannans (Figure 1). Interestingly, in C. albicans, C. tropicalis, C. krusei, and C. auris, O-linked mannans represented about 15% of the total content of cell wall mannan, but in C. guilliermondii these molecules were relatively more abundant than the N-linked mannans, representing around 33% of total mannan content.
Table 1.
Cell wall analysis of C. albicans, C. tropicalis, C. guilliermondii, C. krusei, and C. auris
| Cell wall abundance
|
Phosphomannan content (μg)a | Porosity (%)b | Protein (μg)c | |||
|---|---|---|---|---|---|---|
| Organism | Chitin (%) | Mannan (%) | Glucan (%) | |||
| C. albicans | 2.0±1.3 | 36.1±4.2 | 61.7±4.4 | 114.2±14.3 | 28.4±8.7d | 146.4±14.9 |
| C. tropicalis | 2.3±1.0 | 36.2±3.5 | 62.9±5.0 | 139.5±19.0d | 62.8±8.9 | 129.7±15.3 |
| C. guilliermondii | 2.2±1.0 | 47.8±4.0d | 50.0±5.0d | 131.2±8.9d | 72.3±12.7 | 131.6±18.9 |
| C. krusei | 8.2±1.0d | 23.9±2.0d | 67.8±2.0 | 66.1±9.0d | 57.7±10.5 | 76.4±14.8d |
| C. auris | 4.5±1.6d | 30.8±1.1 | 64.9±2.5 | 110.1±15.2 | 30.9±9.4d | 138.7±18.2 |
Notes:
µg of alcian blue bound/OD600=1.
Relative to DEAE-dextran.
µg of protein/mg of cell wall.
P<0.05, when compared with the values from the other species analyzed. Data presented as mean±SD.
Abbreviations: C. albicans, Candida albicans; C. tropicalis, Candida tropicalis; C. guilliermondii, Candida guilliermondii; C. krusei, Candida krusei; C. auris, Candida auris; DEAE, diethylaminoethyl.
Figure 1.
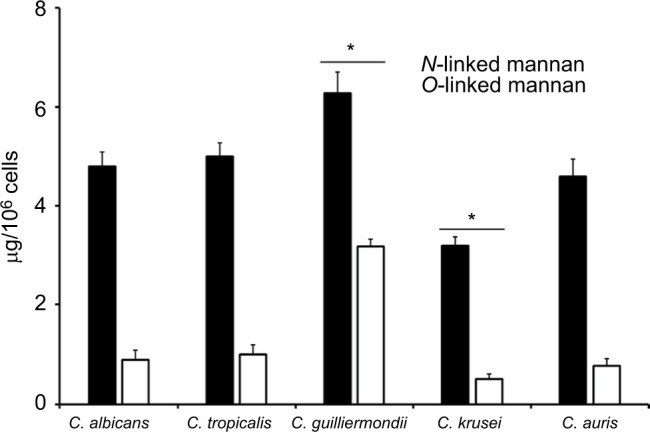
The content of N-linked and O-linked mannan in the cell wall of Candida albicans, Candida tropicalis, Candida guilliermondii, Candida krusei, and Candida auris.
Notes: Yeast cells were treated either with endoglycosidase H or b-eliminated to trim N-linked mannans or O-linked mannans, respectively. The released oligosaccharides were saved and used to measure the mannose content by HPAEC-PAD. Data are mean ± SD of three independent experiments performed in duplicates. *P<0.05, when compared with mannans from the other species analyzed.
Abbreviation: HPAEC-PAD, high-performance anion-exchange chromatography coupled to pulsed amperometric detection.
To provide additional evidence to these observations, we next assessed the content of cell wall phosphomannan and the cell wall porosity to polycations, as these parameters have been associated with the length of cell wall man-nans.39,43,46,47,52–56 The C. albicans and C. auris walls showed similar phosphomannan levels, while C. tropicalis and C. guilliermondii showed higher content of this oligosaccharide, and the C. krusei walls showed lower phosphomannan abundance (Table 1). The cell wall porosity was similar for C. tropicalis, C. guilliermondii, and C. krusei, but this parameter was significantly lower in the walls of C. albicans and C. auris (Table 1).
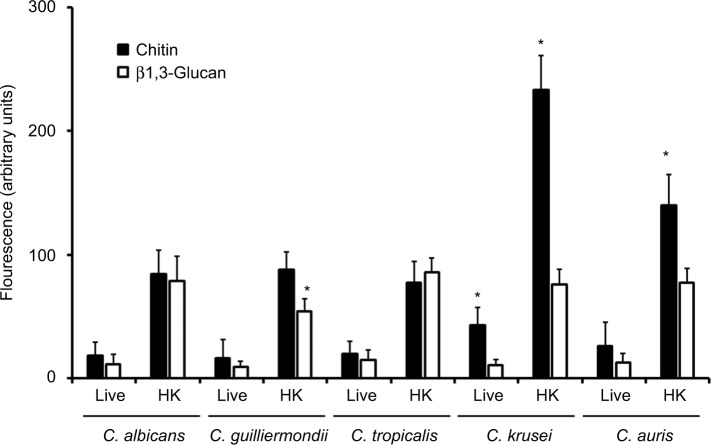
To analyze the organization of the structural polysaccharide within the cell wall, the b1,3-glucan was labeled with the IgG Fc-Dectin-1 chimera,57 and then with FITC-conjugated IgG, whereas WGA-FITC was used for chitin labeling. The five species analyzed were barely stained with both lectins (Figure 2); however, when cells were inactivated by heat, a treatment that artificially exposes chitin and b1,3-glucan on the surface of the cell wall,58 the fluorescence associated with cells from the five species was significantly increased (Figure 2). Therefore, chitin and b1,3-glucan are localized underneath other cell wall components that impair the proper lectin–polysaccharide interaction. It is noteworthy that the labeling of b1,3-glucan in heat-killed (HK) C. guilliermondii cells was significantly lower compared to cells from other species under the same treatment (Figure 2), and HK C. krusei and C. auris cells were more labeled by WGA-FITC, suggesting an increased chitin content (Figure 2). These results correlated with the cell wall composition provided in Table 1. Interestingly, cells from C. krusei showed increased chitin labeling in the live form, suggesting that more of this polysaccharide is naturally exposed on the surface of this organism than in the other species under analysis (Figure 2). Overall, the data indicates that the cell wall of C. tropicalis, C. guilliermondii, C. krusei, and C. auris differs to which has been described in C. albicans.
Figure 2.
Fluorescent labeling of chitin and b1,3-glucan in the cell wall of Candida albicans, Candida tropicalis, Candida guilliermondii, Candida krusei, and Candida auris.
Notes: Live or heat-killed (HK) yeast cells were incubated with either fluorescein isothiocyanate-wheat germ agglutinin conjugate (closed bars, labels chitin) or IgG Fc-Dectin-1 chimera (open bars, labels b1,3-glucan) as described in the “Materials and methods” section, inspected under fluorescence microscopy, and the fluorescence associated to 300 individual cells was recorded. *P<0.05, when compared with cells under the same treatment.
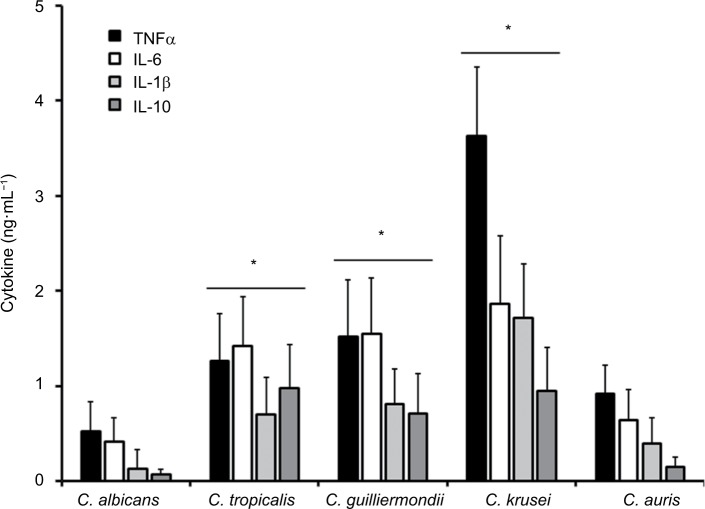
Stimulation of cytokine production by C. albicans, C. tropicalis, C. guilliermondii, C. krusei, and C. auris
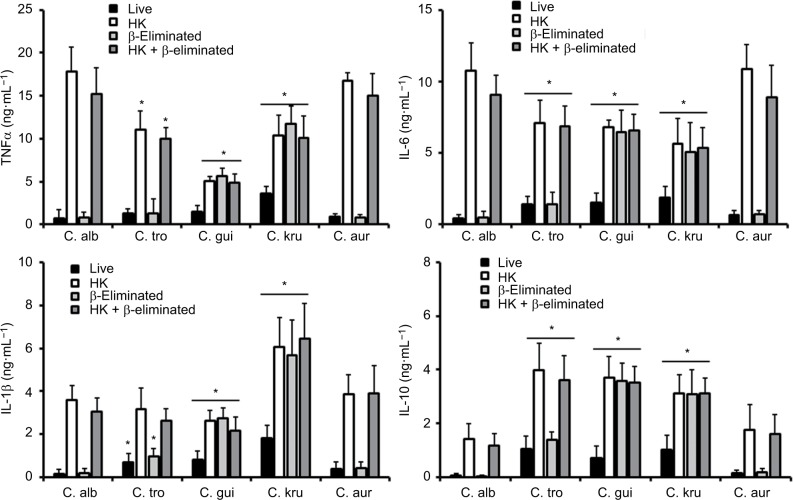
The differences in the cell wall described earlier are likely to affect the Candida–immune cell interaction when compared to that described for C. albicans. Thus, to get some insights about such interaction, yeast cells were coincubated with human PBMCs and the level of secreted cytokines was measured as a read-out of this interaction. C. albicans and C. auris cells barely stimulated the production of TNFα, IL-6, IL-1β, or IL-10 (Figure 3); however, C. tropicalis, C. guilliermondii, and C. krusei stimulated higher levels of the four cytokines analyzed (Figure 3). Among the tested strains, one that is capable of inducing a high cytokine production was C. krusei, and this strain significantly stimulated the secretion of higher levels of TNFα and IL-1b (Figure 3) in comparison with the remaining strains. To assess the role of components of the inner part of the cell wall and O-linked mannans, we compared the cytokine profile stimulated with either live or HK cells with or without b-elimination that trimmed the O-linked mannans from the cell wall.40,58 As previously reported for C. albicans,41,58 HK yeast cells stimulated the production of higher levels of TNFα, IL-6, IL-1β, and IL-10 than live cells (Figure 4). The trimming of O-linked mannan did not affect significantly the cytokine profile stimulated either by live or HK C. albicans cells (Figure 4). The C. auris cells showed similar ability to C. albicans to stimulate cytokine production, as the profiles of the four cytokines tested were similar for both species when live, HK, b-eliminated or HK + β-eliminated cells were used for the interaction with the human PBMCs (Figure 4). For C. tropicalis, C. guilliermondii, and C. krusei, the HK cells stimulated higher cytokine levels than live cells (Figure 4), but some differences were observed when their cytokine profiles were compared with those stimulated by C. albicans. These three species stimulated lower production of TNFα and IL-6 than C. albicans, including when cells were HK and b-eliminated (Figure 4). Interestingly, upon β-elimination, live cells stimulated high levels of these two cytokines, comparable with those obtained with HK cells (Figure 4). In the case of the production of IL-1β, the HK C. tropicalis cells stimulated similar levels of this cytokine to those produced upon interaction of the human cells with C. albicans, but live and b-eliminated cells stimulated higher levels of this cytokine when C. tropicalis cells were used during the coincubation period (Figure 4). For C. guilliermondii, HK and HK + β-eliminated cells stimulated less IL-1β production than C. albicans, but higher levels of this cytokine were observed when β-eliminated cells were used in the interactions (Figure 4). The HK, b-eliminated, and HK + β-eliminated cells from C. krusei stimulated similar IL-1β levels that were significantly higher than those produced by C. albicans cells (Figure 4). A similar observation could be drawn for the stimulation of IL-10 production by cells of C. guilliermondii and C. krusei, whereas only HK and HK + b-eliminated C. tropicalis cells stimulated high production of this anti-inflammatory effector (Figure 4).
Figure 3.
Stimulation of cytokine production by Candida albicans, Candida tropicalis, Candida guilliermondii, Candida krusei, and Candida auris.
Notes: Human PBMCs were coincubated for 24 hours with live yeast cells, and then the supernatant was collected and used to quantify the cytokine levels. *P<0.05, when compared with the cytokine level stimulated by C. albicans cells.
Abbreviations: PBMCs, peripheral blood mononuclear cells; TNFα, tumor necrosis factor alpha.
Figure 4.
Stimulation of cytokine production by heat-killed and b-eliminated cells from Candida albicans, Candida tropicalis, Candida guilliermondii, Candida krusei, and Candida auris.
Notes: Yeast cells were heat-killed (HK), b-eliminated, or subjected to both treatments before being coincubated with human PBMCs for 24 hours. The supernatants of interactions were collected and used to quantify the cytokine levels. * P<0.05, when compared with the cytokine level stimulated by C. albicans cells under the same treatment. Abbreviations: PBMCs, peripheral blood mononuclear cells; TNFα, tumor necrosis factor alpha; C. alb, Candida albicans; C. tro, Candida tropicalis; C. gui, Candida guilliermondii; C. kru, Candida krusei; C. aur, Candida auris.
To explore whether the significant increment in the ability to stimulate cytokine production by HK cells was dependent on the recognition of b1,3-glucan by dectin-1, the human PBMCs were preincubated with laminarin, an antagonist of dectin-1,41,46,47 and then used in the interaction with the yeast cells. Results given in Table 2 indicated that preincubation of the human cells with laminarin did not affect the ability of live C. albicans, C. tropicalis, and C. auris to stimulate cytokine production, but a significant reduction in the cytokine levels were observed when HK cells were used for the interactions. Similar data were obtained when cells were subjected to β-elimination (Table 2). For the case of live and HK cells from C. guilliermondii and C. krusei, similar results were observed, but the cytokine production stimulated by both live β-eliminated and HK + β-eliminated cells was significantly affected by preincubation of human PBMCs with laminarin (Table 2).
Table 2.
Effect of laminarin on the ability of C. albicans, C. tropicalis, C. guilliermondii, C. krusei, and C. auris to stimulate cytokine production
| TNFα (%)a | IL-6 (%)a | IL-1β (%)a | IL-10 (%)a | |
|---|---|---|---|---|
|
| ||||
| C. albicans | ||||
| Live | 100 | 100 | 100 | 100 |
| Live + laminarinb | 99.1±0.1 | 99.2±0.1 | 99.3±0.2 | 95.8±0.6 |
| HK | 100 | 100 | 100 | 100 |
| HK + laminarinb | 15.7±4.4c | 22.8±5.4c | 26.8±6.4c | 23.7±7.8c |
| β-eliminated | 100 | 100 | 100 | 100 |
| β-eliminated + laminarinb | 98.4±1.1 | 97.8±0.9 | 96.8±2.8 | 98.7±0.8 |
| HK + β-eliminated | 100 | 100 | 100 | 100 |
| HK + β-eliminated + laminarinb | 17.2±10.4c | 21.8±6.0c | 22.1±3.2c | 29.3±9.4c |
| C. tropicalis | ||||
| Live | 100 | 100 | 100 | 100 |
| Live + laminarinb | 98.1±0.3 | 97.2±0.1 | 98.8±0.3 | 97.4±0.2 |
| HK | 100 | 100 | 100 | 100 |
| HK + laminarinb | 26.7±6.1c | 28.4±8.1c | 21.8±9.1c | 29.4±10.1c |
| β-eliminated | 100 | 100 | 100 | 100 |
| β-eliminated + laminarinb | 94.2±0.9 | 95.0±2.4 | 98.1±0.7 | 94.6±2.1 |
| HK + β-eliminated | 100 | 100 | 100 | 100 |
| HK + β-eliminated+ laminarinb | 27.9±7.1c | 29.4±4.1c | 30.4±10.5c | 31.7±8.5c |
| C. guilliermondii | ||||
| Live | 100 | 100 | 100 | 100 |
| Live + laminarinb | 99.8±0.1 | 93.0±4.4 | 98.4±0.2 | 90.8±4.4 |
| HK | 100 | 100 | 100 | 100 |
| HK + laminarinb | 34.8±9.4c | 17.5±9.4c | 19.5±4.5c | 20.6±6.1c |
| β-eliminated | 100 | 100 | 100 | 100 |
| β-eliminated + laminarinb | 35.6±7.4c | 27.3±5.6c | 18.5±3.7c | 22.4±8.8c |
| HK + β-eliminated | 100 | 100 | 100 | 100 |
| HK + β-eliminated + laminarinb | 27.2±5.4c | 25.4±8.4c | 20.2±6.1c | 21.9±3.7c |
| C. krusei | ||||
| Live | 100 | 100 | 100 | 100 |
| Live + laminarinb | 93.3±2.4 | 90.4±9.4 | 9.7±1.8 | 93.1±5.2 |
| HK | 100 | 100 | 100 | 100 |
| HK + laminarinb | 23.2±6.1c | 25.8±5.1c | 19.5±4.5c | 26.1±4.8c |
| β-eliminated | 100 | 100 | 100 | 100 |
| β-eliminated + laminarinb | 21.8±5.5c | 16.4±2.4c | 27.4±7.6c | 23.1±6.0c |
| HK + β-eliminated | 100 | 100 | 100 | 100 |
| HK + β-eliminated + laminarinb | 22.2±11.4c | 22.0±5.7c | 30.4±10.5c | 32.5±8.3c |
| C. auris | ||||
| Live | 100 | 100 | 100 | 100 |
| Live + laminarinb | 97.0±0.5 | 97.4±0.4 | 96.9±0.6 | 99.1±0.1 |
| HK | 100 | 100 | 100 | 100 |
| HK + laminarinb | 20.4±8.4c | 27.0±8.1c | 24.1±9.4c | 28.8±7.9c |
| β-eliminated | 100 | 100 | 100 | 100 |
| β-eliminated + laminarinb | 92.8±6.5 | 95.1±1.7 | 92.4±8.0 | 97.2±3.7 |
| HK + β-eliminated | 100 | 100 | 100 | 100 |
| HK + β-eliminated + laminarinb | 21.4±7.5c | 28.0±9.4c | 27.2±6.6c | 21.0±5.6c |
Notes:
The 100% value corresponds to the cytokine concentration produced when laminarin was not included in the stimulation assays.
Human PBMCs were preincubated for 60 minutes at 37°C with 200 µg·mL-1 laminarin before adding the fungal stimuli.
P<0.05, when compared with the assay with no laminarin included. Data presented as mean±SD.
Abbreviations: PBMCs, peripheral blood mononuclear cells; TNFα, tumor necrosis factor alpha; HK, heat-killed; C. albicans, Candida albicans; C. tropicalis, Candida tropicalis; C. guilliermondii, Candida guilliermondii; C. krusei, Candida krusei; C. auris, Candida auris.
Human monocyte-derived macrophages differentially uptake Candida spp
Next, to assess whether other cellular players of the innate immunity differentially interact with the analyzed Candida species and thus affecting the recognition and phagocytosis, we evaluated the ability of human monocyte-derived macrophages to uptake the yeast cells, using a flow cytometry-based protocol that has been used previously to characterize the phagocytic process of fungal cells.37,51,59 Yeast cells were labeled with acridine orange, which emits a green fluorescence that turns reddish once they are in an acidified microenvironment of the mature phagolysosomes.37,59 Results given in Figure 5 shows that most of the fungal species were uptaken and internalized into acid phagolysosomes, but the number of fungal cells undergoing this process was significantly higher for C. tropicalis, C. guilliermondii, and C. krusei than for C. albicans and C. auris, which showed a similar behavior when interacting with these human cells. Similar results were observed for green cells associated to macrophages, ie, recently phagocyted, with low cell numbers associated to C. albicans and C. auris and relatively high cell numbers associated to C. tropicalis, C. guilliermondii, and C. krusei (Figure 5). Collectively, these data indicate a differential ability of human monocyte-derived macrophages to uptake Candida spp. cells.
Figure 5.
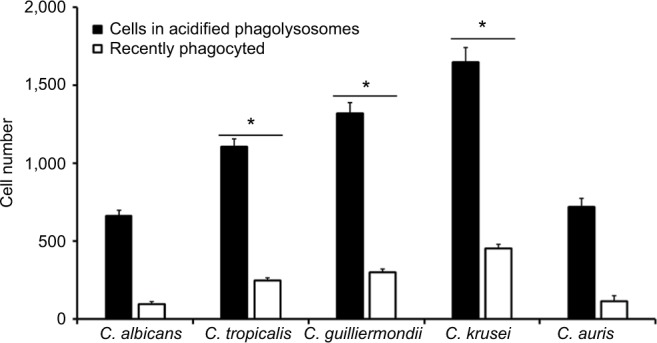
Phagocytosis of Candida albicans, Candida tropicalis, Candida guilliermondii, Candida krusei, and Candida auris by human monocyte-derived macrophages.
Notes: Acridine orange-labeled yeast cells were incubated with the human cells at an MOI ratio of 1:6 for 2.5 hours at 37°C under a CO2 atmosphere. Then, macrophages were gated by FACS system and 50,000 cells were counted/sample. Results represent macrophages interacting with at least one green fluorescent cell (recently phagocyted), and those associated with red fluorescence that were classified as macrophages with yeast cells within acidified phagolysosomes. The data represent the mean ± SD of three independent biological replicates performed in duplicate. * P<0.05, when compared with C. albicans cells.
Abbreviations: MOI, multiplicity of infection; FACS, fluorescence-activated cell sorter.
Discussion
C. albicans is a model organism popular to study fundamental aspects of a human fungal pathogen, and a vast amount of information about its biology is currently available. Because of their taxonomical classification, it is usual to find literature assuming that most of the members of the Candida genus should have biological traits like those described in C. albicans, with minimal differences in the phenotype or the molecular determinants related to virulence.41 Nonetheless, the genomic evidence clearly suggests that these organisms have significant differences in the fitness and metabolic processes,60 which are likely to affect the interaction with the host’s components, such as the immune system. The comparative analysis performed here underscored the fact that even though the species under study had a similar composition of the cell wall, they have differences that are likely to affect the interaction with components of the innate immunity such as PPRs. As a proof of the concept, we have the case of C. guilliermondii cells, which had lower levels of b1,3-glucan and as a consequence, they induced the lowest levels of cytokines when this polysaccharide was exposed on the cell surface. Since the protein content in the C. guilliermondii cell wall was similar to the one obtained from the other species under analysis, we hypothesized that the increment in both N- and O-linked mannans in the cell wall was related to the different variety of cell wall proteins that are likely to contain more sites of these posttranslational modifications. Supporting this notion, a different cell secretome has been predicted for C. guilliermondii, which may affect the kind of proteins associated with the wall.61 The lower C. krusei cell wall protein content fits with the low mannan levels, which is likely to be the reason behind this observation. In agreement, the phosphomannan content in this species was also low. The high porosity of the cell wall has been associated with mannans containing short lateral chains;41,46,47 therefore, it is likely this will be the case of mannans on the surface of C. tropicalis, C. guilliermondii, and C. krusei.
Results presented here clearly demonstrate that in an in vitro setting, the human PMBCs are differentially stimulated to produce cytokines by the analyzed Candida species. It was interesting to observe that C. albicans and C. auris had similar abilities to interact with these immune cells and with monocyte-derived macrophages, even though they are species relatively distant in the phylogeny.62 Thus far, only the interaction of C. auris with neutrophils has been studied, and differences in the ability to evade the neutrophil attack have been documented.34 Nonetheless, our results reported here suggest that if escaped from interaction with these immune cells, the next line of defense, the mononuclear cells and macrophages are likely to control this pathogen. Further experiments are required to confirm this hypothesis.
Live cells of C. tropicalis, C. guilliermondii, and C. krusei stimulated higher cytokine levels than C. albicans cells, which might be related to the increased wall porosity. In addition, these three species showed different wall phosphomannan levels that might also contribute to this observation. It was interesting to note that upon removal of O-linked mannans from live cells from these three species, the ability to stimulate cytokine production changed, which in some cases was comparable to the quantified value when HK cells were used for immune stimulation. These data suggest that O-linked mannans play a negative role during recognition of these cell walls by the human mononuclear cells, and once they are removed, relevant ligands for cytokine production are accessible for immune receptors. Here, we demonstrated that the pathway behind this observation requires dectin-1 engagement with β1,3-glucan. Similar observations have been reported for Candida parapsilosis sensu lato and sensu stricto,41,46 and for the interaction of C. albicans with macrophages.63
The therapeutic strategies to control candidiasis are extremely limited when compared to the wide repertoire of drugs to treat bacterial infections, and this has negatively impacted the sensitivity of fungal cells to the currently available drugs. Moreover, some Candida species are naturally resistant to some antifungal drugs or have the ability to rapidly develop resistance, such as Candida glabrata and C. kruseii.64,65 Therefore, it is imperative to expand the portfolio of antifungal drugs to treat candidiasis. An alternative approach to control these infections has been explored using immunomodulators such as b1,3-glucan. Administration of this cell wall polysaccharide primes PBMCs and monocytes, which acquire prolonged enhanced functional state mediated by epigenetic mechanisms.66 Although these could lead to a promising strategy to help in the control of this disease, our data here demonstrated that this approach might not be beneficial for the treatment of infections caused by C. guilliermondii, as this species does not induce a strong cytokine production via engagement of dectin-1 with β1,3-glucan.
Since the O-linked mannans of the cell wall of the species studied here showed a masking effect on the b1,3-glucan exposure at the cell surface and therefore precluded a proper activation of the cytokine production via dectin-1, it is tempting to speculate that inhibitors of the O-linked mannosylation pathway may have a positive effect on controlling infections caused by these fungal species.
The evaluation of the phagocytic process by human monocyte-derived macrophages pointed out again that C. tropicalis, C. guilliermondii, and C. krusei are species with a different ability to interact with these immune cells when compared to C. albicans. We have recently demonstrated that C. tropicalis is more phagocytosed than C. albicans by a phosphomannan-dependent mechanism,37 and results reported here confirmed this previous observation. Since C. guilliermondii also showed higher phosphomannan content than C. albicans cells, it is tempting to offer a similar explanation for this observation. Although we do not currently have a proper explanation for data generated with C. krusei, a similar observation for this species and C. guilliermondii has been reported when interacting with murine macrophages.28
Conclusion
In conclusion, our study shows that the current knowledge about C. albicans cell wall and its interaction with innate immune cells cannot be extrapolated to C. tropicalis, C. guilliermondii, C. krusei, and C. auris. These species have differences in the cell wall composition and with exception of C. auris, displayed different abilities to stimulate cytokine production by human PBMCs and to be phagocytosed by human monocyte-derived macrophages.
Acknowledgments
We thank Prof Gordon Brown from the University of Aberdeen for the donation of the IgG Fc-Dectin-1 chimera. We acknowledge the technical assistance of Maria F Rodriguez-Preciado (Universidad de Guanajuato) during the acquisition of data related to Candida krusei. This work was supported by Consejo Nacional de Ciencia y Tecnología (ref PDCPN2014-247109 and FC 2015-02-834), Universidad de Guanajuato (ref 1025/2016; CIIC 95/2018), and Red Temática Glicociencia en Salud (CONACYT-México).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Pappas PG, Rex JH, Lee J, et al. A prospective observational study of candidemia: epidemiology, therapy, and influences on mortality in hospitalized adult and pediatric patients. Clin Infect Dis. 2003;37(5):634–643. doi: 10.1086/376906. [DOI] [PubMed] [Google Scholar]
- 2.Brown GD, Denning DW, Gow NA, Levitz SM, Netea MG, White TC. Hidden killers: human fungal infections. Sci Transl Med. 2012;4(165):165rv113. doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- 3.Lass-Flörl C. The changing face of epidemiology of invasive fungal disease in Europe. Mycoses. 2009;52(3):197–205. doi: 10.1111/j.1439-0507.2009.01691.x. [DOI] [PubMed] [Google Scholar]
- 4.Tan TY, Tan AL, Tee NWS, Ng LSY, Chee CWJ. The increased role of non-albicans species in candidaemia: results from a 3-year surveillance study. Mycoses. 2010;53(6):515–521. doi: 10.1111/j.1439-0507.2009.01746.x. [DOI] [PubMed] [Google Scholar]
- 5.Pfaller MA, Boyken L, Hollis RJ, Messer SA, Tendolkar S, Diekema DJ. In vitro activities of anidulafungin against more than 2,500 clinical isolates of Candida spp., including 315 isolates resistant to fluconazole. J Clin Microbiol. 2005;43(11):5425–5427. doi: 10.1128/JCM.43.11.5425-5427.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pfaller MA, Pappas PG, Wingard JR, Pfaller Michael A, Pappas Peter G. Invasive fungal pathogens: current epidemiological trends. Clin Infect Dis. 2006;43(S1):S3–S14. [Google Scholar]
- 7.Krcmery V, Barnes AJ. Non-albicans Candida spp. causing fun-gaemia: pathogenicity and antifungal resistance. J Hosp Infect. 2002;50(4):243–260. doi: 10.1053/jhin.2001.1151. [DOI] [PubMed] [Google Scholar]
- 8.Tortorano A, Kibbler C, Peman J, Bernhardt H, Klingspor L, Grillot R. Candidaemia in Europe: epidemiology and resistance. Int J Antimicrob Agents. 2006;27(5):359–366. doi: 10.1016/j.ijantimicag.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 9.Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis. 2004;39(3):309–317. doi: 10.1086/421946. [DOI] [PubMed] [Google Scholar]
- 10.Chen SCA, Marriott D, Playford EG, et al. Candidaemia with uncommon Candida species: predisposing factors, outcome, antifungal susceptibility, and implications for management. Clin Microbiol Infect. 2009;15(7):662–669. doi: 10.1111/j.1469-0691.2009.02821.x. [DOI] [PubMed] [Google Scholar]
- 11.Savini V, Catavitello C, Onofrillo D, et al. What do we know about Candida guilliermondii? A voyage throughout past and current literature about this emerging yeast. Mycoses. 2011;54(5):434–441. doi: 10.1111/j.1439-0507.2010.01960.x. [DOI] [PubMed] [Google Scholar]
- 12.Bidaud AL, Chowdhary A, Dannaoui E. Candida auris: an emerging drug resistant yeast – A mini-review. J Mycol Med. 2018;28(3):568–573. doi: 10.1016/j.mycmed.2018.06.007. [DOI] [PubMed] [Google Scholar]
- 13.Netea MG, Brown GD, Kullberg BJ, Gow NAR. An integrated model of the recognition of Candida albicans by the innate immune system. Nat Rev Microbiol. 2008;6(1):67–78. doi: 10.1038/nrmicro1815. [DOI] [PubMed] [Google Scholar]
- 14.Martínez-Álvarez JA, Pérez-García LA, Flores-Carreón A, Mora-Montes HM. The immune response against Candida spp. and Sporothrix schenckii. Rev Iberoam Micol. 2014;31(1):62–66. doi: 10.1016/j.riam.2013.09.015. [DOI] [PubMed] [Google Scholar]
- 15.Díaz-Jiménez DF, Pérez-García LA, Martínez-Álvarez JA, Mora-Montes HM. Role of the fungal cell wall in pathogenesis and antifungal resistance. Curr Fungal Infect Rep. 2012;6(4):275–282. [Google Scholar]
- 16.Gow NAR, van de Veerdonk FL, Brown AJP, Netea MG. Candida albicans morphogenesis and host defence: discriminating invasion from colonization. Nat Rev Microbiol. 2012;10(2):112–122. doi: 10.1038/nrmicro2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gow NAR, Hube B. Importance of the Candida albicans cell wall during commensalism and infection. Curr Opin Microbiol. 2012;15(4):406–412. doi: 10.1016/j.mib.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 18.Klis FM, Groot PD, Hellingwerf K. Molecular organization of the cell wall of Candida albicans. Med Mycol. 2001;39(1):1–8. [PubMed] [Google Scholar]
- 19.Brown GD, Gordon S. Immune recognition: a new receptor for β-glucans. Nature. 2001;413(6851):36–37. doi: 10.1038/35092620. [DOI] [PubMed] [Google Scholar]
- 20.Netea MG, Gow NAR, Munro CA, et al. Immune sensing of Candida albicans requires cooperative recognition of mannans and glucans by lectin and Toll-like receptors. J Clin Invest. 2006;116(6):1642–1650. doi: 10.1172/JCI27114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cambi A, Netea MG, Mora-Montes HM, et al. Dendritic Cell Interaction with Candida albicans Critically Depends on N-Linked Mannan. J Biol Chem. 2008;283(29):20590–20599. doi: 10.1074/jbc.M709334200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saijo S, Ikeda S, Yamabe K, et al. Dectin-2 recognition of α-Mannans and induction of Th17 cell differentiation is essential for host defense against Candida albicans. Immunity. 2010;32(5):681–691. doi: 10.1016/j.immuni.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 23.Wells CA, Salvage-Jones JA, Li X, et al. The macrophage-inducible C-type lectin, mincle, is an essential component of the innate immune response to Candida albicans. J Immunol. 2008;180(11):7404–7413. doi: 10.4049/jimmunol.180.11.7404. [DOI] [PubMed] [Google Scholar]
- 24.Zhu L-L, Zhao X-Q, Jiang C, et al. C-type lectin receptors dectin-3 and dectin-2 form a heterodimeric pattern-recognition receptor for host defense against fungal infection. Immunity. 2013;39(2):324–334. doi: 10.1016/j.immuni.2013.05.017. [DOI] [PubMed] [Google Scholar]
- 25.Mora-Montes HM, Netea MG, Ferwerda G, et al. Recognition and blocking of innate immunity cells by Candida albicans chitin. Infect Immun. 2011;79(5):1961–1970. doi: 10.1128/IAI.01282-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wagener J, Malireddi RKS, Lenardon MD, et al. Fungal chitin dampens inflammation through IL-10 induction mediated by Nod2 and TLR9 activation. PLoS Pathog. 2014;10(4):e1004050. doi: 10.1371/journal.ppat.1004050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Richardson MD, Donaldson F. Interaction of Candida krusei with human neutrophils in vitro. J Med Microbiol. 1994;41(6):384–388. doi: 10.1099/00222615-41-6-384. [DOI] [PubMed] [Google Scholar]
- 28.Vecchiarelli A, Bistoni F, Cenci E, Perito S, Cassone A. In-vitro killing of Candida species by murine immunoeffectors and its relationship to the experimental pathogenicity. Med Mycol. 1985;23(5):377–387. doi: 10.1080/00362178585380541. [DOI] [PubMed] [Google Scholar]
- 29.Roilides E, Holmes A, Blake C, Pizzo PA, Walsh TJ. Effects of granulocyte colony-stimulating factor and interferon-gamma on antifungal activity of human polymorphonuclear neutrophils against pseudohyphae of different medically important Candida species. J Leukoc Biol. 1995;57(4):651–656. doi: 10.1002/jlb.57.4.651. [DOI] [PubMed] [Google Scholar]
- 30.Høgåsen AK, Abrahamsen TG, Gaustad P. Various Candida and Torulopsis species differ in their ability to induce the production of C3, factor B and granulocyte-macrophage colony-stimulating factor (GM-CSF) in human monocyte cultures. J Med Microbiol. 1995;42(4):291–298. doi: 10.1099/00222615-42-4-291. [DOI] [PubMed] [Google Scholar]
- 31.Lyman CA, Walsh TJ. Phagocytosis of medically important yeasts by polymorphonuclear leukocytes. Infect Immun. 1994;62(4):1489–1493. doi: 10.1128/iai.62.4.1489-1493.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maródi L, Forehand JR, Johnston RB. Mechanisms of host defense against Candida species. II. Biochemical basis for the killing of Candida by mononuclear phagocytes. J Immunol. 1991;146(8):2790–2794. [PubMed] [Google Scholar]
- 33.Maródi L, Korchak HM, Johnston RB. Mechanisms of host defense against Candida species. I. Phagocytosis by monocytes and monocyte-derived macrophages. J Immunol. 1991;146(8):2783–2789. [PubMed] [Google Scholar]
- 34.Johnson CJ, Davis JM, Huttenlocher A, Kernien JF, Nett JE. Emerging fungal pathogen Candida auris evades neutrophil attack. MBio. 2018;9(4):e01403–e01418. doi: 10.1128/mBio.01403-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gillum AM, Tsay EYH, Kirsch DR. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol Gen Genet. 1984;198(1):179–182. doi: 10.1007/BF00328721. [DOI] [PubMed] [Google Scholar]
- 36.Millerioux Y, Clastre M, Simkin AJ, et al. Development of a URA5 integrative cassette for gene disruption in the Candida guilliermondii ATCC 6260 strain. J Microbiol Methods. 2011;84(2):355–358. doi: 10.1016/j.mimet.2010.12.013. [DOI] [PubMed] [Google Scholar]
- 37.Hernández-Chávez MJ, Franco B, Clavijo-Giraldo DM, Hernández NV, Estrada-Mata E, Mora-Montes HM. Role of protein phosphoman-nosylation in the Candida tropicalis-macrophage interaction. FEMS Yeast Res. 2018;18(5) doi: 10.1093/femsyr/foy053. 01 08 2018. [DOI] [PubMed] [Google Scholar]
- 38.Sharma C, Kumar N, Meis JF, Pandey R, Chowdhary A. Draft genome sequence of a fluconazole-resistant Candida auris strain from a Candidemia patient in India. Genome Announc. 2015;3(4):e00722–15. doi: 10.1128/genomeA.00722-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mora-Montes HM, Bates S, Netea MG, et al. Endoplasmic reticulum alpha-glycosidases of Candida albicans are required for N glycosylation, cell wall integrity, and normal host-fungus interaction. Eukaryot Cell. 2007;6(12):2184–2193. doi: 10.1128/EC.00350-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Díaz-Jiménez DF, Mora-Montes HM, Hernández-Cervantes A, Luna-Arias JP, Gow NAR, Flores-Carreón A. Biochemical characterization of recombinant Candida albicans mannosyltransferases Mnt1, Mnt2 and Mnt5 reveals new functions in O- and N-mannan biosynthesis. Biochem Biophys Res Commun. 2012;419(1):77–82. doi: 10.1016/j.bbrc.2012.01.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Estrada-Mata E, Navarro-Arias MJ, Perez-Garcia LA, et al. Members of the Candida parapsilosis complex and Candida albicans are differentially recognized by human peripheral blood mononuclear cells. Front Microbiol. 2015;6:1527. doi: 10.3389/fmicb.2015.01527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hobson RP, Munro CA, Bates S, et al. Loss of cell wall mannosylphosphate in Candida albicans does not influence macrophage recognition. J Biol Chem. 2004;279(38):39628–39635. doi: 10.1074/jbc.M405003200. [DOI] [PubMed] [Google Scholar]
- 43.de Nobel JG, Klis FM, Munnik T, Priem J, van den Ende H. An assay of relative cell wall porosity in Saccharomyces cerevisiae, Kluyveromyces lactis and Schizosaccharomyces pombe. Yeast. 1990;6(6):483–490. doi: 10.1002/yea.320060605. [DOI] [PubMed] [Google Scholar]
- 44.Graham LM, Tsoni SV, Willment JA, et al. Soluble Dectin-1 as a tool to detect beta-glucans. J Immunol Methods. 2006;314(1-2):164–169. doi: 10.1016/j.jim.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 45.Marakalala MJ, Vautier S, Potrykus J, et al. Differential adaptation of Candida albicans in vivo modulates immune recognition by dectin-1. PLoS Pathog. 2013;9(4):e1003315. doi: 10.1371/journal.ppat.1003315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pérez-García LA, Csonka K, Flores-Carreón A, et al. Role of protein glycosylation in Candida parapsilosis cell wall integrity and host interaction. Front Microbiol. 2016;7(633):306. doi: 10.3389/fmicb.2016.00306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Navarro-Arias MJ, Defosse TA, Dementhon K, et al. Disruption of protein mannosylation affects Candida guilliermondii cell wall, immune sensing, and virulence. Front Microbiol. 1951;2016:7. doi: 10.3389/fmicb.2016.01951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mora-Montes HM, Mckenzie C, Bain JM, et al. Interactions between macrophages and cell wall oligosaccharides of Candida albicans. Methods Mol Biol. 2012;845:247–260. doi: 10.1007/978-1-61779-539-8_16. [DOI] [PubMed] [Google Scholar]
- 49.Endres S, Ghorbani R, Lonnemann G, van der Meer JWM, Dinarello CA. Measurement of immunoreactive interleukin-1β from human mononuclear cells: optimization of recovery, intrasubject consistency, and comparison with interleukin-1α and tumor necrosis factor. Clin Immunol Immunopathol. 1988;49(3):424–438. doi: 10.1016/0090-1229(88)90130-4. [DOI] [PubMed] [Google Scholar]
- 50.Abrams WR, Diamond LW, Kane AB. A flow cytometric assay of neutrophil degranulation. J Histochem Cytochem. 1983;31(6):737–744. doi: 10.1177/31.6.6404983. [DOI] [PubMed] [Google Scholar]
- 51.González-Hernández RJ, Jin K, Hernández-Chávez MJ, et al. Phosphomannosylation and the functional analysis of the extended Candida albicans MNN4-like gene family. Front. Microbiol. 2017;8:2156. doi: 10.3389/fmicb.2017.02156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mora-Montes HM, Bates S, Netea MG, et al. A multifunctional mannosyltransferase family in Candida albicans determines cell wall mannan structure and host-fungus interactions. J Biol Chem. 2010;285(16):12087–12095. doi: 10.1074/jbc.M109.081513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bates S, Maccallum DM, Bertram G, et al. Candida albicans Pmr1p, a secretory pathway P-type Ca2+/Mn2+-ATPase, is required for glycosylation and virulence. J Biol Chem. 2005;280(24):23408–23415. doi: 10.1074/jbc.M502162200. [DOI] [PubMed] [Google Scholar]
- 54.West L, Lowman DW, Mora-Montes HM, et al. Differential virulence of Candida glabrata glycosylation mutants. J Biol Chem. 2013;288(30):22006–22018. doi: 10.1074/jbc.M113.478743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bates S, Hughes HB, Munro CA, et al. Outer chain N-glycans are required for cell wall integrity and virulence of Candida albicans. J Biol Chem. 2006;281(1):90–98. doi: 10.1074/jbc.M510360200. [DOI] [PubMed] [Google Scholar]
- 56.Lopes-Bezerra LM, Lozoya-Perez NE, Lopez-Ramirez LA, et al. Functional characterization of Sporothrix schenckii glycosidases involved in the N-linked glycosylation pathway. Med Mycol. 2015;53(1):60–68. doi: 10.1093/mmy/myu057. [DOI] [PubMed] [Google Scholar]
- 57.Graham LM, Tsoni SV, Willment JA, et al. Soluble Dectin-1 as a tool to detect beta-glucans. J Immunol Methods. 2006;314(1-2):164–169. doi: 10.1016/j.jim.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 58.Gow NAR, Netea MG, Munro CA, et al. Immune recognition of Candida albicans beta-glucan by dectin-1. J Infect Dis. 2007;196(10):1565–1571. doi: 10.1086/523110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lozoya-Pérez NE, Casas-Flores S, Martínez-Álvarez JA, et al. Generation of Sporothrix schenckii mutants expressing the green fluorescent protein suitable for the study of host-fungus interactions. Fungal Biol. 2018;122(10):1023–1030. doi: 10.1016/j.funbio.2018.07.004. [DOI] [PubMed] [Google Scholar]
- 60.Butler G, Rasmussen MD, Lin MF, et al. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature. 2009;459(7247):657–662. doi: 10.1038/nature08064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sorgo AG, Heilmann CJ, Brul S, de Koster CG, Klis FM. Beyond the wall: Candida albicans secret(e)s to survive. FEMS Microbiol Lett. 2013;338(1):10–17. doi: 10.1111/1574-6968.12049. [DOI] [PubMed] [Google Scholar]
- 62.Sharma C, Kumar N, Pandey R, Meis JF, Chowdhary A. Whole genome sequencing of emerging multidrug resistant Candida auris isolates in India demonstrates low genetic variation. New Microbes New Infect. 2016;13:77–82. doi: 10.1016/j.nmni.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mckenzie CGJ, Koser U, Lewis LE, et al. Contribution of Candida albicans cell wall components to recognition by and escape from murine macrophages. Infect Immun. 2010;78(4):1650–1658. doi: 10.1128/IAI.00001-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gong J, Xiao M, Wang H, et al. Genetic differentiation, diversity, and drug susceptibility of Candida krusei. Front Microbiol. 2018;9:2717. doi: 10.3389/fmicb.2018.02717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vale-Silva LA, Sanglard D. Tipping the balance both ways: drug resistance and virulence in Candida glabrata. FEMS Yeast Res. 2015;15(4) doi: 10.1093/femsyr/fov025. [DOI] [PubMed] [Google Scholar]
- 66.van der Meer JWM, Joosten LAB, Riksen N, Netea MG. Trained immunity: a smart way to enhance innate immune defence. Mol Immunol. 2015;68(1):40–44. doi: 10.1016/j.molimm.2015.06.019. [DOI] [PubMed] [Google Scholar]