Abstract
Background
SET and MYND domain-containing protein 2 (SMYD2-OE) plays an important role in cancer development through methylating histone and non-histone proteins. However, little is known about the relevance of SMYD2-OE in colon cancer. Moreover, oxaliplatin (L-OHP) is applied as first line for colon cancer chemotherapy, but drug resistance restricts its efficacy. Unexpectedly, the mechanism of L-OHP resistance in colon cancer remains unclear. In this study, we investigated the relationship of SMYD2-OE expression and L-OHP resistance in colon cancer and further explored the underlying mechanism linking SMYD2-OE, L-OHP resistance, and colon cancer.
Materials and methods
Expression levels of SMYD2-OE in colon cancer tissues of patients were tested. In vitro and in vivo assays were conducted to explore the function and mechanism of SMYD2-OE in colon cancer sensitivity to L-OHP.
Results
SMYD2-OE was overexpressed in colon cancer tissues compared with non-neoplastic tissues and associated with poor prognosis of patients with colon cancer after L-OHP-based chemotherapy. Knockdown of SMYD2-OE increased colon cancer sensitivity to L-OHP in vitro and in vivo. However, SMYD2-OE overexpression promoted L-OHP resistance in colon cancer cell in vitro. In addition, SMYD2-OE could upregulate MDR1/P-glycoprotein expression depending on MEK/ERK/AP-1 signaling pathway activity.
Conclusion
These results imply that SMYD2-OE promotes L-OHP resistance in colon cancer by regulating MDR1/P-glycoprotein through MEK/ERK/AP-1 signaling pathway, providing a potential strategy to sensitize chemotherapy by SMYD2-OE knockdown in colon cancer treatment.
Keywords: SMYD2-OE, oxaliplatin resistance, colon cancer, MEK/ERK/AP-1 signaling pathway
Introduction
Colon adenocarcinoma (COAD) is the third leading cause of tumor-related death all over the world, and it is also one of the most common malignant tumors in P.R. China.1 Once diagnosed, ~80% of patients with non-metastatic COAD can undergo surgical resection. However, recurrence after potentially curative surgery was observed among 30%–50% of patients with poor prognosis.2 Therefore, adjuvant chemotherapy after surgical resection is necessary for patients with high-risk stage II or stage III COAD to reduce the risk of recurrence.3
As a third-generation platinum-based chemotherapy drug for adjuvant treatment, oxaliplatin (L-OHP) mainly induces cell apoptosis by inhibiting DNA replication, followed by inhibition of transcription.4 As a reported OCT2 substrate, L-OHP has effective improvement on outcome of stage III COAD patients.5 In addition, recent studies found L-OHP-resistant cells in different colon cancer cells.6 However, many COAD patients have been shown to be resistant to L-OHP exposure, limiting its application in chemotherapy. L-OHP resistance includes low-efficiency cellular drug uptake and persistent activation of the antioxidant glutathione system for detoxification, as well as DNA repair enhancement.7 L-OHP resistance in COAD is multifactorial and histone methyltransferase could promote L-OHP-based resistance.8 Thus, some compensated signaling can promote L-OHP resistance.9
A SET and MYND domain-containing histone methyltransferase, SMYD2-OE, methylates histone and non-histone proteins, including Rb, HSP90AB1, and p53.10,11 Through lysine methylation of Rb, SMYD2-OE could repress specific Rb/E2F genes and promote cell cycle progression.12 SMYD2-OE could also accelerate the proliferation of cancer cells by methylating HSP90AB1.13 While knockdown of SMYD2-OE enhances DNA damage-induced apoptosis with p53-dependence,14 SMYD2-OE has been reported to be overexpressed in esophageal squamous cell carcinoma,15 hepatocellular carcinoma,16 breast cancer,17 gastric cancer,18 and pediatric acute lymphoblastic leukemia,19 correlated with poor prognosis and patient survival.20 However, there is little progress in research regarding any relationship between SMYD2-OE and L-OHP resistance in COAD.
In this study, we investigated the role of SMYD2-OE in L-OHP-resistant COAD. First, SMYD2-OE was upregulated in primary COAD. Overexpression of SMYD2-OE was an indicator of poor prognosis independent of other prognostic factors in COAD patients. Next, we discovered that knockdown of SMYD2-OE prompted L-OHP sensitivity in COAD cells in vitro and in vivo. Last, we confirmed that SMYD2-OE could increase the MDR1 expression through MEK/ERK/AP-1 signaling pathway, which enhanced chemotherapy resistance in COAD cells.
Materials and methods
Patients and specimens
All 110 specimens were obtained from patients who were diagnosed with colon cancer and underwent resection between 2011 and 2016 in the Third People’s Hospital of Chengdu. The study was approved by the institutional review committee of Chongqing Medical University, and all patients signed informed consent documents. The study was conducted in accordance with the Declaration of Helsinki. Before surgery, the patients in this study did not receive any treatment, such as radiotherapy or chemotherapy. The patient’s follow-up data are described in each patient’s medical record. The clinical pathological factors were generalized in Table 1. Tumor status, histology, and differentiation were assessed and diagnosed independently by two authors (Yuntao Li and Tongtong Zhang), according to the WHO guidelines of classification.
Table 1.
SMYD2-OE expression in colon cancer
| Clinical or molecular feature | Total n | SMYD2-OE
|
||
|---|---|---|---|---|
| High | Low | P | ||
|
| ||||
| Age | 0.989 | |||
| <60 | 56 | 30 | 26 | |
| ≥60 | 54 | 29 | 25 | |
| Gender | 0.268 | |||
| Male | 52 | 25 | 27 | |
| Female | 58 | 34 | 24 | |
| Location | 0.655 | |||
| Colon | 82 | 45 | 37 | |
| Rectum | 28 | 14 | 14 | |
| Grade | 0.847 | |||
| Low | 81 | 43 | 38 | |
| High | 29 | 16 | 13 | |
| Stage | 0.016 | |||
| III | 94 | 46 | 48 | |
| IV | 16 | 13 | 3 | |
| Large | 0.033 | |||
| ≤14 cm3 | 57 | 25 | 32 | |
| >14 cm3 | 53 | 34 | 19 | |
| T | 0.564 | |||
| T3 | 95 | 52 | 43 | |
| T4 | 15 | 7 | 8 | |
| N | 0.016 | |||
| N0 | 94 | 46 | 48 | |
| N1+N2 | 16 | 13 | 3 | |
| M | 0.246 | |||
| M0 | 106 | 58 | 48 | |
| M1 | 4 | 1 | 3 | |
L-OHP preparation
L-OHP was purchased from Hengrui Medicine (Lianyungang, Jiangsu Province, P.R. China), dissolved in DMSO. The aliquots were stored at 4°C for use.
Cell culture and clinical samples
Colon cancer cells (SW480 and SW620) were purchased from the Institute of Cell Biology, Chinese Academy of Sciences (Shanghai, P.R. China) and cultured in RPMI 1640 medium supplemented with 10% FBS, l-glutamine, and antibiotics at 37°C with 5% CO2.
Human colon cancer tissues and their paired adjacent normal mucosa tissues were provided by the Third People’s Hospital of Chengdu. All samples were collected with patients’ informed consent, and the study was approved by the Institutional Review Board of the Third People’s Hospital of Chengdu.
L-OHP treatment in vitro
Cells were exposed to 20 µM L-OHP in culture medium for 24 hours, then washed twice with PBS according to previously described criteria.21 Cells were collected for the following analysis.
Measurement of mRNA expression
Total RNA was extracted from COAD and the matched non-tumorous tissues using TRIzol (Thermo Fisher Scientific, Waltham, MA, USA). Total RNA (1 µg) was used for RT reactions in a 20-µL reaction to synthesize cDNA using the RevertAid first-strand cDNA synthesis kit (Thermo Scientific, Fermentas, Lithuania). RNA expression profiles were analyzed by real-time PCR using iTaq SYBR Green Super-mix with ROX (BioRad) in an icycleriQTM Real-time PCR detection system. SMYD2-OE and MDR1 transcripts were amplified using primers described previously.22,23
Immunohistochemistry staining
Immunohistochemical analysis was performed as previously described.24 In brief, colon cancer tissues were subjected to deparaffinization, antigen retrieval, and blockage of nonspecific binding in turn. And then, the sections were specifically incubated with primary anti-SMYD2-OE antibody (1:100; Abcam, MA, USA) or anti-p-ERK1/2 antibody (1:1,000; Cell Signaling Technology, Danvers, MA, USA) at 4°C overnight followed by incubation with biotinylated secondary antibody and 3,3′ diaminobenzidine. Immunostaining assessment was determined using composite scores by multiplying the percentage of immunoreactive cells (10%–25% as 1, 26%–50% as 2, 51%–75% as 3, and 76%–100% as 4) and the staining intensity (no staining as 0, weak staining as 1, moderate staining as 2, and strong staining as 3). The final staining score was divided into a low expression group (≤6) and a high expression group (>7).
Proliferation assay
The cell proliferation assay was performed using CCK-8 assay according to the manufacturer’s protocol (Sigma-Aldrich, St Louis, MO, USA). Briefly, 1,000 cells were seeded per well in 96-well plates, incubated with the supplied reagent for 1 hour, and the absorbance value of each well was measured using a microplate spectrophotometer at 490 nm from day 1 to day 5. The experiment was repeated in triplicate.
Western blot analysis
Samples were prepared from the cells lysed with CelLytic™ M cell lysis reagent (Sigma-Aldrich, St Louis, MO, USA) supplemented with a protease inhibitor (cOmplete™ protease inhibitor cocktail, Roche Applied Science). Fifty micrograms of protein was loaded for the gel electrophoresis and transferred to a polyvinylidene difluoride membrane. Membranes were blocked with 5% nonfat milk for 1 hour at room temperature, then probed with ERK1/2, p-ERK1/2, MEK1/2, p-MEK1/2 (1:1,000; Cell Signaling Technology), SMYD2-OE, GAPDH, and P-gp antibodies (1:1,000; Abcam, MA, USA) at 4°C. After getting rid of non-conjugated primary antibodies with washing buffer, the membranes were further incubated with goat anti-rabbit or anti-mouse IgG secondary antibodies conjugated to alkaline phosphatase, specifically. The protein bands were detected by chemiluminescence (Amersham Pharmacia Corp, Piscataway, NJ, USA).
Colony formation assays
Cells (500 cells/well) were placed in 6-well plates and maintained in media containing 10% FBS. The medium was replaced every 4 days; after 14 days, cells were fixed with methanol and stained with 0.1% crystal violet (Sigma-Aldrich). Visible colonies were then counted. For each treatment group, wells were assessed in triplicate.
Sphere formation assay
Two thousand to three thousand single cells were cultured in 1640 medium in one well of an ultra-low attachment 6-well plate. Medium was changed every 3 days by centrifuging the spheres formed at 1,000 rpm for 2 minutes. The spheres were counted after 10–14 day culture. To determine the self-renewal ability, spheres were trypsinized and replated at the concentration of 1,000 cells/mL for another 10 days. After that, spheres were fixed by 70% ethanol, both sphere numbers and diameters were calculated.
siRNA transfection
siRNA oligonucleotide duplexes and siNegative control (control) were synthesized by GenePharma, Suzhou Co, Ltd (Suzhou, P.R. China). Control: UUCCCGAACGUGUC ACGU; siSMYD2: CAGGAACGACCGGTTAAGAGA. Briefly, HVJ 10 µL (HVJ-E) (Ishikawa Sangyo Kaisha Ltd, Osaka, Japan) was mixed with 5 µL of siRNA solution and 3 µL of Reagent B. After centrifugation, the pellet was resuspended in 27 µL of buffer, followed by the addition of 8 µL of Reagent C. The siRNA–HVJ-E mixture was combined with HCC cells and centrifuged at 12,000 rpm for 20 minutes at 4°C. The cells were resuspended in 1 mL of culture media and incubated for 72 hours.
Luciferase reporter gene assay
An AP-1 luciferase reporter gene vector with AP-1 consensus oligonucleotide was constructed and transfected into colon cancer cells. For luciferase reporter assays, cells were seeded at a density of 2×105 cells per 6-well plate, 24 hours prior to transfection. Total DNA (3.5 µg) plus 10.5 µL of FuGENE6 reagent was used per well. Luciferase assays were performed using the Dual-Luciferase Reporter Assay System kit (Pro-mega) according to the manufacturer’s directions. Samples were read using a Dynex microtiter luminometer.
Assessment of drug efficacy in vivo
All animals were maintained and used in accordance with the guidelines of the Institutional Animal Care and Use Committee of the Chongqing Medical University. All of the experimental procedures were approved by the Chongqing Medical University ethics commission. In vivo, tumor formation was established by subcutaneous injection of cancer cells suspended in 200 µL 1:1 RPMI 1640 and Matrigel into the dorsal flank of 5- to 7-week-old male Nu/Nu mice; 2.5×106 colon cancer cells were injected once. Mice were weighed and randomly sorted into treatment groups (six to eight mice per group). Treatment began when tumors reached 100–150 mm3. L-OHP dissolved in PBS was administered to tumor-bearing mice with 5 mg/kg body weight via tail intravenous injection once every 3 days.25 The control mice were only treated with the same volume of PBS. Measurements were recorded every 3 days using a digital caliper. Tumor volumes were estimated by measuring two perpendicular diameters, a and b, using the formula (V=0.5 × a × b2 [a and b indicate the long and short perpendicular diameters, respectively]). The relative tumor size was defined by V/V0, where V represents the tumor volume over time and V0 represents the initial tumor volume. Mice were euthanized when the tumor volume reached five times of the initial volume.
Statistical analysis
All experiments were performed at least three times. The results were shown as mean ± SEM. All statistical analyses were performed using SPSS 22.0 software unless otherwise specified. Two-tailed unpaired Student’s t-tests or Fisher’s exact tests were used to evaluate the data. P-values <0.05 were considered significant.
Results
High expression of SMYD2-OE was associated with poor clinical outcome in COAD patients undergoing L-OHP chemotherapy
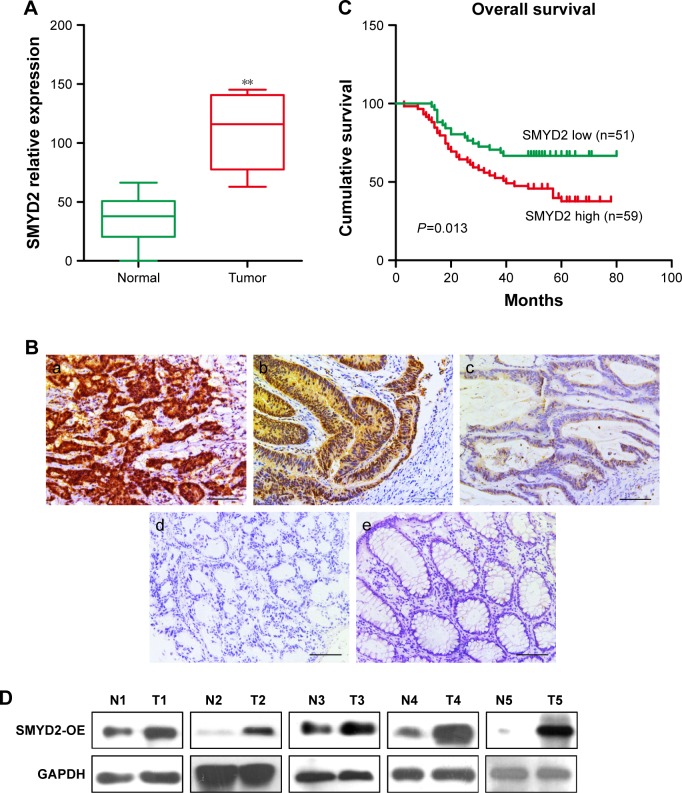
First, we detected the mRNA expression of SMYD2-OE in paired colon and matching non-neoplastic colon tissues, showing that SMYD2-OE was upregulated in COAD tissues compared with non-neoplastic tissues (Figure 1A). Consistently, we analyzed the level of SMYD2-OE in human COAD tissues using published data sets from TCG mRNA A database. In silico analysis demonstrated that SMYD2-OE was upregulated in COAD tissues compared with normal tissues. To explore the prevalence and clinical significance of SMYD2-OE overexpression, we quantified the expression of SMYD2-OE by immunohistochemistry in a cohort of 110 COAD patients with postoperative L-OHP chemotherapy (Figure 1B). Tumors with moderate and strong immunostaining were classified as high expression, while those with negative and weak as low expression. Interestingly, SMYD2-OE was expressed in only 2.5% (5/110) of the normal colon epithelium samples, but a significantly high expression level of SMYD2-OE was observed in 53.6% (59/110) of the COAD samples (P<0.001, Table 1). Moreover, Kaplan–Meier analysis revealed that SMYD2-OE overexpression was frequently observed in L-OHP exposure patients with shorter disease-free survival times (Figure 1C). Through analyzing the relationship between SMYD2-OE expression and clinicopathological characteristics of patients receiving postoperative L-OHP chemotherapy (Table 2), we discovered that SMYD2-OE overexpression was more frequent in tumors with distant metastases compared with those without distant metastases (Figure 1D).
Figure 1.
High expression of SMYD2-OE was associated with poor clinical outcome in COAD patients undergoing L-OHP chemotherapy.
Notes: (A) Expression levels of SMYD2-OE were determined by qRT-PCR in COAD and matching non-neoplastic colon tissues. (B) Expression analysis of SMYD2-OE protein in normal colorectal mucosa and COAD tissues by immunohistochemistry (magnification 400×). (a) Strong expression of SMYD2-OE in COAD. (b) Moderate expression of SMYD2-OE in COAD. (c) Weak expression of SMYD2-OE in COAD. (d) Negative expression of SYMD2 in COAD. (e) Negative expression of COAD in normal colorectal mucosa. (C) The expression of SMYD2-OE predicted DFS in patients with L-OHP exposure determined by the Cox proportional hazards model and log-rank analysis (Table 1). (D) Western blot for paired COAD tissues (T) and normal adjacent tissues (N) with indicated antibodies. The experiments were all repeated three times, and the representative results were shown. Scale bars, 100 µm. **P<0.01.
Abbreviations: COAD, colon adenocarcinoma; DFS, disease-free survival; L-OHP, oxaliplatin.
Table 2.
Univariate and multivariate analysis for relapse-free survival (Cox proportional hazards regression model)
| Characteristics | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| HR | 95% CI | P-value | HR | 95% CI | P-value | |
|
| ||||||
| Gender (males vs females) | 0.866 | 0.494–1.518 | 0.615 | 0.707 | 0.379–1.319 | 0.276 |
| Age (≤60 vs >60 years) | 0.940 | 0.549–1.611 | 0.822 | 0.761 | 0.401–1.443 | 0.403 |
| Grade (low vs high) | 2.977 | 1.645–5.387 | <0.001 | 3.002 | 1.598–5.638 | 0.001 |
| Large (≤14 vs >14 cm3) | 1.030 | 0.600–1.766 | 0.915 | 0.599 | 0.287–1.251 | 0.173 |
| Stage (phage) | 2.840 | 1.500–5.379 | 0.001 | – | – | – |
| T (T2 + T3 vs T4) | 1.917 | 0.961–3.825 | 0.065 | 1.585 | 0.624–4.026 | 0.333 |
| N (N0 vs N1 + N2) | 0.718 | 0.175–2.951 | 0.646 | 0.819 | 0.183–3.674 | 0.795 |
| M (M0 vs M1) | 2.840 | 1.500–5.379 | 0.001 | 2.184 | 0.977–4.883 | 0.057 |
| SETBP1 (high vs low expression) | 2.086 | 1.171–3.718 | 0.013 | 1.865 | 1.028–3.384 | 0.040 |
Note: P-values <0.05 are indicated in bold.
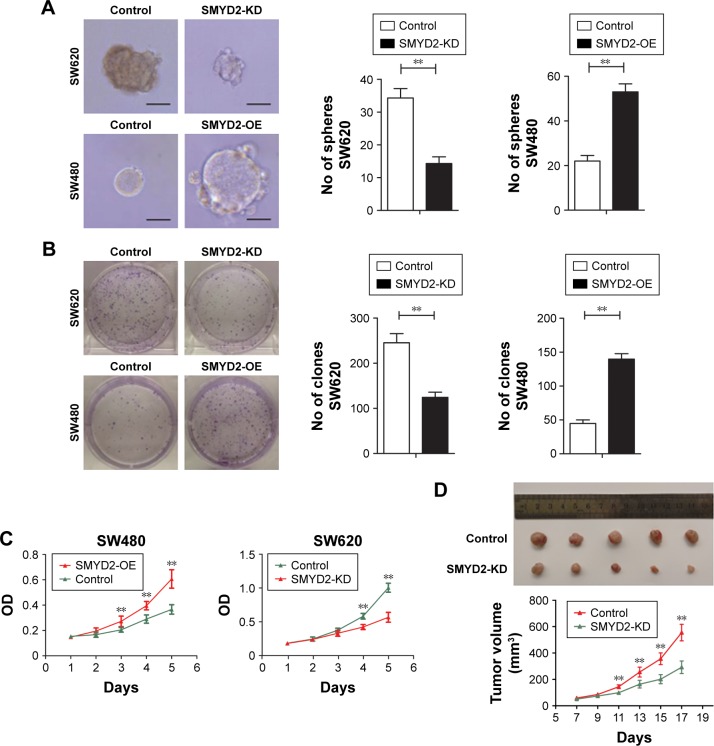
SMYD2-OE induced L-OHP resistance of colon cancer cells in vitro and in vivo
To investigate the biological association of SMYD2-OE with chemotherapy resistance in COAD, we assessed the effects of SMYD2-OE silencing or overexpression on cell sensitivity on exposure to L-OHP. Our previous studies found that SMYD2-OE was highly expressed in SW620 cells and lowly expressed in SW480 cells. Correspondingly, we established SMYD2-OE knockdown using specific siRNA and high-expression in COAD cells using an overexpression plasmid. Interestingly, we observed that SMYD2-OE silencing promoted sensitivity of SW620 cells to L-OHP treatment. However, SW480 cells with SMYD2-OE overexpression exhibited increased resistance to L-OHP treatment (Figure 2A–C). Therefore, these results suggested that SMYD2-OE induces L-OHP resistance of colon cancer cells.
Figure 2.
SMYD2-OE induced L-OHP resistance of colon cancer cells in vitro and in vivo.
Notes: (A) SW620 cells with SMYD2-OE knockdown and SW480 cells with SMYD2-OE overexpression were seeded and observed for cell proliferation, (B) clone formation, and (C) mammosphere formation assays. Scale bars, 40 µm. Error bars, mean ± SD (n=3 independent experiments). (D) The tumor weight and tumor growth curve of SMYD2-OE knockdown cells were compared with the control cells after L-OHP treatment. The results were obtained from three independent experiments. Error bars, mean ± SD (n=5). **P<0.01.
Abbreviations: KD, knockdown; L-OHP, oxaliplatin.
To verify the effect of SMYD2-OE knockdown on L-OHP-exposed colon tumor in vivo, we assessed tumorigenicity of SW620 cells in a nude mouse model. Tumor xenografts of SW620 cells with SMYD2-OE knockdown were markedly more sensitive to L-OHP compared with their counterparts after 2 weeks, and a similar trend was maintained until study termination (Figure 2D). Collectively, these results indicated that SMYD2-OE knockdown sensitized SW620 cells to L-OHP exposure in vivo, suggesting that SMYD2-OE downregulation may promote the curative effects of L-OHP on tumor growth.
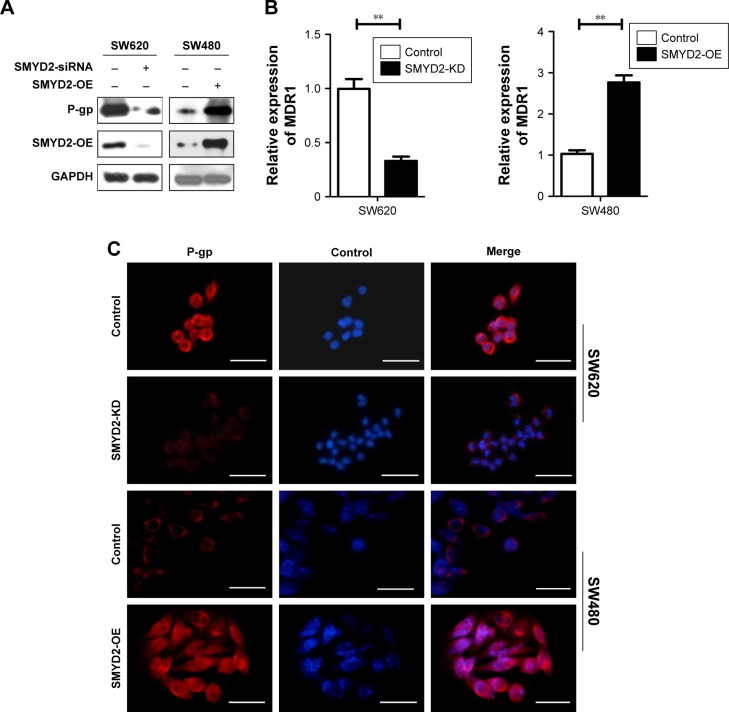
P-glycoprotein was a key player in SMYD2-OE-mediated L-OHP resistance
To further investigate the mechanism underlying SMYD2-OE-mediated L-OHP resistance in COAD, we explored the expression of MDR1 encoded P-gp at the transcriptional level, and at the protein level by Western blot, as well as performed immunofluorescence staining after SMYD2-OE knockdown in SW620 cells and SMYD2-OE overexpression in SW480 cells. As shown, our data revealed that P-gp downregulation was consistent with inhibition of SMYD2-OE expression in SW620 cells, and accordingly, the P-gp upregulation with the increased SMYD2-OE in SW480 cells (Figure 3A–C). Previous study has indicated that P-gp expression was upregulated26 in COAD and overexpression of P-gp promoted L-OHP resistance in colon cancer cells. These data indicated that P-gp was very important for SMYD2-OE-mediated L-OHP resistance in COAD.
Figure 3.
P-glycoprotein was a key downstreamer of SMYD2-OE in colon cancer cells.
Notes: (A) P-gp expression was analyzed using Western blot in SW620 and SW480 cells transfected with the indicated vectors or siRNA. (B) MDR1 transcription was analyzed using qRT-PCR in SW620 and SW480 cells transfected with the indicated vectors or siRNA. (C) The expression of P-gp was determined by immunofluorescence assay in SW620 and SW480 cells transfected with the indicated vectors or siRNA. Scale bars, 40 µm. **P<0.01.
Abbreviation: L-OHP, oxaliplatin.
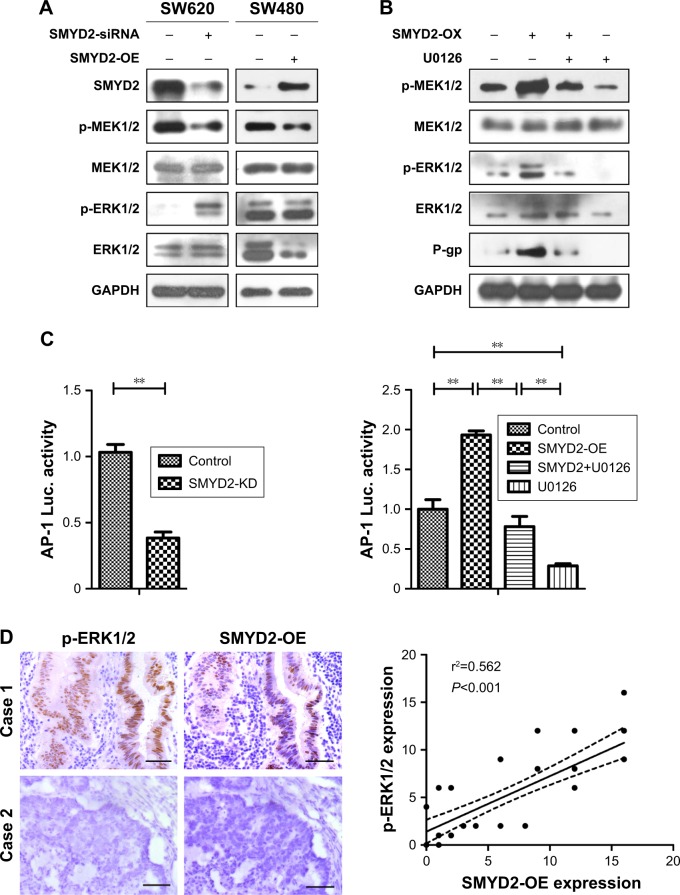
SMYD2-OE promoted P-glycoprotein expression through MEK/ERK/AP-1 signaling pathway
Increasing evidences suggested that P-gp was involved in MEK/ERK/AP-1 signaling pathway with P-gp as one of target genes, and so we measured the activity of MEK/ERK/AP-1 pathway in COAD. The results showed that MEK/ERK pathway was inhibited in SMYD2-OE-downregulated SW620 cells (Figure 4A). On the contrary, the expression of p-ERK1/2 and p-MEK1/2 was both upregulated in SMYD2-OE-overexpressed SW480 cells. And also, ERK1/2 inhibitor U0126 could partially reverse the P-gp upregulation in SW480 cells (Figure 4B). The AP-1 binding ability had the same trend (Figure 4C). These results suggested SMYD2-OE promoted P-gp expression through activation of MEK/ERK/AP-1 signaling pathway in COAD. To further explore the clinical implication of the above findings, we analyzed the relationship between SMYD2-OE and p-ERK1/2 expression in COAD tissues by immunohistochemistry. We observed that 25/40 (62.5%) tumors had more intense staining of SMYD2-OE, and 19/40 (47.5%) tumors with high expression of p-ERK1/2 (Figure 4D). Both SMYD2-OE and p-ERK1/2 were located in the nucleus of cancer cells. Statistical analysis revealed a significant positive correlation between SMYD2-OE and p-ERK protein expression in COAD (Figure 4D, P=0.0). So, these results indicated that SMYD2-OE might induce resistance to L-OHP by upregulating MEK/ERK/AP-1 signaling pathway.
Figure 4.
SMYD2-OE promoted P-glycoprotein expression through MEK/ERK/AP-1 signaling pathway.
Notes: SMYD2-OE promoted P-gp expression through MEK/ERK/AP-1 signaling pathway. (A) The expression of MEK, ERK and their phosphorylation levels in SW620 cells with SMYD2-OE knockdown and SW480 cells with SMYD2-OE overexpression using Western blot. (B) The expression of MEK, ERK and their phosphorylation levels, as well as P-gp in SW480 cells with or without SMYD2-OE-overexpression and SMYD2-OE-inhibition using U0126. (C) AP-1 binding activity detection using luciferase reporter gene assay. (D) The correlation between SMYD2-OE and p-ERK1/2 in COAD by immunohistochemistry assay. Scale bars, 100 µm. Representative images (left) and summarized graph (right). The relationship between these two variables was determined by Pearson’s correlation coefficient. The dotted lines indicate 95% confidence band of the best fitted solid line.
Abbreviations: COAD, colon adenocarcinoma; L-OHP, oxaliplatin.
Discussion
L-OHP, a third-generation platinum antitumor drug, prevents DNA replication and transcription by forming crosslinks in DNA, thus leading to cell death.27 L-OHP is often the first-line clinical therapy for COAD, and about 50% of patients benefit from L-OHP treatment. However, resistance to L-OHP occurs frequently.28 The L-OHP resistance is caused by multiple mechanisms, including chemotherapy drug resistance associated protein overexpression, DNA-Pt adduct formation, and defects in signaling transduction pathways.8 And also, the L-OHP resistance is closely related with the recurrence rate of patients with COAD after radical resection.29
Previous study reported that SMYD2-OE, as a clinically relevant prognostic marker, was overexpressed in various cancer tissues including hepatocellular cancer,16 triple negative breast cancer,17 gastric cancer,18 lung cancer,30 and bladder cancer.31 In addition, SMYD2-OE played an important role in cancer chemotherapy.32 In this study, we examined the role of SMYD2-OE in COAD with L-OPH exposure. SMYD2-OE was overexpressed in COAD tissues and associated with poor prognosis for L-OHP-based chemotherapy in COAD patients. Our data also showed that knockdown of SMYD2-OE expression sensitized colon cancer cells to L-OPH exposure in vivo and in vitro. Therefore, previous results and our data both indicated that SMYD2-OE might play an important role in L-OHP resistance of COAD. And for all we know, this is the first study to estimate the potential of SMYD2-OE as a therapeutic target to sensitize COAD cells to L-OHP.
More importantly, we provided a potential molecular mechanism to elucidate the SMYD2-OE-mediated resistance to L-OHP. P-gp is a membrane transporter, as a member of ATP-binding cassette superfamily, encoded by the MDR1 gene.33 Previous study indicated that P-gp deregulation in COAD tissues predicated poor diagnosis,34 while P-gp overexpression induced L-OHP resistance in COAD cells.35 Here, we found that P-gp was suppressed when SMYD2-OE expression was inhibited in SW620 cells, and also, P-gp was upregulated when SMYD2-OE expression was increased in SW480 cells, indicating that P-gp was a key player in SMYD2-OE-mediated L-OHP resistance.
For the mechanism of SMYD2-OE-mediated L-OHP resistance, the upstream MEK/ERK/AP-1 signaling pathway of MDR1 gene was revealed to regulate P-gp transcription. Activated ERK1/2 phosphorylates transcription factor AP-1, contributing to AP-1 nuclear translocation, and then promoting MDR1 expression. Our data showed that SMYD2-OE overexpression enhanced MEK/ERK/AP-1 activation while SMYD2-OE inhibition blocked their activities. Consistently, both SMYD2-OE and p-ERK1/2 were located in the nucleus of cancer cells. Moreover, there was a significant positive correlation between SMYD2-OE and p-ERK1/2 protein expression in COAD, using statistical analysis. On the basis of the results, the MEK/ERK/AP-1 pathways may mediate the activity of SMYD2-OE in inducing L-OHP resistance in colon cancer. This in turn may interfere with receptors and signal transduction pathways associated with drug metabolism and may have an impact on chemotherapeutic sensitivity of COAD cells. We analyzed the mechanisms and functions of SMYD2-OE in L-OPH resistant COAD using specific siRNA. Further research is also needed to confirm whether small molecule inhibitors of SMYD2-OE could promote L-OHP sensitivity in COAD and its possible mechanisms and, in addition, how SMYD2-OE could regulate MEK/ERK/AP-1 signaling pathway in vitro and in vivo.
Taken together, SMYD2-OE promoted L-OPH resistance in colon cancer cells by mediating P-gp upregulation through MEK/ERK/AP-1 signaling pathway, suggesting that inhibition of SMYD2-OE could be a therapeutic strategy candidate for colon cancer with L-OHP resistance.
Conclusion
SMYD2-OE promotes L-OHP resistance in colon cancer by regulating MDR1/P-glycoprotein through MEK/ERK/AP-1 signaling pathway, providing a potential strategy to sensitize chemotherapy by SMYD2-OE knockdown in colon cancer treatment.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (81502075), the Foundation of Science and Technology of Sichuan Province (2014SZ0055, 2015JY0095), and the Science and Technology Project of the Healthy Planning Committee of Sichuan (17PJ382).
Footnotes
Author contributions
All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Miller KD, Siegel RL, Lin CC, et al. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin. 2016;66(4):271–289. doi: 10.3322/caac.21349. [DOI] [PubMed] [Google Scholar]
- 3.Fang SH, Efron JE, Berho ME, Wexner SD. Dilemma of stage II colon cancer and decision making for adjuvant chemotherapy. J Am Coll Surg. 2014;219(5):1056–1069. doi: 10.1016/j.jamcollsurg.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 4.Lea MA. Recently identified and potential targets for colon cancer treatment. Future Oncol. 2010;6(6):993–1002. doi: 10.2217/fon.10.53. [DOI] [PubMed] [Google Scholar]
- 5.Bonetti A, Giuliani J, Muggia F. Targeted agents and oxaliplatin-containing regimens for the treatment of colon cancer. Anticancer Res. 2014;34(1):423–434. [PubMed] [Google Scholar]
- 6.Pirpour Tazehkand A, Akbarzadeh M, Velaie K, Sadeghi MR, Samadi N. The role of Her2-Nrf2 axis in induction of oxaliplatin resistance in colon cancer cells. Biomed Pharmacother. 2018;103:755–766. doi: 10.1016/j.biopha.2018.04.105. [DOI] [PubMed] [Google Scholar]
- 7.Raymond E, Chaney SG, Taamma A, Cvitkovic E. Oxaliplatin: a review of preclinical and clinical studies. Ann Oncol. 1998;9(10):1053–1071. doi: 10.1023/a:1008213732429. [DOI] [PubMed] [Google Scholar]
- 8.Martinez-Balibrea E, Martínez-Cardús A, Ginés A, et al. Tumor-related molecular mechanisms of oxaliplatin resistance. Mol Cancer Ther. 2015;14(8):1767–1776. doi: 10.1158/1535-7163.MCT-14-0636. [DOI] [PubMed] [Google Scholar]
- 9.Rejhová A, Opattová A, Čumová A, Slíva D, Vodička P. Natural compounds and combination therapy in colorectal cancer treatment. Eur J Med Chem. 2018;144:582–594. doi: 10.1016/j.ejmech.2017.12.039. [DOI] [PubMed] [Google Scholar]
- 10.Huang J, Perez-Burgos L, Placek BJ, et al. Repression of p53 activity by Smyd2-mediated methylation. Nature. 2006;444(7119):629–632. doi: 10.1038/nature05287. [DOI] [PubMed] [Google Scholar]
- 11.Zhang X, Huang Y, Shi X. Emerging roles of lysine methylation on non-histone proteins. Cell Mol Life Sci. 2015;72(22):4257–4272. doi: 10.1007/s00018-015-2001-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho HS, Hayami S, Toyokawa G, et al. RB1 methylation by SMYD2 enhances cell cycle progression through an increase of Rb1 phosphorylation. Neoplasia. 2012;14(6):476–486. doi: 10.1593/neo.12656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamamoto R, Toyokawa G, Nakakido M, Ueda K, Nakamura Y. SMYD2-dependent Hsp90 methylation promotes cancer cell proliferation by regulating the chaperone complex formation. Cancer Lett. 2014;351(1):126–133. doi: 10.1016/j.canlet.2014.05.014. [DOI] [PubMed] [Google Scholar]
- 14.Sajjad A, Novoyatleva T, Vergarajauregui S, et al. Lysine methyltransferase SMYD2 suppresses p53-dependent cardiomyocyte apoptosis. Biochim Biophys Acta Mol Cell Res. 2014;1843(11):2556–2562. doi: 10.1016/j.bbamcr.2014.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Komatsu S, Imoto I, Tsuda H, et al. Overexpression of SMYD2 relates to tumor cell proliferation and malignant outcome of esophageal squamous cell carcinoma. Carcinogenesis. 2009;30(7):1139–1146. doi: 10.1093/carcin/bgp116. [DOI] [PubMed] [Google Scholar]
- 16.Zuo SR, Zuo XC, He Y, et al. Positive expression of SMYD2 is associated with poor prognosis in patients with primary hepatocellular carcinoma. J Cancer. 2018;9(2):321–330. doi: 10.7150/jca.22218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li LX, Zhou JX, Calvet JP, et al. Lysine methyltransferase SMYD2 promotes triple negative breast cancer progression. Cell Death Dis. 2018;9(3):326. doi: 10.1038/s41419-018-0347-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Komatsu S, Ichikawa D, Hirajima S, et al. Overexpression of SMYD2 contributes to malignant outcome in gastric cancer. Br J Cancer. 2015;112(2):357–364. doi: 10.1038/bjc.2014.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sakamoto LH, Andrade RV, Felipe MS, Motoyama AB, Pittella Silva F. Smyd2 is highly expressed in pediatric acute lymphoblastic leukemia and constitutes a bad prognostic factor. Leuk Res. 2014;38(4):496–502. doi: 10.1016/j.leukres.2014.01.013. [DOI] [PubMed] [Google Scholar]
- 20.Ohtomo-Oda R, Komatsu S, Mori T, et al. SMYD2 overexpression is associated with tumor cell proliferation and a worse outcome in human papillomavirus-unrelated nonmultiple head and neck carcinomas. Hum Pathol. 2016;49:145–155. doi: 10.1016/j.humpath.2015.08.025. [DOI] [PubMed] [Google Scholar]
- 21.Zhang X, Luo H. Effects of thalidomide on growth and VEGF-A expression in SW480 colon cancer cells. Oncol Lett. 2018;15(3):3313–3320. doi: 10.3892/ol.2017.7645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu W, Chen F, Fei X, Yang X, Lu X. Overexpression of SET and MYND domain-containing protein 2 (SMYD2) is associated with tumor progression and poor prognosis in patients with papillary thyroid carcinoma. Med Sci Monit. 2018;24:7357–7365. doi: 10.12659/MSM.910168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen S, Wang H, Li Z, et al. Interaction of WBP2 with ERα increases doxorubicin resistance of breast cancer cells by modulating MDR1 transcription. Br J Cancer. 2018;119(2):182–192. doi: 10.1038/s41416-018-0119-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Munkanatta Godage DNP, Vanhecke GC, Samarasinghe KTG, et al. SMYD2 glutathionylation contributes to degradation of sarcomeric proteins. Nat Commun. 2018;9(1):4341. doi: 10.1038/s41467-018-06786-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang C, Liu HZ, Lu WD, Fu ZX. PEG-liposomal oxaliplatin potentialization of antitumor efficiency in a nude mouse tumor-xenograft model of colorectal carcinoma. Oncol Rep. 2011;25(6):1621–1628. doi: 10.3892/or.2011.1238. [DOI] [PubMed] [Google Scholar]
- 26.Siddiqui L, Mishra H, Mishra PK, Iqbal Z, Talegaonkar S. Novel 4-in-1 strategy to combat colon cancer, drug resistance and cancer relapse utilizing functionalized bioinspiring lignin nanoparticle. Med Hypotheses. 2018;121:10–14. doi: 10.1016/j.mehy.2018.09.003. [DOI] [PubMed] [Google Scholar]
- 27.Rabik CA, Dolan ME. Molecular mechanisms of resistance and toxicity associated with platinating agents. Canc Treat Rev. 2007;33(1):9–23. doi: 10.1016/j.ctrv.2006.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bahrami A, Amerizadeh F, Hassanian SM, et al. Genetic variants as potential predictive biomarkers in advanced colorectal cancer patients treated with oxaliplatin-based chemotherapy. J Cell Physiol. 2018;233(3):2193–2201. doi: 10.1002/jcp.25966. [DOI] [PubMed] [Google Scholar]
- 29.Sipos F, Constantinovits M, Műzes G. Intratumoral functional heterogeneity and chemotherapy. World J Gastroenterol. 2014;20(10):2429–2432. doi: 10.3748/wjg.v20.i10.2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang R, Deng X, Yoshioka Y, et al. Effects of SMYD2-mediated EML4-ALK methylation on the signaling pathway and growth in non-small-cell lung cancer cells. Cancer Sci. 2017;108(6):1203–1209. doi: 10.1111/cas.13245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hamamoto R, Nakamura Y. Dysregulation of protein methyltransferases in human cancer: an emerging target class for anticancer therapy. Cancer Sci. 2016;107(4):377–384. doi: 10.1111/cas.12884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu J, Cheung T, Grande C, et al. Biochemical characterization of human SET and MYND domain-containing protein 2 methyltransferase. Biochemistry. 2011;50(29):6488–6497. doi: 10.1021/bi200725p. [DOI] [PubMed] [Google Scholar]
- 33.Nies AT, Magdy T, Schwab M, Zanger UM. Role of ABC transporters in fluoropyrimidine-based chemotherapy response. Adv Cancer Res. 2015;125:217–243. doi: 10.1016/bs.acr.2014.10.007. [DOI] [PubMed] [Google Scholar]
- 34.de Iudicibus S, de Pellegrin A, Stocco G, et al. ABCB1 gene polymorphisms and expression of P-glycoprotein and long-term prognosis in colorectal cancer. Anticancer Res. 2008;28(6B):3921–3928. [PubMed] [Google Scholar]
- 35.Plisson F, Huang XC, Zhang H, Khalil Z, Capon RJ. Lamellarins as inhibitors of P-glycoprotein-mediated multidrug resistance in a human colon cancer cell line. Chem Asian J. 2012;7(7):1616–1623. doi: 10.1002/asia.201101049. [DOI] [PubMed] [Google Scholar]