Abstract
Maternal inflammation and diabetes increase the risk for psychiatric disorders in offspring. We hypothesized that these co-occurring risk factors with may potentiate each other. To test this, we maternally exposed developing mice in utero to gestational diabetes mellitus (GDM) and/or maternal immune activation (MIA). Fetal mouse brains were exposed to either vehicle, GDM, MIA, or GDM+MIA. At gestational day (GD) 12.5, GDM produced a hyperglycemic, hyperleptinemic maternal state whereas MIA produced significant increases in pro-inflammatory cytokines and chemokines. Each condition alone resulted in an altered, inflammatory and neurodevelopmental transcriptome profile. In addition, GDM+ MIA heightened the maternal inflammatory state and gave rise to a new, specific transcriptional response. This exacerbated response was associated with pathways implicated in psychiatric disorders, including dopamine neuron differentiation and innate immune response. Based on these data, we hypothesize that children born to GDM mothers and exposed to mid-gestation infections have an increased vulnerability to psychiatric disorder later in life, and this should be tested in follow-up epidemiological studies.
Introduction
It is increasingly recognized that the prenatal environment influences the risk for developing disease later in life 1. Multiple studies have defined links between diverse stressors during pregnancy and the risk to offspring, including neurocognitive problems 2, 3. The majority of studies linking prenatal stress to developmental origins of disease in offspring have focused on a single type of stress (e.g. diabetes or infection). However, human populations are routinely subjected to more than one potential developmental stressor at a time, and recent studies suggest that such interactions are important in the context of future health outcomes (reviewed in 4).
Increased risk for psychiatric disease is a result of genetic liability interacting with a variety of environmental perturbations. Numerous gene x environment (G*E) interactions have been identified, such as BDNF variants co-occurring with serious obstetric complications increasing risk for schizophrenia 2. Yet environment x environment (E*E) interactions on the developing fetus are greatly understudied in comparison. Two commonly co-occurring psychiatric disorder risk factors that alter the in utero environment are maternal immune activation (MIA) and gestational diabetes mellitus (GDM) 5–8.
Both animal and epidemiologic studies support that midgestation influenza infection increases risk for autism and schizophrenia 5, 9, 10. However, unlike some viruses (e.g. Rubella, Zika), influenza virus does not cross the placenta, suggesting that the maternal immune response (and not the pathogen) is the key to pathology in this setting 11, 12. This process of viral-induced MIA is modeled in pregnant mice using viral mimetics such as polyinosinic:polycytidylic acid (poly(I:C)), a synthetic, double-stranded RNA agonist of Toll-like receptor 3 11, 12. In fact, it has been repeatedly established with multiple models that MIA disrupts normal fetal brain development 10, 13 and that mice born to mothers exposed to MIA in adulthood display behavioral abnormalities that are relevant to both schizophrenia and ASD 12, 14.
GDM is the most common metabolic disorder during pregnancy, causing considerable morbidity and long-term complications for both mother and child 15. Recent birth cohort studies have demonstrated that diabetes during pregnancy increases risk for autism spectrum disorder and potentially schizophrenia 8, 16, 17, and offspring of GDM rodent models exhibit a wide-range of neurodevelopmental, behavioral, and cognitive abnormalities 18, 19. Furthermore, hyperglycemia and hyperinsulinemia can increase systemic inflammation and exaggerate and/or prolong responsiveness to pro-inflammatory stimuli, which supports a possible interaction of GDM and MIA 20–24.
Disruptions to the normal immune homeostasis within the fetus or placenta can disrupt neurodevelopment and predispose to psychiatric disorders 7, 25. This inflammation does not have to be chronic or excessive to predispose to psychiatric disorders but depends greatly on the contextual factors it is combined with 26, 27. Both MIA and GDM have their peak influence at midgestation 8, 28, and previous studies suggest that GDM may interact with the pro-inflammatory state produced by MIA 20–23, 29, 30. Resulting changes in the maternal state can potentially disrupt the in utero environment, such as increases in interleukin (IL)-6, which can alter placental function and development and, in turn, might negatively impact fetal brain development 31–33. The purpose of this study was to interrogate the interaction between MIA and GDM during prenatal development, hypothesizing that these two common and potentially co-occurring factors may potentiate each other.
Methods
Animal procedures
All animal procedures were approved by the Vanderbilt Animal Care and Use Committee. Mice were housed in ventilated cages under standard laboratory conditions and allowed ad libitum access to food and water. Female and male C57Bl/6J mice were obtained from The Jackson Laboratory (Bar Harbor, ME, USA) for timed pregnant breeding. Control fed female mice received standard chow throughout the experiment (5LOD, Lab Diet, St. Louis, MO, USA). To induce GDM, females received a 60% calories-by-fat diet (58Y1, Test Diet, St. Louis, MO, USA) from 4 weeks of age until 10 weeks of age, continuing the high fat diet throughout pregnancy. At 10 weeks of age, mice were mated overnight, and the presence of a vaginal plug marked gestational day 0.5 (GD0.5). Pregnant females were left undisturbed except for cage changes at GD9.5 and weight measurements.
GDM and control females were assigned to either saline or poly(I:C) treatment groups. Samples were balanced between MIA and control each week, providing randomization within a constraints of the breeding scheme. This design created 4 groups: control fed saline (SAL or Control), high fat fed saline (GDM), control fed poly(I:C) (MIA), and high fat fed poly(I:C) (GDM+MIA). GD12.5 was chosen as the midgestation timepoint in mice, which is neurodevelopmentally distinct from midgestation in humans. At GD12.5, pregnant females were injected intraperitoneally with either sterile saline or 20 mg/kg poly(I:C) potassium salt (Sigma Aldrich, St. Louis, MO, USA) in sterile saline, which was based on the weight of the poly(I:C) itself. Pregnant mice were sacrificed 3 hours after injection at GD12.5. Data from a second cohort of pregnant mice sacrificed 4 days after injection at GD16.5 were used to evaluate the evolution of these effects after the immune stimulus passed. Sex genotyping was performed for each embryo using a previously published protocol 34. Each group at each time point (4 groups, 2 time points) contained 9 pregnant females with the exception of GD12.5 MIA, which only contained 8 pregnant females, making a total of 71 females. Sample size was chosen based on empirical data from previous literature findings, as well as power calculations.
Glucose tolerance testing and body composition analysis
A separate cohort of female mice underwent the same feeding, mating, and treatment regime as described above for the purposes of confirming the presence of GDM via metabolic testing and glucose tolerance testing (n= 6 per group). At GD12.5, pregnant control fed and high fat fed females were fasted for 6 hours (7 AM–1 PM). Body composition was then measured via whole body NMR (Minispec Model mq7.5, Bruker Instruments) with the assistance of the Vanderbilt Mouse Metabolic Phenotyping Center (supported by U24 DK059637). At 1 PM, fasting glucose and insulin samples were taken followed by intraperitoneal glucose tolerance testing with 2g dextrose/kg body weight, which was performed according to standardized procedures 35. To evaluate the effects of time on high fat diet in the absence of pregnancy, one 10–12 week old non-pregnant control group (n=8) was utilized as the control for non-pregnant high fat fed females age 10 weeks (n=7) and age 12 weeks (n=8).
Antibody-conjugated bead array immune profiling
Blood was collected via cardiac puncture and allowed to clot at room temperature for 30 minutes. Samples were centrifuged at 8,000 rpm for 10 minutes at 4°C. Supernatants were centrifuged again at 8,000 RPM at 4°C for 10 minutes to remove residual RBC’s. Serum was stored at −80°C until antibody-conjugated bead array analysis. Luminex® analysis was performed by the Vanderbilt Hormone Core (funded by DK059637 and DK020593). The Luminex® multiplex panels (EMD Millipore, Darmstadt, Germany) used for analysis of the maternal serum were the mouse metabolic hormone panel (insulin and leptin – Cat# MMHMAG-44K), mouse adiponectin (Cat # MADPNMAG-70K-01), and mouse cytokine/chemokine panel 14 plex (eotaxin, IFNγ, IL1α, IL1β, IL4, IL6, IL10, IL12p40, IL13, IL17, KC, MCP1, RANTES, and TNFα-Cat# MCYTOMAG-70K). When values fell outside the normal range of the assay (which occurred for some serum samples for MCP1 and IL-6) they were analyzed as the highest detectable or lowest detectable value as appropriate. Since MCP1 levels were the most consistently upregulated, 3 poly(I:C) treated dams sacrificed at 3 hours post exposure below 2 standard deviation from the MCP1 mean were excluded from the study as non-responders or poor injections.
RNA isolation and gene expression analysis
The right hemisphere from each flash frozen embryonic brain (telencephalon and diencephalon) was utilized for RNA isolation. RNA was isolated in 1 mL Trizol, following the protocol given by the manufacturer. Samples then underwent a Qiagen RNeasy clean-up, following the manufacturer instructions. RNA concentration was measured with a Thermo Scientific Nanodrop 2000. Equal amounts of RNA (1250 ng) from each embryo were pooled for a litter sample. These pooled samples again underwent a Qiagen RNeasy clean-up to ensure absence of contaminants. Nanodrop was utilized to measure the final concentration and ensure purity (260/280 values above 2 and 260/230 values above 1.5). Agilent 2100 Bioanalyzer was utilized to ensure RNA integrity, with all RNA integrity numbers between 9.5 and 10.
150 ng of purified RNA from each pooled litter was used for unblinded Nanostring nCounter gene expression analysis (http://www.nanostring.com/applications/technology). Two panels (mouse inflammation V2 panel-254 genes- and a custom inflammation and neurodevelopment panel – 46 genes-Supplementary Table 1) were utilized to determine gene expression, which was performed according to manufacturer instructions. Target genes were prioritized based upon three criteria: (a) importance in innate and adaptive immune responses to infection, (b) importance in neural development, and (c) existing knowledge regarding changes in gene expression reported in previous studies of MIA or GDM alone. The raw “count”- the number of times the reporter probe was detected in each sample- was normalized to positive and negative spike-ins as well as 5 housekeeping genes (Cltc, Gapdh, Gusb, Hprt, and Tubb5). After normalization, the sum of all counts was calculated and counts greater than log2(3.5) were further used for comparison.
Statistical Approaches
Power analyses were performed using the NCSS sample size software PASS (www.ncss.com/software/pass/). We had >90% power to detect one unit difference in outcome with an estimated standard deviation of 0.2 with a false discovery rate of 0.05. The power of study further improved due to pooling RNA from each litter, thus reducing the effects of individual variability.
To investigate the effects of each exposure, a one-way ANOVA with Bonferonni post hoc analysis was used for the measured fetal or maternal parameters, with p<0.05 considered significant. For transcriptional analysis, unpaired student t-test was utilized to compare values between control and each condition, and significant genes (p<0.05 and false discovery analysis (FDR)<0.01) were used for differential expression analysis. Comparisons between multiple conditions utilized two-way ANOVA with Tukey multiple comparisons correction with significance of p<0.05. Statistics analysis was performed using Graphpad prizm. Gene ontology analysis and weighted functional gene network mapping was performed for each condition with genes at least 1.15-fold change compared to control mice using DAVID Bioinformatics resources and Cytoscape’s Genemania application, respectively 36–39. Venn diagram analyses were generated using Venny. Data represent average of eight or nine biological replicates.
Results
High fat diet and poly(I:C) treatment affect dam weight
Poly(I:C) treatment has been shown to cause a transient increase in cytokine production, accompanied by anorexia, fever, and sickness behaviors that usually resolves in 24–48 hours. The poly(I:C) treated dams in our study either lost weight or failed to gain weight compared to saline treated dams (weight change±SEM: SAL = 0.60±0.33, MIA = −0.83±0.34; GDM = 1.33±0.25, GDM+MIA = −0.67±0.31), p<0.05. Neither diet nor treatment altered the average number of embryos per litter or the male:female ratio (Supplementary Table 2).
High fat diet leads to GDM mid-gestation
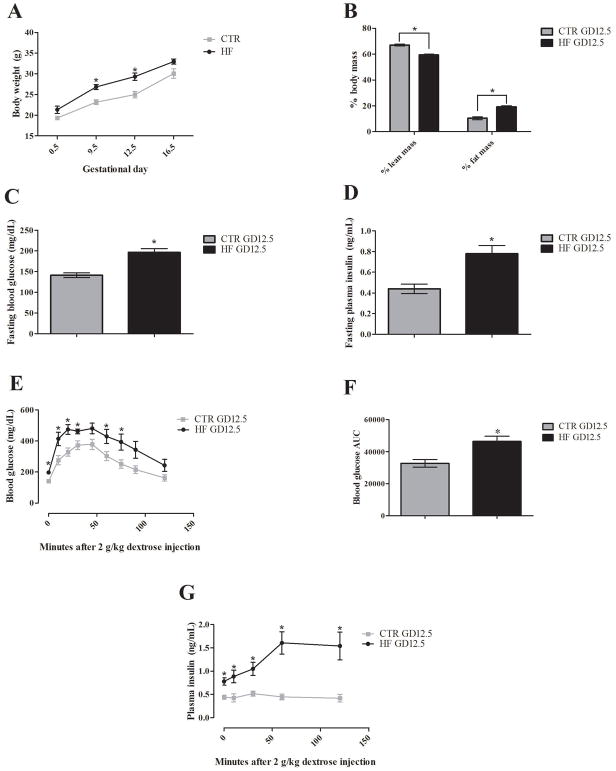
Although the dams were on a 60% by calorie fat diet for 6 weeks prior to mating and throughout pregnancy, these mice were not obese compared the controls. Compared to control diet fed dams, the high fat fed dams had increased body weight (Weight±SEM (g): control = 25±0.8, high fat = 29.3±0.9), p<0.01, and percent fat mass (% ± SEM: control = 10.3±1.1, high fat = 19.2±0.9), p<0.0001, at GD12.5 (Figure 1A/B). Even so, this model successfully produced a GDM phenotype. To demonstrate this, glucose tolerance testing was conducted on mice fed normal or high fat diets. The high fat diet fed dams had both impaired glucose (glucose ± SEM (mg/dL): control = 141±6, high fat = 197±9), p<0.0001, and increased insulin (insulin ± SEM: control = 0.44±0.1, high fat = 0.78±0.1), p<0.01, levels after a 6 hour fast (Figure 1C/D). When challenged with 2 g/kg dextrose after the 6 hour fast, high fat diet fed dams displayed significant glucose intolerance (AUC±SEM: control = 32653±2361, high fat = 46528±3172), p<0.01, and exaggerated glucose-induced insulin secretion that remained high even at 2 hours post-injection (insulin±SEM (ng/mL): control = 0.42±0.1, high fat = 1.54±0.3), p<0.01 (Figure 1E/F/G). Confirming that this phenotype is truly gestation-dependent, neither high fat nor control diet fed dams demonstrated this phenotype at GD0.5 (Supplementary Figure 1). Similarly, age-matched, non-pregnant high fat and control diet fed dams at GD12.5 did not report differences in these GD measures (Supplementary Figure 2).
Figure 1. High fat diet produces a diabetic phenotype midgestation.
Body composition, 6 hour fasting glucose and insulin, and glucose tolerance measurements were performed on a cohort of high fat and control fed GD12.5 pregnant dams (n=6 per group, HF and CTR in graph, respectively). (A) High fat fed dams showed significant increases in body weight at GD9.5 and GD12.5. (B) Whole body NMR at GD12.5 showed significant changes in % body mass in high fat dams with increased % fat mass and decreased % lean body mass. (C–D) 6 hour fasting blood glucose and serum insulin were both significantly increased in high fat dams. (E–G) After a 6 hour fast, dams were injected intraperitoneally with 2 g dextrose/kg body weight. High fat fed dams demonstrated a higher peak and prolonged increase in blood glucose with significant differences from control dams observable at 0 10, 20, 30, 60, and 75 minutes post-injection, which is supported by a significantly increased area under the curve. A significant heightened insulin response that was maintained up until the 2 hour end point was also observed in high fat dams. Error bars represent SEM. Significance determined by unpaired student t-test with Welch’s correction in all assays. (* = p <0.05, n=6 dams per group)
MIA produces an acute increase in maternal cytokines and chemokines
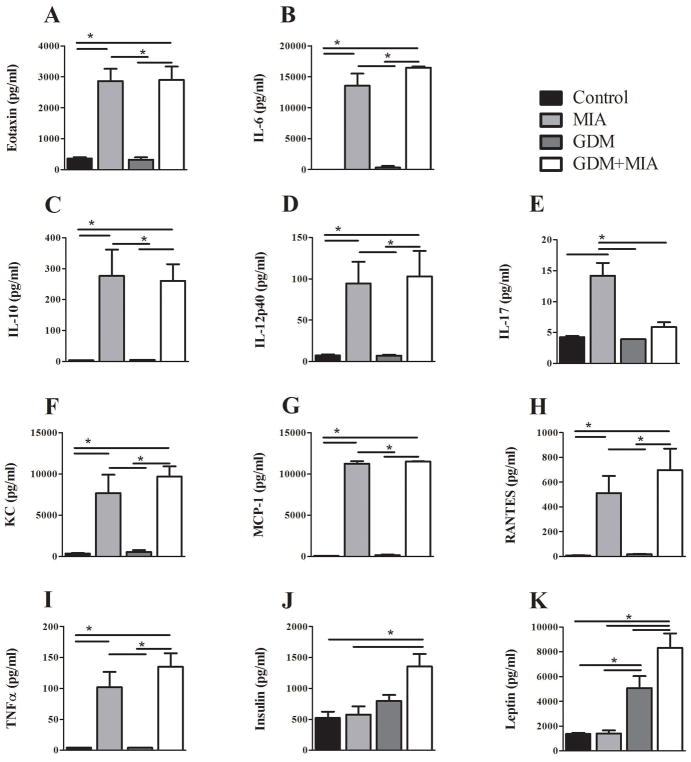
To assess the poly(I:C) induced immune response in pregnant dams as well as differences between normal and GDM mice, cytokines and chemokines were measured in maternal serum at the time of sacrifice. Similar to previous observations of the acute response to poly(I:C) 40, 41 three hours post poly(I:C) exposure MIA and GDM+MIA dams demonstrated increases in eotaxin, IL-1β, IL-6, IL-10, IL-12p40, IL-17, KC, TNF-α, MCP1 (CCL2), and RANTES (CCL5) (p<0.05) (Figure 2) No significant differences were found with IFNγ, IL1-α, IL1-β, or IL-4 (Supplementary Table 3). Perhaps not surprisingly, by GD16.5 almost no serum factor changes with MIA were observed, suggesting that the effects of single poly(I:C) injection had largely subsided four by days post exposure (Supplementary Figure 3).
Figure 2. MIA and GDM together produce metabolic and inflammatory disruption of maternal serum factors.
Maternal serum collected 3 hours post injection on GD12.5 was analyzed for chemokine, cytokine, and adipokine levels via antibody-conjugated bead assay. (AI) Significant increases were observed with MIA and MIA+GDM in most cytokines/chemokines measured without any difference between MIA and MIA+ GDM or attributable to GDM alone. Only IL-17 (E) was significantly increased by GDM alone. (J) GDM alone did not alter non-fasting serum insulin levels, but GDM+MIA produced a significant increase. (K) The pro-inflammatory satiety hormone leptin was significantly increased by GDM, and this increase was even more prominent in the GDM+MIA group. Error bars represent SEM. Significance determined by one-way ANOVA with Bonferonni post hoc analysis in all assays. Serum was collected from dams whose embryos were utilized for gene expression analysis (* = p<0.05, SAL n=9, MIA n=8, GDM n=9, GDM+MIA n=9).
Maternal metabolic hormones are altered by GDM+MIA
Non-fasting serum insulin levels reported only a non-significant increase in the GDM dams, while the combination of GDM and MIA exposure produced a strong, significant increase in serum insulin levels (insulin ± SEM (pg/mL): SAL = 523±102, MIA = 575±135; GDM = 800±94, GDM+MIA = 1353±203), p<0.05 (Figure 2J). Interestingly, the immune-activating, satiety-inducing hormone leptin was significantly increased by GDM and further increased by combination of GDM and MIA exposure, but was not altered by MIA alone (leptin ± SEM (pg/mL): SAL = 1358±94, MIA = 1389 ±256; GDM = 5064±977, GDM+MIA = 8316±1173), p<0.05 (Figure 2K). This increase persisted in the GDM+MIA group at GD16.5 (leptin ± SEM (pg/mL): SAL = 1320±203, MIA = 1184±188; GDM = 3288±569, GDM+MIA = 3940±1177), p<0.05 (Supplementary Figure 3). In contrast, MIA exposure alone did not significantly alter leptin or insulin levels at either GD12.5 or GD16.5.
Inflammation and development related gene expression in the embryonic brain is disrupted by GDM and MIA
Our gene expression investigation focused on a panel of literature- and hypothesis-derived transcripts that encompassed genes involved in inflammation, neurodevelopment, and neuronal glucose transporters. Both high fat diet-induced GDM and MIA resulted in altered gene expression in the fetal brain at GD12.5. When comparing significantly different (p<0.05, FDR<0.01) gene expression changes between SAL, MIA, GDM, or GDM+MIA in an unsupervised hierarchical clustering analysis, each of the four treatment groups demonstrated specific gene expression profiles (Supplementary Figure 4, Supplementary Table 4).
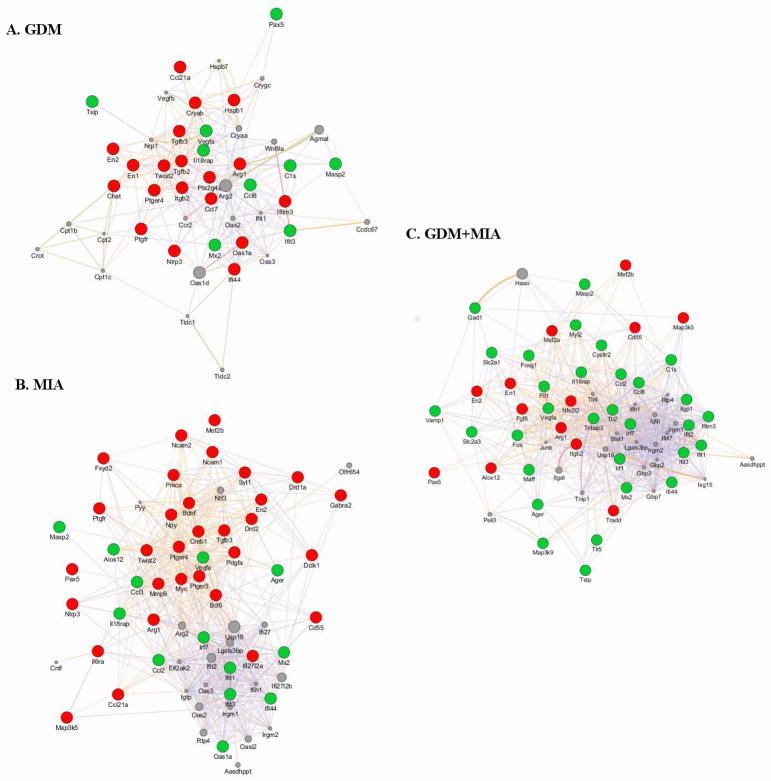
GDM alone displayed significant differences in gene expression compared to SAL. Neurodevelopmental genes involved in patterning (En1, En2) and cholinergic signaling (Chat) were repressed 42. A subset of inflammation-associated genes were induced (e.g. Cls, Ccl8, Ifit3, Mx2) while others were repressed (e.g. Ccl21a, Ifi44, Tgfb2, Tgfb3), with these genes functioning in IFN/antiviral response, growth, cell cycle regulation, and apoptosis 43. Moreover, GDM increased Vegfa expression, which is associated with a hypoxic environment (Figure 3A) 44.
Figure 3. MIA, GDM, and GDM+MIA produce unique transcriptional changes in the developing fetal brain.
Genes significantly altered in each condition compared to control were utilized for functional gene network diagrams. Predicted functional gene networks were generated with Genemania’s algorithm available within Cytoscape 38, 39. For each condition in comparison to control, genes significantly induced are shown in green whereas genes significantly repressed are shown in red. Gray nodes represent additional genes strongly associated with the predicted network, with node size proportional to strength of association of gene within the network. Lines demonstrate functional association with line thickness indicating association strength. Significant genes were determined by one-way ANOVA with post hoc Bonferonni correction (p<0.05, FDR<0.01). Gene symbols are listed according to HUGO Gene Nomenclature Committee 75.
Our lab has previously characterized MIA-triggered gene expression changes in the fetus after maternal poly(I:C) treatment at GD12.5 10. Here again, we report altered inflammation responsive gene expression with MIA alone compared to SAL (Figure 3B). We noted both induction of antiviral/IFN response genes (e.g. Ifi44, Ifit3, Mx2, Oasl1) and Alox12, which produces a lipid peroxidation enzyme 43, 45, 46. Repressed inflammation genes observed participate in cell cycle regulation/apoptosis (Bcl6, Cd55, Map3k5, Mef2b, Myc), cell growth (Pdgfa, Tgfb3), and intracellular signaling pathways (Creb1, Fxyd2) 47–49. Yet, the MIA- and GDM-dependent expression profiles were also distinct, as MIA repressed many unique neurodevelopment-associated transcripts (e.g. Bdnf, Dckl1, Gabra2, Ncam1, Syt1) 50. Furthermore, Vegfa was induced by MIA, suggesting alteration in oxygen and nutrients reaching the fetal brain 44, 51.
GDM potentiates MIA induced gene expression changes
Three hours following injection, poly(I:C) treatment in the context of GDM produced marked induction of many inflammation and neurodevelopment genes (Figure 3C). GDM+MIA both induced (Foxg1, Gad1, Vamp1) and repressed (En1, En2, Fgf8, Pax5) subsets of neurodevelopment genes involved in patterning and migration 50, 52–54. Moreover, both the constitutively expressed GLUT1 and high affinity neuronal specific GLUT3 genes (Slc2a1, Slc2a3) and hypoxia-associated Vegfa were induced in GDM+MIA. Inflammation genes induced by GDM+MIA are mostly involved in antiviral/IFN response (e.g. Ifi44, Ifit3, Ifitm3, Mx2, Oasl1) and the generalized innate immune response (e.g. Ager, Ccl2, Ccl8)43, 45, 49. Contrastingly, several apoptosis/cell cycle regulation (e.g. Mef2a, Mef2b, Tradd) associated genes were repressed by GDM+MIA 43, 47, 48. GDM+MIA also repressed genes critical for intracellular signaling pathways associated with pro- and anti-inflammatory processes (Map3k5, Nfe2l2), immune response (Cd55, Itgb2), and neurodevelopment (En1, En2, Fgf8, Pax5) 55. Altogether, our data suggest further disruption of the fetal brain by the complex metabolic, inflammatory derangement produced by GDM+MIA, including both unique and differentially altered MIA- and/or GDM-associated transcription changes (Figure 4). This unique phenotype is enriched in dopamine neuron differentiation and multiple inflammatory pathways (Figure 5).
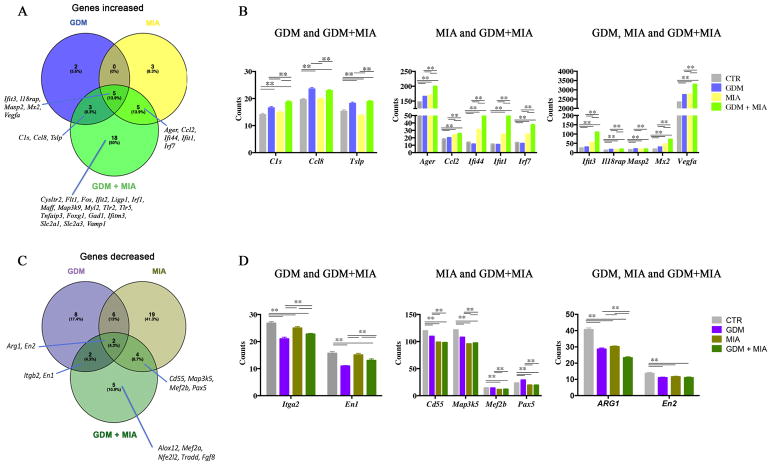
Figure 4. GDM and MIA alter expression of neurodevelopmental and inflammatory genes at GD12.5.
Venn diagrams denote the number of genes increased (A) or decreased (B) in GDM, MIA, or GDM+MIA when compared to control mice. The graphics represent gene counts increased (C) or decreased (D) in GDM and GDM+MIA, MIA and GDM+MIA or GDM, MIA and GDM+MIA as indicated. (A and B) Significant changes in gene expression were determined by multiple t tests, and genes with p<0.05, FDR<0.01 were selected. (C and D) Tukey’s multiple comparisons 2way ANOVA was used to determine significance among the groups. Gene symbols are listed according to HUGO Gene Nomenclature Committee 75.
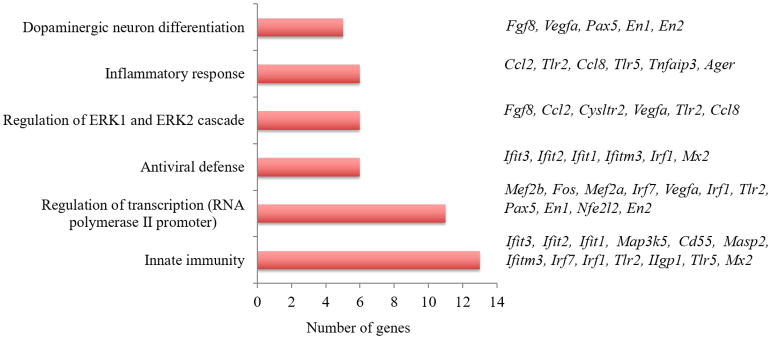
Figure 5. Differentially expressed transcripts in GDM+MIA condition at GD12.5.
Gene Ontology (GO) enrichment analysis was performed using genes significantly altered in GDM+MIA when compared to control mice fed a normal diet and treated with saline. Clustering was performed using DAVID bioinformatics database analysis (p<0.05) 36.
At GD16.5, smaller and fewer changes in relative gene expression were detected with MIA and GDM+MIA, which should be expected since the acute immune response induced by poly(I:C) exposure passes after 24–48 hours (Supplementary Figure 5, Supplementary Table 5). In contrast, we observed a similar number of gene expression changes produced by GDM alone at GD16.5 compared with those observed at GD12.5, including induction of neurodevelopment genes (En2, Pax5) and both repression (Ccr1, Cd163, Itgb2) and induction (Hras1, Ptger3) of inflammation genes. This supports continued effects of GDM on the fetal brain. This evolving phenotype reflected ongoing changes in neurodevelopment and immune response in the developing brain.
Discussion
There is abundant epidemiological evidence supporting an association between either GDM or infection-associated MIA and adverse neurocognitive outcomes in offspring, including autism and schizophrenia 5, 8, 9, 16. However, the extent to which these two stressors interact has not been closely evaluated to date. Potential interactions between GDM and MIA are of great clinical relevance in developed countries such as the United States, where the incidence of GDM is increasing 15, and where infections pose a persistent threat to pregnant mothers. In low- or middle-income countries this may be even more important in light of high rates of infectious diseases and recent transitions from undernutrition to excess adiposity 4.
Herein we present new evidence for a significant interaction between GDM and MIA in the developing fetal brain. These two environmental insults commonly occur during gestation and are associated with increased risk for psychiatric disorders later in life 3, 5, 8, 9, 56. The combination of these insults demonstrate that 1) GDM and MIA individually alter circulating factors in maternal serum and alter inflammation-associated and neurodevelopmental embryonic gene expression, 2) GDM and MIA have a complex maternal interaction that produces a novel transcriptional phenotype, and 3) these novel transcriptional changes are associated with pathways implicated in psychiatric disorders like dopamine neuron differentiation and innate immune response. These findings provide insight into prenatal interactions that may increase risk for psychiatric disorder development later in life.
Our maternal serum data confirm that MIA induces a pro-inflammatory state. We do not find this altered by co-occurrence with GDM. Yet, results from fetal brain gene expression suggest that metabolic stress (e.g., GDM) primes the fetus for an altered response to MIA. GDM has been associated with low-grade inflammation 29, which might have the potential to produce a “primed” state, altering and potentially exacerbating the response to additional inflammatory stimuli 25, 29. In fact, multiple environmental stressors such as midgestation maternal infection, perinatal hypoxia, or early childhood trauma- are often considered co-factors or triggers for this “primed” state, which may have arisen from genetic and/or environmental insults 57, 58.
In addition, these studies confirmed previous findings in pregnant mice exposed to MIA, where similar transient increases in cytokine and chemokine levels were observed during the acute response to poly(I:C) 40, 41, 59. In contrast to the acute phase, 1 – 8 days post injection most studies have reported subtle or no increases in cytokines or chemokines in maternal serum. This is perhaps not surprising given the transient maternal response to poly(I:C) 40, 41, 60. Altogether, our results support that poly(I:C) administered at GD12.5 produces an acute, strong maternal increase in multiple cytokines and chemokines, including IL-6, that is no longer present 4 days post injection. Combined with the gene expression changes in the brains of offspring, we believe that the data argue that this pro-inflammatory cytokine environment poses a significant risk for adverse health outcomes in later life, which should be investigated with additional TLR agonists in future studies 14, 61.
Beyond inflammatory mediators, we investigated the extent to which GDM and MIA induced changes in metabolic hormone levels in the maternal serum. Increased serum leptin and insulin levels, as well as leptin and insulin insensitivity have been reported in adult mice fed a high fat diet 62, 63. In our studies, non-fasting serum insulin levels were significantly affected at GD12.5, regardless of MIA exposure, with no increase noted at GD16.5. In addition, the significant increase in pro-inflammatory leptin levels in observed in GDM increased further with MIA at GD12.5 but diminished by GD16.5 62. Although the effects of GDM on metabolic hormones appeared to remain consistent from GD12.5 to GD16.5, the interaction of MIA and GDM did lessen. We propose that (by a temporary disruption of typical developmental trajectory) this combined acute MIA+GDM insult did leave a lasting consequence on brain development.
Of the transcriptional effects produced by GDM and MIA alone, the hypoxic response caused by both insults is of particular interest. We propose that a MIA-induced hypoxia is a consequence of mounting an antiviral response in the activated fetal brain. In addition, a hypoxia-related expression signature might also reflect the increased oxygen demand due to a heightened metabolic demand associated with maternal hyperglycemia in GDM 64, 65. Regardless of the exact mechanism of Vegfa and Flt1 induction, it is likely to result in vascular remodeling that occurs with chronic hypoxia, which might persist across the lifespan 44, 66, 67. This finding might have important clinical implications, as microvascular and cerebral blood volume alterations appear to increase risk for developing schizophrenia 68, 69.
One of the more important questions we sought to answer was if GDM interacts with MIA to produce a novel phenotype. We did find that MIA differentially affected inflammation and neurodevelopment associated gene expression in the context of GDM. The altered levels of neurodevelopmental transcripts included crucial patterning genes that are part of neuronal fate differentiation of dopaminergic neurons in the developing midbrain 70–72, and altered dopaminergic neuron development has been noted in both GDM and MIA 60, 73. Our data support that disruption of dopaminergic neuronal development can be strongly exacerbated by a combination of multiple deleterious environmental factors, including MIA and GDM. This process might predispose to increased risk of developing neuropsychiatric disorders in later life and should be investigated in future studies.
In summary, we have shown that combined disruptions to the in utero environment interacted to result in an exacerbated transcriptional response in the developing brain. Importantly, neither of these insults (alone or combined) are specific to a particular outcome in later life, but they do predispose to a variety of brain disorders, with diagnosis-specificity being genetic context dependent 74. More specifically, the ultimate effects of co-occurring GDM and infections during pregnancy most likely depend on the genetic makeup of the developing mother and fetus (or both), likely leading to complex GMOTHER*GFETUS*EGDM*EMIA interactions that may be critical for determining specific disorder phenotypes. Regardless of this, based on our data we hypothesize that children born to mothers with GDM, exposed to mid-gestation infections/immune activation, have increased vulnerability to psychiatric disorders later in life, which could and should be tested in large-scale epidemiologic studies.
Supplementary Material
Acknowledgments
The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or other funding agencies. The authors declare no conflict of interest related to the performed work. This work was supported by R01MH079299 (KM) T32MH064913 (KMM), T32GM007347 (KMM), F31DK108652 (TLB), ADA grant 1-16-IBS-100 (MG), JDRF 1-INO-2014-177-A-V (MG), and a Pilot & Feasibility Program of the Vanderbilt Diabetes Research and Training Center (TLB/DMA).
Footnotes
Supplementary information is available at Molecular Psychiatry’s website
References
- 1.Rees S, Harding R, Walker D. An adverse intrauterine environment: implications for injury and altered development of the brain. Int J Dev Neurosci. 2008;26(1):3–11. doi: 10.1016/j.ijdevneu.2007.08.020. [DOI] [PubMed] [Google Scholar]
- 2.Nicodemus KK, Marenco S, Batten AJ, Vakkalanka R, Egan MF, Straub RE, et al. Serious obstetric complications interact with hypoxia-regulated/vascular-expression genes to influence schizophrenia risk. Mol Psychiatry. 2008;13(9):873–877. doi: 10.1038/sj.mp.4002153. [DOI] [PubMed] [Google Scholar]
- 3.Gardener H, Spiegelman D, Buka SL. Prenatal risk factors for autism: comprehensive meta-analysis. Br J Psychiatry. 2009;195(1):7–14. doi: 10.1192/bjp.bp.108.051672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goldstein JA, Norris SA, Aronoff DM. DOHaD at the intersection of maternal immune activation and maternal metabolic stress: a scoping review. Journal of Developmental Origins of Health and Disease. 2017 doi: 10.1017/S2040174417000010. in press. [DOI] [PubMed] [Google Scholar]
- 5.Atladóttir H, Thorsen P, Østergaard L, Schendel D, Lemcke S, Abdallah M, et al. Maternal Infection Requiring Hospitalization During Pregnancy and Autism Spectrum Disorders. J Autism Dev Disord. 2010;40(12):1423–1430. doi: 10.1007/s10803-010-1006-y. [DOI] [PubMed] [Google Scholar]
- 6.Lee BK, Magnusson C, Gardner RM, Blomstrom A, Newschaffer CJ, Burstyn I, et al. Maternal hospitalization with infection during pregnancy and risk of autism spectrum disorders. Brain Behav Immun. 2015;44:100–105. doi: 10.1016/j.bbi.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Lieshout RJ, Voruganti LP. Diabetes mellitus during pregnancy and increased risk of schizophrenia in offspring: a review of the evidence and putative mechanisms. J Psychiatry Neurosci. 2008;33(5):395–404. [PMC free article] [PubMed] [Google Scholar]
- 8.Xiang AH, Wang X, Martinez MP, Walthall JC, Curry ES, Page K, et al. Association of maternal diabetes with autism in offspring. JAMA. 2015;313(14):1425–1434. doi: 10.1001/jama.2015.2707. [DOI] [PubMed] [Google Scholar]
- 9.Brown AS, Begg MD, Gravenstein S, Schaefer CA, Wyatt RJ, Bresnahan M, et al. Serologic evidence of prenatal influenza in the etiology of schizophrenia. Arch Gen Psychiatry. 2004;61(8):774–780. doi: 10.1001/archpsyc.61.8.774. [DOI] [PubMed] [Google Scholar]
- 10.Garbett KA, Hsiao EY, Kalman S, Patterson PH, Mirnics K. Effects of maternal immune activation on gene expression patterns in the fetal brain. Transl Psychiatry. 2012;2:e98. doi: 10.1038/tp.2012.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi L, Tu N, Patterson PH. Maternal influenza infection is likely to alter fetal brain development indirectly: the virus is not detected in the fetus. Int J Dev Neurosci. 2005;23(2–3):299–305. doi: 10.1016/j.ijdevneu.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 12.Shi L, Fatemi SH, Sidwell RW, Patterson PH. Maternal Influenza Infection Causes Marked Behavioral and Pharmacological Changes in the Offspring. The Journal of Neuroscience. 2003;23(1):297–302. doi: 10.1523/JNEUROSCI.23-01-00297.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meyer U. Prenatal Poly(I:C) Exposure and Other Developmental Immune Activation Models in Rodent Systems. Biol Psychiatry. 2014;75(4):307–315. doi: 10.1016/j.biopsych.2013.07.011. [DOI] [PubMed] [Google Scholar]
- 14.Smith SE, Li J, Garbett K, Mirnics K, Patterson PH. Maternal immune activation alters fetal brain development through interleukin-6. J Neurosci. 2007:27. doi: 10.1523/JNEUROSCI.2178-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeSisto CL, Kim SY, Sharma AJ. Prevalence estimates of gestational diabetes mellitus in the United States, Pregnancy Risk Assessment Monitoring System (PRAMS), 2007–2010. Prev Chronic Dis. 2014;11:E104. doi: 10.5888/pcd11.130415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krakowiak P, Walker CK, Bremer AA, Baker AS, Ozonoff S, Hansen RL, et al. Maternal metabolic conditions and risk for autism and other neurodevelopmental disorders. Pediatrics. 2012;129(5):e1121–1128. doi: 10.1542/peds.2011-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cannon M, Jones PB, Murray RM. Obstetric Complications and Schizophrenia: Historical and Meta-Analytic Review. Am J Psychiatry. 2002;159(7):1080–1092. doi: 10.1176/appi.ajp.159.7.1080. [DOI] [PubMed] [Google Scholar]
- 18.Chandna AR, Kuhlmann N, Bryce CA, Greba Q, Campanucci VA, Howland JG. Chronic maternal hyperglycemia induced during mid-pregnancy in rats increases RAGE expression, augments hippocampal excitability, and alters behavior of the offspring. Neuroscience. 2015;303:241–260. doi: 10.1016/j.neuroscience.2015.06.063. [DOI] [PubMed] [Google Scholar]
- 19.Golalipour MJ, Kafshgiri SK, Ghafari S. Gestational diabetes induced neuronal loss in CA1 and CA3 subfields of rat hippocampus in early postnatal life. Folia Morphol (Warsz) 2012;71(2):71–77. [PubMed] [Google Scholar]
- 20.Gyurko R, Siqueira CC, Caldon N, Gao L, Kantarci A, Van Dyke TE. Chronic hyperglycemia predisposes to exaggerated inflammatory response and leukocyte dysfunction in Akita mice. J Immunol. 2006;177(10):7250–7256. doi: 10.4049/jimmunol.177.10.7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ling PR, Smith RJ, Bistrian BR. Acute effects of hyperglycemia and hyperinsulinemia on hepatic oxidative stress and the systemic inflammatory response in rats. Crit Care Med. 2007;35(2):555–560. doi: 10.1097/01.CCM.0000253310.02180.C2. [DOI] [PubMed] [Google Scholar]
- 22.Stegenga ME, van der Crabben SN, Dessing MC, Pater JM, van den Pangaart PS, de Vos AF, et al. Effect of acute hyperglycaemia and/or hyperinsulinaemia on proinflammatory gene expression, cytokine production and neutrophil function in humans. Diabet Med. 2008;25(2):157–164. doi: 10.1111/j.1464-5491.2007.02348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krogh-Madsen R, Moller K, Dela F, Kronborg G, Jauffred S, Pedersen BK. Effect of hyperglycemia and hyperinsulinemia on the response of IL-6, TNF-alpha, and FFAs to low-dose endotoxemia in humans. Am J Physiol Endocrinol Metab. 2004;286(5):E766–772. doi: 10.1152/ajpendo.00468.2003. [DOI] [PubMed] [Google Scholar]
- 24.Lian Q, Dheen ST, Liao D, Tay SS. Enhanced inflammatory response in neural tubes of embryos derived from diabetic mice exposed to a teratogen. J Neurosci Res. 2004;75(4):554–564. doi: 10.1002/jnr.20006. [DOI] [PubMed] [Google Scholar]
- 25.Leboyer M, Oliveira J, Tamouza R, Groc L. Is it time for immunopsychiatry in psychotic disorders? Psychopharmacology (Berl) 2016;233(9):1651–1660. doi: 10.1007/s00213-016-4266-1. [DOI] [PubMed] [Google Scholar]
- 26.Derry HM, Padin AC, Kuo JL, Hughes S, Kiecolt-Glaser JK. Sex Differences in Depression: Does Inflammation Play a Role? Curr Psychiatry Rep. 2015;17(10):78. doi: 10.1007/s11920-015-0618-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berk M, Williams LJ, Jacka FN, O’Neil A, Pasco JA, Moylan S, et al. So depression is an inflammatory disease, but where does the inflammation come from? BMC Med. 2013;11:200. doi: 10.1186/1741-7015-11-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buka SL, Tsuang MT, Torrey EF, Klebanoff MA, Bernstein D, Yolken RH. Maternal infections and subsequent psychosis among offspring. Arch Gen Psychiatry. 2001;58(11):1032–1037. doi: 10.1001/archpsyc.58.11.1032. [DOI] [PubMed] [Google Scholar]
- 29.Abell SK, De Courten B, Boyle JA, Teede HJ. Inflammatory and Other Biomarkers: Role in Pathophysiology and Prediction of Gestational Diabetes Mellitus. Int J Mol Sci. 2015;16(6):13442–13473. doi: 10.3390/ijms160613442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deverman BE, Patterson PH. Cytokines and CNS development. Neuron. 2009;64(1):61–78. doi: 10.1016/j.neuron.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 31.Li HP, Chen X, Li MQ. Gestational diabetes induces chronic hypoxia stress and excessive inflammatory response in murine placenta. Int J Clin Exp Pathol. 2013;6(4):650–659. [PMC free article] [PubMed] [Google Scholar]
- 32.Hsiao EY, Patterson PH. Activation of the maternal immune system induces endocrine changes in the placenta via IL-6. Brain, Behav, Immun. 2011;25(4):604–615. doi: 10.1016/j.bbi.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goeden N, Velasquez J, Arnold KA, Chan Y, Lund BT, Anderson GM, et al. Maternal Inflammation Disrupts Fetal Neurodevelopment via Increased Placental Output of Serotonin to the Fetal Brain. The Journal of Neuroscience. 2016;36(22):6041–6049. doi: 10.1523/JNEUROSCI.2534-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jacobs MM, Fogg RL, Emeson RB, Stanwood GD. ADAR1 and ADAR2 expression and editing activity during forebrain development. Dev Neurosci. 2009;31(3):223–237. doi: 10.1159/000210185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ayala JE, Samuel VT, Morton GJ, Obici S, Croniger CM, Shulman GI, et al. Standard operating procedures for describing and performing metabolic tests of glucose homeostasis in mice. Dis Model Mech. 2010;3(9–10):525–534. doi: 10.1242/dmm.006239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.St Laurent G, Shtokalo D, Tackett MR, Yang Z, Vyatkin Y, Milos PM, et al. On the importance of small changes in RNA expression. Methods. 2013;63(1):18–24. doi: 10.1016/j.ymeth.2013.03.027. [DOI] [PubMed] [Google Scholar]
- 38.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38(Web Server issue):W214–220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mandal M, Donnelly R, Elkabes S, Zhang P, Davini D, David BT, et al. Maternal immune stimulation during pregnancy shapes the immunological phenotype of offspring. Brain, Behav, Immun. 2013;33:33–45. doi: 10.1016/j.bbi.2013.04.012. [DOI] [PubMed] [Google Scholar]
- 41.Arrode-Bruses G, Bruses JL. Maternal immune activation by poly I:C induces expression of cytokines IL-1beta and IL-13, chemokine MCP-1 and colony stimulating factor VEGF in fetal mouse brain. J Neuroinflammation. 2012;9:83. doi: 10.1186/1742-2094-9-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sadler TW, Liu ET, Augustine KA. Antisense targeting of engrailed-1 causes abnormal axis formation in mouse embryos. Teratology. 1995;51(5):292–299. doi: 10.1002/tera.1420510505. [DOI] [PubMed] [Google Scholar]
- 43.Liu SY, Sanchez DJ, Cheng G. New developments in the induction and antiviral effectors of type I interferon. Curr Opin Immunol. 2011;23(1):57–64. doi: 10.1016/j.coi.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pearce WJ, Butler SM, Abrassart JM, Williams JM. Fetal cerebral oxygenation: the homeostatic role of vascular adaptations to hypoxic stress. Adv Exp Med Biol. 2011;701:225–232. doi: 10.1007/978-1-4419-7756-4_30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou X, Michal JJ, Zhang L, Ding B, Lunney JK, Liu B, et al. Interferon induced IFIT family genes in host antiviral defense. Int J Biol Sci. 2013;9(2):200–208. doi: 10.7150/ijbs.5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dobrian AD, Lieb DC, Cole BK, Taylor-Fishwick DA, Chakrabarti SK, Nadler JL. Functional and pathological roles of the 12- and 15-lipoxygenases. Prog Lipid Res. 2011;50(1):115–131. doi: 10.1016/j.plipres.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, et al. Comprehensive Identification of Cell Cycle–regulated Genes of the Yeast Saccharomyces cerevisiae by Microarray Hybridization. Mol Biol Cell. 1998;9(12):3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Demoulin J-B, Essaghir A. PDGF receptor signaling networks in normal and cancer cells. Cytokine Growth Factor Rev. 2014;25(3):273–283. doi: 10.1016/j.cytogfr.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 49.Barton GM, Medzhitov R. Toll-Like Receptor Signaling Pathways. Science. 2003;300(5625):1524–1525. doi: 10.1126/science.1085536. [DOI] [PubMed] [Google Scholar]
- 50.Marin O. Cellular and molecular mechanisms controlling the migration of neocortical interneurons. Eur J Neurosci. 2013;38(1):2019–2029. doi: 10.1111/ejn.12225. [DOI] [PubMed] [Google Scholar]
- 51.Salihagić-Kadić A, Medić M, Jugović D, Kos M, Latin V, Kušan Jukić M, et al. Fetal cerebrovascular response to chronic hypoxia—implications for the prevention of brain damage. The Journal of Maternal-Fetal & Neonatal Medicine. 2006;19(7):387–396. doi: 10.1080/14767050600637861. [DOI] [PubMed] [Google Scholar]
- 52.Ohtsuka N, Badurek S, Busslinger M, Benes FM, Minichiello L, Rudolph U. GABAergic neurons regulate lateral ventricular development via transcription factor Pax5. Genesis. 2013;51(4):234–245. doi: 10.1002/dvg.22370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Walshe J, Mason I. Unique and combinatorial functions of Fgf3 and Fgf8 during zebrafish forebrain development. Development. 2003;130(18):4337–4349. doi: 10.1242/dev.00660. [DOI] [PubMed] [Google Scholar]
- 54.Danesin C, Houart C. A Fox stops the Wnt: implications for forebrain development and diseases. Curr Opin Genet Dev. 2012;22(4):323–330. doi: 10.1016/j.gde.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 55.Janeway CA, Jr, Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 56.Yamashita Y, Fujimoto C, Nakajima E, Isagai T, Matsuishi T. Possible association between congenital cytomegalovirus infection and autistic disorder. J Autism Dev Disord. 2003;33(4):455–459. doi: 10.1023/a:1025023131029. [DOI] [PubMed] [Google Scholar]
- 57.Millan MJ, Andrieux A, Bartzokis G, Cadenhead K, Dazzan P, Fusar-Poli P, et al. Altering the course of schizophrenia: progress and perspectives. Nat Rev Drug Discov. 2016 doi: 10.1038/nrd.2016.28. advance online publication. [DOI] [PubMed] [Google Scholar]
- 58.Maes M, Yirmyia R, Noraberg J, Brene S, Hibbeln J, Perini G, et al. The inflammatory & neurodegenerative (I&ND) hypothesis of depression: leads for future research and new drug developments in depression. Metab Brain Dis. 2009;24(1):27–53. doi: 10.1007/s11011-008-9118-1. [DOI] [PubMed] [Google Scholar]
- 59.Khan D, Fernando P, Cicvaric A, Berger A, Pollak A, Monje FJ, et al. Long-term effects of maternal immune activation on depression-like behavior in the mouse. Transl Psychiatry. 2014;4:e363. doi: 10.1038/tp.2013.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Meyer U, Engler A, Weber L, Schedlowski M, Feldon J. Preliminary evidence for a modulation of fetal dopaminergic development by maternal immune activation during pregnancy. Neuroscience. 2008;154(2):701–709. doi: 10.1016/j.neuroscience.2008.04.031. [DOI] [PubMed] [Google Scholar]
- 61.Pratt L, Ni L, Ponzio NM, Jonakait GM. Maternal inflammation promotes fetal microglial activation and increased cholinergic expression in the fetal basal forebrain: role of interleukin-6. Pediatr Res. 2013;74(4):393–401. doi: 10.1038/pr.2013.126. [DOI] [PubMed] [Google Scholar]
- 62.Gallou-Kabani C, Vige A, Gross MS, Rabes JP, Boileau C, Larue-Achagiotis C, et al. C57BL/6J and A/J mice fed a high-fat diet delineate components of metabolic syndrome. Obesity (Silver Spring) 2007;15(8):1996–2005. doi: 10.1038/oby.2007.238. [DOI] [PubMed] [Google Scholar]
- 63.Van Heek M, Compton DS, France CF, Tedesco RP, Fawzi AB, Graziano MP, et al. Diet-induced obese mice develop peripheral, but not central, resistance to leptin. J Clin Invest. 1997;99(3):385–390. doi: 10.1172/JCI119171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cerf ME, Williams K, van Rooyen J, Esterhuyse AJ, Muller CJ, Louw J. Gestational 30% and 40% fat diets increase brain GLUT2 and neuropeptide Y immunoreactivity in neonatal Wistar rats. Int J Dev Neurosci. 2010;28(7):625–630. doi: 10.1016/j.ijdevneu.2010.07.226. [DOI] [PubMed] [Google Scholar]
- 65.Bastian TW, Santarriaga S, Nguyen TA, Prohaska JR, Georgieff MK, Anderson GW. Fetal and neonatal iron deficiency but not copper deficiency increases vascular complexity in the developing rat brain. Nutr Neurosci. 2015;18(8):365–375. doi: 10.1179/1476830515Y.0000000037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Osada-Oka M, Ikeda T, Imaoka S, Akiba S, Sato T. VEGF-enhanced proliferation under hypoxia by an autocrine mechanism in human vascular smooth muscle cells. J Atheroscler Thromb. 2008;15(1):26–33. doi: 10.5551/jat.e533. [DOI] [PubMed] [Google Scholar]
- 67.Nilsson I, Shibuya M, Wennstrom S. Differential activation of vascular genes by hypoxia in primary endothelial cells. Exp Cell Res. 2004;299(2):476–485. doi: 10.1016/j.yexcr.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 68.Talati P, Rane S, Skinner J, Gore J, Heckers S. Increased hippocampal blood volume and normal blood flow in schizophrenia. Psychiatry Res. 2015;232(3):219–225. doi: 10.1016/j.pscychresns.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Meier MH, Shalev I, Moffitt TE, Kapur S, Keefe RS, Wong TY, et al. Microvascular abnormality in schizophrenia as shown by retinal imaging. AJ Psychiatry. 2013;170(12):1451–1459. doi: 10.1176/appi.ajp.2013.13020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sonnier L, Le Pen G, Hartmann A, Bizot JC, Trovero F, Krebs MO, et al. Progressive loss of dopaminergic neurons in the ventral midbrain of adult mice heterozygote for Engrailed1. J Neurosci. 2007;27(5):1063–1071. doi: 10.1523/JNEUROSCI.4583-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schwarz M, Alvarez-Bolado G, Urbanek P, Busslinger M, Gruss P. Conserved biological function between Pax-2 and Pax-5 in midbrain and cerebellum development: evidence from targeted mutations. Proc Natl Acad Sci U S A. 1997;94(26):14518–14523. doi: 10.1073/pnas.94.26.14518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Simon HH, Bhatt L, Gherbassi D, Sgado P, Alberi L. Midbrain dopaminergic neurons: determination of their developmental fate by transcription factors. Ann N Y Acad Sci. 2003;991:36–47. [PubMed] [Google Scholar]
- 73.Plagemann A, Harder T, Lindner R, Melchior K, Rake A, Rittel F, et al. Alterations of hypothalamic catecholamines in the newborn offspring of gestational diabetic mother rats. Brain Res Dev Brain Res. 1998;109(2):201–209. doi: 10.1016/s0165-3806(98)00083-2. [DOI] [PubMed] [Google Scholar]
- 74.Horvath S, Mirnics K. Immune system disturbances in schizophrenia. Biol Psychiatry. 2014;75(4):316–323. doi: 10.1016/j.biopsych.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yates B, Braschi B, Gray KA, Seal RL, Tweedie S, Bruford EA. Genenames.org: the HGNC and VGNC resources in 2017. Nucleic Acids Res. 2017;45(D1):D619–d625. doi: 10.1093/nar/gkw1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.