Key Points
Question
What is the prevalence of FOXC1 variants, which are associated with Axenfeld-Rieger syndrome and secondary glaucoma, in individuals with a suspected diagnosis of primary congenital glaucoma?
Findings
Among a cohort of 166 Australian and Italian affected individuals, predicted deleterious FOXC1 variants were observed in approximately 1 in 20 participants. On reexamination or reinvestigation, all of these individuals had at least 1 detectable ocular and/or systemic feature associated with Axenfeld-Rieger syndrome.
Meaning
These findings highlight the genetic and phenotypic heterogeneity of childhood glaucoma and suggest that FOXC1 sequencing may aid its diagnosis.
This cohort study investigates the prevalence of FOXC1 variants in participants with a suspected diagnosis of primary congenital glaucoma.
Abstract
Importance
Both primary and secondary forms of childhood glaucoma have many distinct causative mechanisms, and in many cases a cause is not immediately clear. The broad phenotypic spectrum of secondary glaucoma, particularly in individuals with variants in FOXC1 or PITX2 genes associated with Axenfeld-Rieger syndrome, makes it more difficult to diagnose patients with milder phenotypes. These cases are occasionally classified and managed as primary congenital glaucoma.
Objective
To investigate the prevalence of FOXC1 variants in participants with a suspected diagnosis of primary congenital glaucoma.
Design, Setting, and Participants
Australian and Italian cohorts were recruited from January 1, 2007, through March 1, 2016. Australian individuals were recruited through the Australian and New Zealand Registry of Advanced Glaucoma and Italian individuals through the Genetic and Ophthalmology Unit of l’Azienda Socio–Sanitaria Territoriale Grande Ospedale Metropolitano Niguarda in Milan, Italy. We performed exome sequencing, in combination with Sanger sequencing and multiplex ligation-dependent probe amplification, to detect variants of FOXC1 in individuals with a suspected diagnosis of primary congenital glaucoma established by their treating specialist. Data analysis was completed from June 2015 to November 2017.
Main Outcome and Measures
Identification of single-nucleotide and copy number variants in FOXC1, along with phenotypic characterization of the individuals who carried them.
Results
A total of 131 individuals with a suspected diagnosis of primary congenital glaucoma were included. The mean (SD) age at recruitment in the Australian cohort was 24.3 (18.1) years; 37 of 84 Australian participants (44.0%) were female, and 71 of 84 (84.5%) were of European ancestry. The mean (SD) age at recruitment was 22.5 (18.4) years in the Italian cohort; 21 of 47 Italian participants (44.7%) were female, and 45 of 47 (95.7%) were of European ancestry. We observed rare, predicted deleterious FOXC1 variants in 8 of 131 participants (6.1%), or 8 of 166 participants (4.8%) when including those explained by variants in CYP1B1. On reexamination or reinvestigation, all of these individuals had at least 1 detectable ocular and/or systemic feature associated with Axenfeld-Rieger syndrome.
Conclusions and Relevance
These data highlight the genetic and phenotypic heterogeneity of childhood glaucoma and support the use of gene panels incorporating FOXC1 as a diagnostic aid, especially because clinical features of Axenfeld-Rieger syndrome can be subtle. Further replication of these results will be needed to support the future use of such panels.
Introduction
Glaucoma is a heterogeneous group of optic neuropathies associated with irreversible visual field loss.1 Childhood glaucoma (defined here as glaucoma occurring with an age at onset younger than 18 years)2 is generally a more severe disease than its adult counterpart, and it often requires multiple surgical interventions to control high intraocular pressure (IOP). Causes of childhood glaucoma are typically classified as primary or secondary2; primary childhood glaucoma includes primary congenital glaucoma (PCG) and juvenile open-angle glaucoma, while secondary causes include Axenfeld-Rieger anomaly (ARA), Peters anomaly, aniridia, and ectopia lentis.
Primary congenital glaucoma is the most common nonsyndromic cause of childhood glaucoma and is characterized by elevated IOP and associated sequelae, including optic disc cupping, buphthalmos, Haab striae, enlarged corneal diameter, corneal edema, and/or increased axial length. The disease has an approximate incidence of 1:18 500 in Great Britain3 and 1:30 000 in Australia.4 There is an increased incidence of PCG in ethnic groups in which parental consanguinity is more common (such as Pakistani British populations),3 or where there is a strong founder effect (eg, an incidence of 1:1250 among Romani Slovakian individuals),5,6 which is consistent with autosomal recessive inheritance.
Axenfeld-Rieger anomaly is a rare condition defined by a collection of 1 or more ocular features, including posterior embryotoxon, iris stromal hypoplasia, peripheral anterior synechiae (PAS), corectopia, or pseudopolycoria.7 This condition may also be associated with 1 or more systemic features, in which case it is referred to as Axenfeld-Rieger Syndrome (ARS). Glaucoma is present in approximately 50% of individuals with ARS.8
Both primary and secondary childhood glaucoma may be associated with variants in one of several genes. Cases of PCG are most commonly associated with autosomal recessive variants in CYP1B19 or less commonly with heterozygous variants of TEK10 or ANGPT1.11 Causes of secondary glaucoma include microspherophakia and/or ectopia lentis associated with variants in LTBP2,12,13,14 aniridia owing to variants in PAX6,15 Peters anomaly owing to variants in PAX6, CYP1B1, PITX2, FOXC1, or other genes,16 and ARA or ARS owing to variation in PITX217 or FOXC1.18
Key phenotypic differences exist between ARS associated with FOXC1 or PITX2 variants. Cases of ARS associated with PITX2 typically present with dental and umbilical anomalies,7,19,20 while sensorineural hearing loss, intellectual disability, and congenital heart defects are more characteristic of patients with variants in FOXC1.21 Facial dysmorphism, including maxillary hypoplasia, a broad and flat nasal bridge, telecanthus, or hypertelorism, can be observed in both. FOXC1-associated ARS typically presents with glaucoma at a younger age than PITX2-associated ARS, with 23% of FOXC1 carriers diagnosed before the age of 3 years compared with 7% of PITX2 carriers.21
Ocular or systemic features of ARS are not always readily apparent; posterior embryotoxon, for example, may not always be visible on slitlamp examination,22 and systemic features may not be apparent until later in life, if at all. In our previous study,21 1 FOXC1 carrier displayed systemic features in the first few years of life, compared with all of the PITX2 carriers. These ambiguities, combined with the difficulties associated with thorough pediatric ophthalmic examination (often requiring general anesthesia), can occasionally lead to a diagnosis of PCG. Overall, this suggests that FOXC1 variants might account for suspected diagnoses of PCG. In this study, we used sequencing and copy-number analysis of the FOXC1 locus to refine the diagnostic classification of 166 participants with a suspected diagnosis of PCG.
Methods
Participant Recruitment
Australian individuals with PCG and their families were recruited through the Australian and New Zealand Registry of Advanced Glaucoma (ANZRAG),23 with Italian individuals with PCG and their families recruited at the Genetic and Ophthalmology Unit of l’Azienda Socio–Sanitaria Territoriale Grande Ospedale Metropolitano Niguarda (Milan, Italy). Patients with a suspected diagnosis of PCG made by their treating ophthalmologist were included from the Australian cohort (n = 84) and the Italian cohort (n = 47). Control individuals (n = 106) were confirmed to not have glaucoma or anterior segment dysgenesis on ocular examination.
Informed written consent was obtained from participants or their guardians, and blood or saliva samples collected along with clinical details from the referring ophthalmologist. Ethics approval was obtained from the Southern Adelaide Clinical Human Research Ethics Committee (Australia), the Medical Faculty of the University of Erlangen-Nürnberg (Germany), and l’Azienda Socio–Sanitaria Territoriale Grande Metropolitano Niguarda Hospital (Italy).
Sequencing
For exome sequencing, genomic DNA was extracted from blood samples using a QIAamp Blood Maxi Kit (Qiagen) and subjected to exome capture (Agilent SureSelect, version 4). Saliva samples were collected with Oragene DNA Self-Collection Kits (DNA Genotek Inc) and DNA extracted per the manufacturer’s instructions. All paired-end libraries were sequenced on a HiSeq 2000 (Illumina), with the exception of a single Italian trio that was sequenced on the SOLiD 4 System (Life Technologies). Reads were mapped to the human reference genome (hg19) using Burrows-Wheeler Aligner (http://bio-bwa.sourceforge.net), and duplicates marked and removed with Picard Tools (Broad Institute). Variants were called using Sequence Alignment/Map (SAM) tools and annotated with ANNOVAR (http://annovar.openbioinformatics.org/en/latest/). All variants were annotated using the genome aggregation database (gnomAD) variant database (release 2.0.2; http://gnomad.broadinstitute.org), and with Sorting Intolerant From Tolerant (SIFT) and PolyPhen-2 scores. FOXC1 variants were annotated against the consensus transcript (NM_001453.2) and protein (NP_001444.2) sequences. For capillary sequencing, genomic DNA was prepared from 200 μL of venous blood collected in EDTA-coated tubes and extracted using an Illustra Blood Genomic Prep MiniSpin kit (GE Healthcare), or Flexi-Gene kit (Qiagen), according to the manufacturer’s instructions. Primers for amplification of FOXC1 genomic DNA are available on request. All variants are publicly available at the ClinVar database (accession numbers SCV000494251, SCV000494255, SCV000494257, SCV000494258, SCV000494261, SCV000494267, SCV000693681, and SCV000693680).
Exome Copy Number Variant Analysis
Coverage depth across the FOXC1 locus and neighboring loci was extracted from exome Binary Alignment/Map files using SAMtools. For copy number variations analysis using Copy Number Inference From Exome Reads (CoNIFER; version 0.2.2), the same interval was analyzed in 343 contemporaneously sequenced exomes using the parameters singular value decomposition (SVD) version 5 and the z score of the reads per kilobase per million mapped reads (ZRPKM) version 1.5.
Multiplex Ligation-Dependent Probe Amplification
FOXC1 copy number was measured by multiplex ligation-dependent probe amplification (MLPA) using the SALSA MLPA P054-B2 FOXL2-TWIST1 probemix (MRC Holland) according to the manufacturer’s instructions. Ligation sites for the FOXC1 probes (FOXC1-A [02561-L02029]) and FOXC1-B [02562-L02030]) were located at 925 base pairs (bp) and 1532 bp, respectively, of NM_001453.2. The copy number present in the original DNA specimen was determined from the relative amplitude of each amplicon product detected using the ABI 3130xl Genetic Analyzer, and data analyzed using Peak Scanner version 2.0 (ThermoFisher Scientific).
Results
Genetic Analysis
In the Australian cohort, mean (SD) age at recruitment was 24.3 (18.1) years; 37 of 84 (44.0%) were female, and 71 of 84 (84.5%) were of European ancestry. In the Italian cohort, the mean (SD) age at recruitment was 22.5 (18.4) years; 21 of 47 (44.7%) were female, and 45 of 47 (95.7%) were of European ancestry.
From a collection of Australian individuals with a suspected diagnosis of PCG, we selected 65 probands whose conditions were not explained by homozygous or compound heterozygous variants of CYP1B1. All 65 individuals underwent exome sequencing, and variation at the FOXC1 locus was examined. Rare FOXC1 variants predicted to be deleterious were identified in 2 of the 65 individuals, including frameshift deletion c.718_719delCT, p.(Leu240ValfsTer65) in participant PCG128 and missense variant c.269C>A, p.(Ala90Asp) in participant PCG140 (Figure 1A).
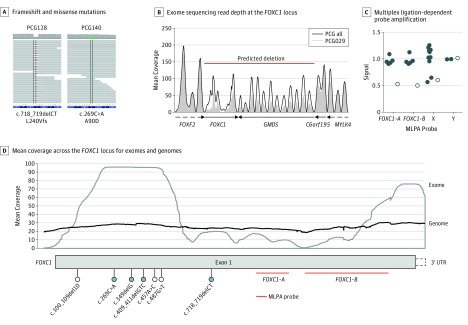
Figure 1. FOXC1 Variants Associated With Suspected Primary Congenital Glaucoma (PCG).
A, Frameshift and missense variants identified by exome sequencing. B, Exome sequencing read depth at the FOXC1 locus (chr6:1390204-2679449, hg19) for participant PCG029 compared with the mean read depth across all suspected PCG samples. C, Multiplex ligation-dependent probe amplification (MLPA) using probes within FOXC1 (chr6) and on both sex chromosomes. Signal intensity was measured in duplicate, normalized, and plotted as the mean for participant PCG029 and all other samples processed in the same batch (control participants). D, Mean coverage across the FOXC1 locus for exomes (gray) and genomes (black) from the gnomAD release 2.0.2 collection, indicating location of variants identified by exome (blue fill) or capillary (no fill) sequencing. Multiplex ligation-dependent probe amplification probe locations are indicated in red. Most gnomAD exome sequences (approximately 77%) were processed with the same or similar exome capture kits as those used in the current study (Agilent SureSelect).
By examining the depth of coverage across the FOXC1 locus, a large deletion spanning FOXC1 and the neighboring GMDS gene was identified in participant PCG029 (Figure 1B). It also became clear that portions of FOXC1 were poorly covered by exome capture and sequencing (Figure 1B and C), likely because of a high guanine-cytosine content.24 The single FOXC1 coding exon was therefore resequenced in all 65 proband individuals by capillary sequencing, alongside an additional 19 PCG probands whose conditions were not explained by CYP1B1 variants. Multiplex ligation-dependent probe amplification was performed on 73 of these individuals to detect FOXC1 copy number variations (both gains and losses). An additional 3 rare protein-coding variants were identified by capillary sequencing, including nonsense variant c.487G>T, p.(Glu163Ter) in participant PCG033, missense variant c.457A>C, p.(Thr153Pro) in participant PCG057, and frameshift deletion c.100_109del10, p.(Gly34ThrfsTer8) in participant PCG125 (Figure 1C). All variants and deletions identified by exome sequencing were validated by capillary sequencing or MLPA (Figure 1D).
None of the 6 variants identified in the Australian cohort were present in 106 controls exomes sequenced in parallel, nor were they reported in the gnomAD variant collection. In total, FOXC1 variants accounted for 6 of 84 participants (7.1%) in the Australian cohort or 6 of 104 participants (5.8%) when including those whose conditions were explained by variants in CYP1B1.
In a separate cohort of Italian participants with suspected PCG, 22 family trios whose conditions were not explained by variants in CYP1B1 underwent exome sequencing. FOXC1 was also covered by capillary sequencing in all 22 probands plus an additional 25 Italian probands in which CYP1B1 variants had been excluded by capillary sequencing. Predicted pathogenic FOXC1 variants were identified in 2 of 47 individuals (4.2%) in the Italian cohort. De novo in-frame deletion c.409_411del, p.(Val137del) in participant PCG402-IT, identified by exome sequencing, has previously been described.25 A second de novo frameshift deletion, c.349delG, p.(Asp117ThrfsTer64), was identified in participant PCG297-IT by capillary sequencing. Neither variant was present in 680 European control participants without glaucoma, and neither was reported in gnomAD. When including patients whose conditions were explained by variants in CYP1B1, we identified FOXC1 variants in 2 of 62 participants (3.2%) of the Italian PCG cohort (Figure 1C and D).
Across both cohorts, we identified predicted pathogenic FOXC1 variants in 8 of 131 participants (6.1%) who had conditions that were not explained by CYP1B1 variants, and 8 of 166 participants (4.8%) when including individuals whose conditions were explained by CYP1B1 variants (Table 1).
Table 1. Heterozygous FOXC1 Variants Identified in Affected Individuals.
| Proband Identification No. | Cohort | Coordinates (hg19) | Method | FOXC1 Complementary DNAa | FOXC1 Protein | PolyPhen 2, HumVar | SIFT |
|---|---|---|---|---|---|---|---|
| PCG402-IT | Italian | 6:1611089-1611091 | Exome | c.409_411del | p.(Val137del) | NA | NA |
| PCG297-IT | Italian | 6:1611029-1611029 | Capillary | c.349delG | p.(Asp117ThrfsTer64) | NA | NA |
| PCG029 | Australian | NA | Exome | c.902-?_1560+?del | NA | NA | NA |
| PCG033 | Australian | 6:1611167-1611167 | Capillary | c.487G>T | p.(Glu163Ter) | NA | NA |
| PCG057 | Australian | 6:1611137-1611137 | Capillary | c.457A>C | p.(Thr153Pro) | 0.978 (Probably damaging) | 0 (Damaging) |
| PCG125 | Australian | 6:1610780-1610789 | Capillary | c.100_109del10 | p.(Gly34ThrfsTer8) | NA | NA |
| PCG128 | Australian | 6:1611398-1611399 | Exome | c.718_719delCT | p.(Leu240ValfsTer65) | NA | NA |
| PCG140 | Australian | 6:1610949-1610949 | Exome | c.269C>A | p.(Ala90Asp) | 0.999 (Probably damaging) | 0 (Damaging) |
Abbreviations: NA, not available; SIFT, Sorting Intolerant From Tolerant.
Coordinates of complementary DNA and protein variants are with respect to consensus complementary DNA and amino acid sequences (NM_001453.2 and NP_001444.2, respectively).
Clinical Reinvestigation
Having uncovered potentially pathogenic FOXC1 variants, all probands were reinvestigated for evidence of ocular and systemic features of ARS (Table 2). Family pedigrees were ascertained, with variant segregation testing performed where possible (Figure 2). Ocular features of the probands and their examined family members are shown in eFigure in the Supplement.
Table 2. Clinical Details of the Probands.
| Proband Identification No. | Age at Diagnosis | Diagnosis | Ocular Features | Facial Dysmorphism | Systemic Features | Best-Corrected Visual Acuity | Maximum Intraocular Pressure | Cup-Disc Ratio |
|---|---|---|---|---|---|---|---|---|
| PCG402-IT | 14 y | Congenital glaucoma | Buphthalmos, posterior embryotoxon, peripheral anterior synechiae, megalocornea | Yes | NA | 20/30:20/30 | 50/50 | 0.6/0.7 |
| PCG297-IT | 1 mo | Congenital glaucoma | Buphthalmos, posterior embryotoxon, iris stromal hypoplasia, peripheral anterior synechiae, corectopia, corneal opacity, megalocornea | Yes | NA | 20/40:20/30 | 23/22 | 0.6/0.3 |
| PCG029 | 0 y | Primary congenital glaucoma | Corneal opacity, nystagmus | NA | Hearing loss, hydrocephalus, inguinal hernia, heart murmur | Count fingers:20/40 | 22/27 | No view |
| PCG033 | 1 y | Congenital glaucoma | Posterior embryotoxon, peripheral anterior synechiae, corneal opacity, megalocornea | NA | Hearing loss, congenital hip dislocation | 20/200:20/70 | 30/30 | 0.9/0.9 |
| PCG057 | 0 y | Primary congenital glaucoma | Haab striae, iris stromal hypoplasia, corectopia | Yes | Short stature | 20/40:20/30 | 48/40 | 0.6/0.8 |
| PCG125 | 8 y | Primary congenital glaucoma | Buphthalmos, Haab striae, posterior embryotoxon, iris stromal hypoplasia, peripheral anterior synechiae, corneal decompensation | Yes | Developmental delay | 20/40:20/25 | 40/30 | 0.95/0.6 |
| PCG128 | 0 y | Primary congenital glaucoma | Buphthalmos, Haab striae, peripheral anterior synechiae | Yes | Developmental delay | NA | 50/50 | 0.75/0.8 |
| PCG140 | 0 y | Primary congenital glaucoma | Accommodative esotropia | NA | Mitral valve replacement, short stature | 20/40:20/40 | 23/21 | 0.3/0.6 |
Abbreviation: NA, not applicable.
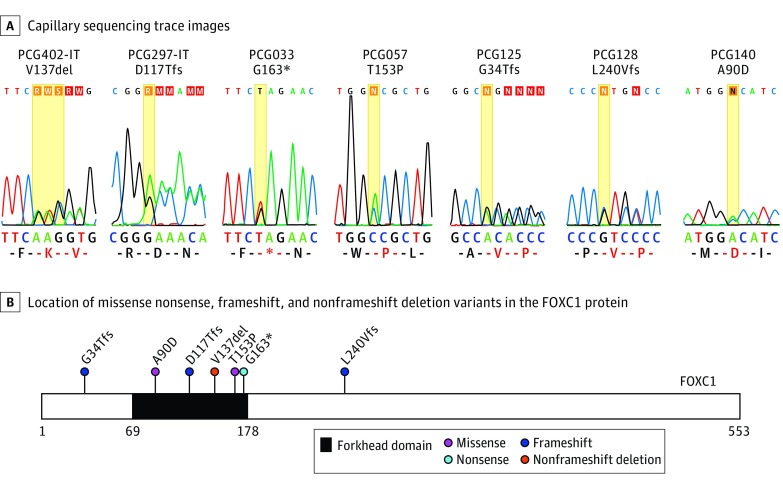
Figure 2. Segregation of FOXC1 Variants.
A, Capillary sequencing trace images. B, Location of missense, nonsense, frameshift, and nonframeshift deletion variants within the FOXC1 protein; the forkhead domain is indicated by black shading.
Detailed ophthalmological reexamination of participant PCG402-IT revealed prominent posterior embryotoxon and PAS on gonioscopy.25 Facial dysmorphism included a flat philtrum, a flat nasal bridge, a prominent forehead, lateral bushy eyebrows, hypertelorism, and a thin upper lip. No other systemic features were present. These features all suggested a revised diagnosis of ARS.
Review of the clinical records of participant PCG297-IT led to a revised diagnosis of glaucoma with ARA by 1 year of age. Posterior embryotoxon, PAS, optic disc cupping, and corneal edema were present in both eyes, with corneal opacity and sclerocornea in the right eye. Other systemic features associated with ARS were not present, with the exception of potential midface dysmorphism, including hypertelorism, a thin upper lip, a flat philtrum, dysmorphic ears, and lateral bushy eyebrows with medial flare.
Review of participant PCG029 did not reveal any anterior segment anomalies consistent with ARA. A more detailed examination by the individual’s current ophthalmologist was complicated by bilateral corneal opacity, making a formal exclusion of ARA difficult. Possible syndromic associations included hydrocephalus and hearing loss. The same FOXC1 deletion was absent in all family members tested (ie, 1 parent and 5 siblings), and although the other parent could not be tested, this suggested a de novo origin.
For participant PCG033, corneal opacity again complicated detailed anterior segment examination for ARA. Review of previous medical records indicated prominent posterior embryotoxon and PAS, with a possible systemic association of hearing loss. Both 1 parent of participant PCG033 and that parent’s parent were affected by glaucoma, although a detailed medical history (including ages at diagnosis) could not be obtained.
The diagnosis for participant PCG057 was revised to ARS with secondary glaucoma after the referral of 2 of the participant’s relatives, of whom 1 was diagnosed with ARA or the other was diagnosed with ARS. Participant PCG057 had no dental, umbilical, or cardiac anomalies or hearing loss but was of short stature. An additional relative of participant PCG057 was initially diagnosed with juvenile open-angle glaucoma at age 13 years. On reexamination, this relative was found to have posterior embryotoxon, mild PAS, and hearing loss and so was given a revised diagnosis of ARS. Midface abnormalities were evident in at least 3 affected family members (including participant PCG057). The same FOXC1 variant was present in all 4 affected family members.
Reexamination of the anterior segment of participant PCG125 was complicated by corneal opacity in the right eye. After reviewing past records, participant PCG125 was found to have been diagnosed with ARA at age 8 years. This participant also had midface abnormalities and mild intellectual disability. One of this participant’s parents had also been diagnosed with ARA at a similar time, and both the parent and the participant were found to carry the same FOXC1 variant.
Participant PCG128 was reexamined several times following genetic testing. Apart from mild PAS in the left eye on gonioscopy, no features of ARA were evident. Facial dysmorphism included a flat nasal bridge and possible telecanthus or hypertelorism, but dental, umbilical, and cardiac abnormalities were all absent. There was an extensive history on 1 side of the family of adult-onset glaucoma, despite that testing of the parent on that side of the family of PCG128 did not reveal the same FOXC1 variant as the participant. Although the other parent of PCG128 could not be tested, an absence of reported glaucoma or features consistent with ARS suggested a de novo origin of the FOXC1 variant.
Finally, participant PCG140 had no evidence of ARA on repeated examination (Figure 4). Associated mitral valve disease and short stature may be systemic manifestations of this individual’s FOXC1 variant. One parent of this individual did not carry the same FOXC1 variant. Testing of the other parent was not performed, although family history suggested a de novo origin of the FOXC1 variant.
Discussion
Among a total of 131 individuals with a suspected diagnosis of PCG not explained by CYP1B1 variants, 8 of the 131 participants (6.1%) were found to carry a predicted disease-causing heterozygous variant of FOXC1. Including individuals whose conditions were previously explained by CYP1B1 variants, the prevalence of FOXC1 variants in individuals with PCG from an Australian and Italian cohort was 8 of 166 participants (4.8%). The true frequency could indeed be higher, given that MLPA failed in 11 of the 84 Australian samples and was not performed on any of the Italian PCG cohort samples.
None of the 8 variants described here have been formally tested at a functional level, although many are consistent with a revised clinical diagnosis of ARA or ARS. No variant was present in the gnomAD database, and 5 were predicted protein-truncating variants. Both missense variants were predicted to be deleterious by PolyPhen-2 and Sorting Intolerant From Tolerant algorithms and were located in the conserved DNA-binding forkhead domain of FOXC1, while the in-frame deletion removed a highly conserved amino acid.25
Of the 8 probands found to carry potentially pathogenic FOXC1 variants, 6 were subsequently discovered to have features consistent with ARA, either on reexamination or after review of medical records. For the remaining probands, detailed examination of the anterior segment was either hindered by corneal opacity in 1 person (participant PCG029) or there was no evidence of ARA after repeated examination (participant PCG140). At least 4 probands had systemic features consistent with ARS, including hearing loss, hydrocephalus, short stature, or congenital heart disease. Developmental delay was present in a further 2 probands, although this is not typically associated with ARS. All 5 of the probands examined for facial features had some degree of facial dysmorphism consistent with features reported in ARS. Based on their newly ascertained genetic evidence, all 8 probands were therefore given a revised diagnosis of ARS.
Axenfeld-Rieger anomaly and Axenfeld-Rieger syndrome are rare conditions that can be difficult to diagnose in children, especially without detailed ophthalmic examination under anesthesia. Ocular signs such as posterior embryotoxon or PAS may only be visible by gonioscopy,22 and even then, these may only become apparent with age. Diagnosis may also be complicated in adults, particularly if corneal opacity obscures a thorough anterior segment examination. Both PITX2 and FOXC1 show phenotypic variation within families, and only approximately 30% of individuals with FOXC1 variants have extraocular features.21 When compared with PITX2, FOXC1 is less likely to be associated with more striking systemic features, such as dental or umbilical anomalies,20 and is therefore more likely to resemble PCG. Systemic features of ARS, particularly those in FOXC1-associated disease, often become apparent only later in life (eg, hearing loss, developmental delay, and short stature), making an accurate diagnosis of FOXC1-associated ARA or ARS even more challenging. Furthermore, FOXC1 variants can be inherited de novo (as was confirmed in 2 families and suspected in a further 3 participants in this study), making clinical diagnosis more difficult than probands with a known family history of ARS (as reported in 3 pedigrees). All of these issues highlight the difficulties in reaching a precise clinical diagnosis of ARA or ARS at an early age. These data show the value of early genetic testing in presentations of suspected PCG cases and the revision of several diagnoses as a direct result.
Accurate diagnosis of PCG has a number of important implications for genetic counseling. Biallelic variants of CYP1B1 are the most common known genetic cause of PCG, accounting for 22% of affected individuals in the Australian population.26 Therefore families with children affected by PCG are often counseled for autosomal recessive inheritance. Revising this diagnosis to ARA or ARS caused by autosomal dominant FOXC1 variants therefore requires a revision of counseling: rather than a maximum risk to siblings of 25%, first-degree relatives of individuals with inherited disease-causing FOXC1 variants have a risk of 50% for ARA and/or ARS, with approximately 50% of these expected to develop glaucoma. Importantly, in families with de novo inheritance, the risk of recurrence for siblings of affected individuals is low. Glaucoma is also not the only concern for a family with ARA; the presence of multiple associated systemic features should also be closely investigated and managed. Reaching the correct clinical and genetic diagnosis allows all family members the opportunity for genetic testing, appropriate referrals to be made for surveillance and management, and discussion of reproductive options.
Our data support the importance of genetic information in a clinical context, especially for conditions, such as childhood glaucoma, that carry a broad phenotypic spectrum and genetic heterogeneity. Indeed, one might consider sequencing not only FOXC1, but all genes associated with childhood glaucoma. This might be achieved with exome or genome sequencing; however, exome sequencing is likely to be less reliable, given the uneven capture and coverage of FOXC1 and the limited ability to detect copy number variations.27 At the very least, exome and other capture-based sequencing tests should be interpreted cautiously when excluding variants in poorly captured genes such as FOXC1.
Limitations
These cohorts were predominantly of European ancestry, and these findings may therefore not be generalizable to other populations, although lower prevalence of FOXC1 variants has been reported in PCG cohorts from India (0% to 2.4%).28,29 Additionally, MLPA failed in 11 of 84 participants from the Australian cohort, which means that the true prevalence of FOXC1 variants could be higher.
Conclusions
In summary, our data support the use of FOXC1 sequencing as a diagnostic aid in cases of suspected PCG, although further replication will be needed to support the future use of such panels. Timely sequencing has the potential to accelerate diagnosis, minimize excessive examinations under anesthesia, refine genetic counseling, guide appropriate therapy, and maximize long-term patient benefit.
eFigure. Spectrum of ocular features associated with FOXC1 variants in individuals with suspected primary congenital glaucoma.
References
- 1.Quigley HA. Glaucoma. Lancet. 2011;377(9774):1367-1377. doi: 10.1016/S0140-6736(10)61423-7 [DOI] [PubMed] [Google Scholar]
- 2.Weinreb RN, Grajewski AL, Papadopoulos M, Grigg J, Freedman S. Childhood Glaucoma: Vol 9 Amsterdam, the Netherlands: Kugler Publications; 2013. [Google Scholar]
- 3.Papadopoulos M, Cable N, Rahi J, Khaw PT; BIG Eye Study Investigators . The British Infantile and Childhood Glaucoma (BIG) eye study. Invest Ophthalmol Vis Sci. 2007;48(9):4100-4106. doi: 10.1167/iovs.06-1350 [DOI] [PubMed] [Google Scholar]
- 4.MacKinnon JR, Giubilato A, Elder JE, Craig JE, Mackey DA. Primary infantile glaucoma in an Australian population. Clin Exp Ophthalmol. 2004;32(1):14-18. doi: 10.1046/j.1442-9071.2004.00750.x [DOI] [PubMed] [Google Scholar]
- 5.Genĉík A. Epidemiology and genetics of primary congenital glaucoma in Slovakia: description of a form of primary congenital glaucoma in gypsies with autosomal-recessive inheritance and complete penetrance. Dev Ophthalmol. 1989;16:76-115. [PubMed] [Google Scholar]
- 6.Plásilová M, Stoilov I, Sarfarazi M, Kádasi L, Feráková E, Ferák V. Identification of a single ancestral CYP1B1 mutation in Slovak Gypsies (Roms) affected with primary congenital glaucoma. J Med Genet. 1999;36(4):290-294. [PMC free article] [PubMed] [Google Scholar]
- 7.Alward WL. Axenfeld-Rieger syndrome in the age of molecular genetics. Am J Ophthalmol. 2000;130(1):107-115. doi: 10.1016/S0002-9394(00)00525-0 [DOI] [PubMed] [Google Scholar]
- 8.Fitch N, Kaback M. The Axenfeld syndrome and the Rieger syndrome. J Med Genet. 1978;15(1):30-34. doi: 10.1136/jmg.15.1.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stoilov I, Akarsu AN, Sarfarazi M. Identification of three different truncating mutations in cytochrome P4501B1 (CYP1B1) as the principal cause of primary congenital glaucoma (Buphthalmos) in families linked to the GLC3A locus on chromosome 2p21. Hum Mol Genet. 1997;6(4):641-647. doi: 10.1093/hmg/6.4.641 [DOI] [PubMed] [Google Scholar]
- 10.Souma T, Tompson SW, Thomson BR, et al. Angiopoietin receptor TEK mutations underlie primary congenital glaucoma with variable expressivity. J Clin Invest. 2016;126(7):2575-2587. doi: 10.1172/JCI85830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomson BR, Souma T, Tompson SW, et al. Angiopoietin-1 is required for Schlemm’s canal development in mice and humans. J Clin Invest. 2017;127(12):4421-4436. doi: 10.1172/JCI95545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ali M, McKibbin M, Booth A, et al. Null mutations in LTBP2 cause primary congenital glaucoma. Am J Hum Genet. 2009;84(5):664-671. doi: 10.1016/j.ajhg.2009.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Désir J, Sznajer Y, Depasse F, et al. LTBP2 null mutations in an autosomal recessive ocular syndrome with megalocornea, spherophakia, and secondary glaucoma. Eur J Hum Genet. 2010;18(7):761-767. doi: 10.1038/ejhg.2010.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khan AO, Aldahmesh MA, Alkuraya FS. Congenital megalocornea with zonular weakness and childhood lens-related secondary glaucoma—a distinct phenotype caused by recessive LTBP2 mutations. Mol Vis. 2011;17:2570-2579. [PMC free article] [PubMed] [Google Scholar]
- 15.Jordan T, Hanson I, Zaletayev D, et al. The human PAX6 gene is mutated in two patients with aniridia. Nat Genet. 1992;1(5):328-332. doi: 10.1038/ng0892-328 [DOI] [PubMed] [Google Scholar]
- 16.Reis LM, Semina EV. Genetics of anterior segment dysgenesis disorders. Curr Opin Ophthalmol. 2011;22(5):314-324. doi: 10.1097/ICU.0b013e328349412b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Semina EV, Reiter R, Leysens NJ, et al. Cloning and characterization of a novel bicoid-related homeobox transcription factor gene, RIEG, involved in Rieger syndrome. Nat Genet. 1996;14(4):392-399. doi: 10.1038/ng1296-392 [DOI] [PubMed] [Google Scholar]
- 18.Nishimura DY, Swiderski RE, Alward WL, et al. The forkhead transcription factor gene FKHL7 is responsible for glaucoma phenotypes which map to 6p25. Nat Genet. 1998;19(2):140-147. doi: 10.1038/493 [DOI] [PubMed] [Google Scholar]
- 19.D’haene B, Meire F, Claerhout I, et al. Expanding the spectrum of FOXC1 and PITX2 mutations and copy number changes in patients with anterior segment malformations. Invest Ophthalmol Vis Sci. 2011;52(1):324-333. doi: 10.1167/iovs.10-5309 [DOI] [PubMed] [Google Scholar]
- 20.Tümer Z, Bach-Holm D. Axenfeld-Rieger syndrome and spectrum of PITX2 and FOXC1 mutations. Eur J Hum Genet. 2009;17(12):1527-1539. doi: 10.1038/ejhg.2009.93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Souzeau E, Siggs OM, Zhou T, et al. Glaucoma spectrum and age-related prevalence of individuals with FOXC1 and PITX2 variants. Eur J Hum Genet. 2017;25(7):839-847. doi: 10.1038/ejhg.2017.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shields MB. Axenfeld-Rieger syndrome: a theory of mechanism and distinctions from the iridocorneal endothelial syndrome. Trans Am Ophthalmol Soc. 1983;81:736-784. [PMC free article] [PubMed] [Google Scholar]
- 23.Souzeau E, Goldberg I, Healey PR, et al. Australian and New Zealand registry of advanced glaucoma: methodology and recruitment. Clin Exp Ophthalmol. 2012;40(6):569-575. doi: 10.1111/j.1442-9071.2011.02742.x [DOI] [PubMed] [Google Scholar]
- 24.Meienberg J, Zerjavic K, Keller I, et al. New insights into the performance of human whole-exome capture platforms. Nucleic Acids Res. 2015;43(11):e76. doi: 10.1093/nar/gkv216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pasutto F, Mauri L, Popp B, et al. Whole exome sequencing reveals a novel de novo FOXC1 mutation in a patient with unrecognized Axenfeld-Rieger syndrome and glaucoma. Gene. 2015;568(1):76-80. doi: 10.1016/j.gene.2015.05.015 [DOI] [PubMed] [Google Scholar]
- 26.Dimasi DP, Hewitt AW, Straga T, et al. Prevalence of CYP1B1 mutations in Australian patients with primary congenital glaucoma. Clin Genet. 2007;72(3):255-260. doi: 10.1111/j.1399-0004.2007.00864.x [DOI] [PubMed] [Google Scholar]
- 27.Siggs OM, Javadiyan S, Sharma S, et al. Partial duplication of the CRYBB1-CRYBA4 locus is associated with autosomal dominant congenital cataract. Eur J Hum Genet. 2017;25(6):711-718. doi: 10.1038/ejhg.2017.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chakrabarti S, Kaur K, Rao KN, et al. The transcription factor gene FOXC1 exhibits a limited role in primary congenital glaucoma. Invest Ophthalmol Vis Sci. 2009;50(1):75-83. doi: 10.1167/iovs.08-2253 [DOI] [PubMed] [Google Scholar]
- 29.Tanwar M, Kumar M, Dada T, Sihota R, Dada R. MYOC and FOXC1 gene analysis in primary congenital glaucoma. Mol Vis. 2010;16:1996-2006. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eFigure. Spectrum of ocular features associated with FOXC1 variants in individuals with suspected primary congenital glaucoma.