Key Points
Question
What is the prevalence of microsatellite instability in prostate cancer and its association with response to immune checkpoint blockade?
Findings
In this case series of 1346 patients with prostate cancer who underwent paired tumor and germline sequencing, 32 of 1033 (3.1%) had microsatellite instability–high or mismatch repair deficient disease, of whom 7 (21.9%) carried a germline mutation in a Lynch syndrome–associated gene. Five of 11 patients who received an anti–PD-1/PD-L1 agent had durable clinical benefit.
Meaning
The microsatellite instability–high/mismatch repair deficient phenotype is uncommon but clinically important in prostate cancer and can be somatically acquired during disease evolution.
Abstract
Importance
The anti–programmed cell death protein 1 (PD-1) antibody pembrolizumab is approved by the US Food and Drug Administration for the treatment of microsatellite instability–high (MSI-H) or mismatch repair–deficient (dMMR) solid tumors, but the prevalence of MSI-H/dMMR prostate cancer and the clinical utility of immune checkpoint blockade in this disease subset are unknown.
Objective
To define the prevalence of MSI-H/dMMR prostate cancer and the clinical benefit of anti–PD-1/programmed cell death 1 ligand 1 (PD-L1) therapy in this molecularly defined population.
Design, Setting, and Participants
In this case series, 1551 tumors from 1346 patients with prostate cancer undergoing treatment at Memorial Sloan Kettering Cancer Center were prospectively analyzed using a targeted sequencing assay from January 1, 2015, through January 31, 2018. Patients had a diagnosis of prostate cancer and consented to tumor molecular profiling when a tumor biopsy was planned or archival tissue was available. For each patient, clinical outcomes were reported, with follow-up until May 31, 2018.
Main Outcomes and Measures
Tumor mutation burden and MSIsensor score, a quantitative measure of MSI, were calculated. Mutational signature analysis and immunohistochemistry for MMR protein expression were performed in select cases.
Results
Among the 1033 patients who had adequate tumor quality for MSIsensor analysis (mean [SD] age, 65.6 [9.3] years), 32 (3.1%) had MSI-H/dMMR prostate cancer. Twenty-three of 1033 patients (2.2%) had tumors with high MSIsensor scores, and an additional 9 had indeterminate scores with evidence of dMMR. Seven of the 32 MSI-H/dMMR patients (21.9%) had a pathogenic germline mutation in a Lynch syndrome–associated gene. Six patients had more than 1 tumor analyzed, 2 of whom displayed an acquired MSI-H phenotype later in their disease course. Eleven patients with MSI-H/dMMR castration-resistant prostate cancer received anti–PD-1/PD-L1 therapy. Six of these (54.5%) had a greater than 50% decline in prostate-specific antigen levels, 4 of whom had radiographic responses. As of May 2018, 5 of the 6 responders (5 of 11 total [45.5%]) were still on therapy for as long as 89 weeks.
Conclusions and Relevance
The MSI-H/dMMR molecular phenotype is uncommon yet therapeutically meaningful in prostate cancer and can be somatically acquired during disease evolution. Given the potential for durable responses to anti–PD-1/PD-L1 therapy, these findings support the use of prospective tumor sequencing to screen all patients with advanced prostate cancer for MSI-H/dMMR. Because not all patients with the MSI-H/dMMR phenotype respond, further studies should explore mechanisms of resistance.
This case series study assesses the prevalence of microsatellite instability–high or mismatch repair–deficient prostate cancer and the clinical benefit of immune checkpoint blockade with anti–PD-1/PD-L1 therapy in this molecularly defined population.
Introduction
Immune checkpoint blockade has shown limited benefit in prostate cancer in several studies.1,2,3 Nonetheless, durable objective responses have been reported, suggesting that patients with molecularly defined subsets of prostate cancer may benefit from this therapeutic approach.4,5,6 Pembrolizumab, an antibody targeting the programmed cell death protein 1 (PD-1) receptor, recently earned accelerated approval by the US Food and Drug Administration for the treatment of microsatellite instability–high (MSI-H) or mismatch repair deficient (dMMR) solid tumors, independent of site of origin. Detection of MSI thus represents the first clinical indication for prospective tumor profiling in patients with prostate cancer.7 However, the optimal method for determining MSI-H/dMMR status in patients with prostate cancer and the clinical implications of broader screening for this phenotype remain unknown.
The prevalence of MSI-H/dMMR prostate cancer is unclear, with frequencies ranging from 1.2% to 12.0% in prior reports.8,9 Recent sequencing studies of metastatic castration–resistant prostate cancer (mCRPC) showed that 2% to 3% of tumors have a higher mutation burden that is often associated with genomic alterations in MMR-associated genes, suggesting that tumor sequencing may be an efficient method for identifying MSI-H/dMMR prostate cancer.10,11 Herein, we leveraged a prospectively generated genomic data set of 1551 prostate tumors from 1346 patients to define the frequency of MSI-H/dMMR prostate cancer and report outcomes for patients with MSI-H/dMMR mCRPC who were treated with anti–PD-1/programmed cell death 1 ligand 1 (PD-L1) therapy.
Methods
Patients and Samples
From January 1, 2015, through January 31, 2018, 1551 tumors from 1346 patients with prostate cancer treated at Memorial Sloan Kettering Cancer Center, New York, New York, underwent clinical genomic profiling with a hybridization capture–based next-generation sequencing assay (Integrated Mutation Profiling of Actionable Cancer Targets [MSK-IMPACT]).12,13 Patients were given the option to consent to secondary germline analysis. Immunohistochemical (IHC) analysis for MSH2 (OMIM 609309), MSH6 (OMIM 600678), MLH1 (OMIM 600678), and PMS2 (OMIM 600259) was performed on select tumors. Medical record review for patient clinical characteristics and outcomes, with follow-up until May 31, 2018, was performed under a protocol approved by the institutional review board of Memorial Sloan Kettering Cancer Center, with a waiver of consent for the analysis of deidentified data.
Sequencing and Analysis
Tumors and matched normal blood samples were sequenced as previously described.10,13 An algorithm for the detection of somatic microsatellite changes using paired tumor-normal sequence data (MSIsensor)14,15 was applied to all tumors, yielding a quantitative MSIsensor score. Tumors deemed to have inadequate tumor content or quality (<200 × median exon coverage, <10% median exonic variant allele frequency, or no mutations with ≤20% tumor content on pathologic review) were flagged, and their MSIsensor scores were excluded from the primary analysis. Tumor mutation burden (TMB) was calculated as mutations per megabase (mut/Mb). Mutational signature analysis16,17 (eMethods in the Supplement) was performed for tumors with somatic mutation counts of at least 10. Alterations in MMR genes were considered deleterious and likely oncogenic if they were pathogenic or likely pathogenic alterations present in the germline18,19 or if they were somatic truncating mutations or deletions.20 For statistical analysis, comparison of MSI-H/dMMR frequency between disease subsets was performed using a 2-tailed Fisher exact text.
Results
Identification of MSI-H/dMMR in Prostate Cancer Using Next-Generation Sequencing
A total of 1551 tumors from 1346 patients with prostate cancer underwent prospective sequencing.10,13 Forty-seven tumors (3.0%) were hypermutated, defined herein as a TMB of at least 10 mut/Mb.10,11 To assess for MSI, we calculated MSIsensor scores14,15 for all samples. Notably, 384 tumors (24.8% of the total) were excluded from the primary MSIsensor analysis owing to low tumor purity (eTable 1 in the Supplement), which can affect MSIsensor score reliability.14 Thus, 1167 tumors from 1033 patients (mean [SD] age, 65.6 [9.3] years) were considered adequate for MSIsensor assessment (Figure 1A).
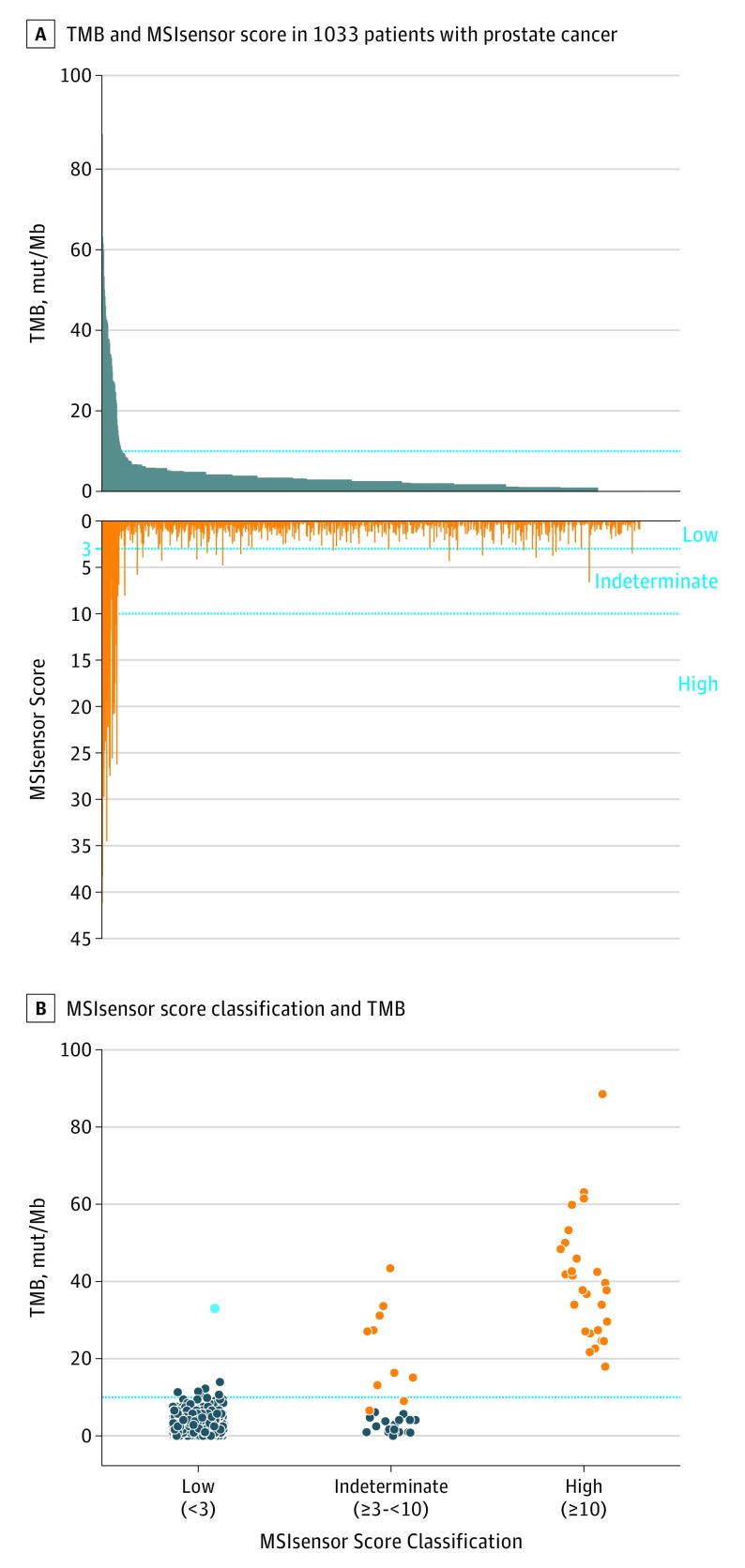
Figure 1. Tumor Mutation Burden (TMB) and Microsatellite Instability (MSI) in Prostate Cancer.
A, Tumor mutation burden in mutations per megabase (mut/Mb) and MSIsensor score, a measure of microsatellite instability derived from sequencing data, are given for 1167 tumors from 1033 patients. B, MSIsensor scores were classified as high (≥10), indeterminate (≥3 to <10), or low (<3). Tumors with high MSIsensor scores invariably had high TMB and were considered to have high MSI (MSI-H) (orange). Tumors with MSIsensor indeterminate scores were classified as MSI-H or mismatch repair deficient (dMMR) if they had deleterious MMR gene alterations or MMR protein loss on immunohistochemical analysis (orange). Tumors with MSIsensor low scores typically had lower TMBs, with 1 exception that harbored a hotspot mutation in POLE (cyan).
We classified MSIsensor scores as high (≥10), indeterminate (≥3 to <10), or low (<3) based on a large, retrospective pancancer analysis comparing MSIsensor score with orthogonal methods14 (Figure 1B). Twenty-three of 1033 patients (2.2%) had tumors with MSIsensor scores of at least 10 and were thus classified as having MSI-H disease. All MSI-H tumors had a TMB of at least 10 mut/Mb and evidence of MMR protein loss by IHC or a deleterious alteration in an MMR gene with the exception of 1 patient (P-0010034), who had a high TMB (22.6 mut/Mb) but no MMR gene alteration and insufficient tissue for IHC (Figure 2).
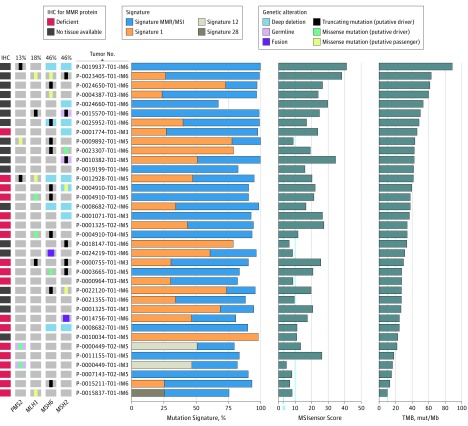
Figure 2. Integrative Analysis of Microsatellite Instability (MSI), Tumor Mutation Burden (TMB), Mutational Signature Decomposition, and Mismatch Repair (MMR) Gene and Protein Status.
Tumors with the highest TMB (in mutations per megabase [mut/Mb]) and indeterminate (≥3 to <10) and high (≥10) MSIsensor scores are shown. Mutation signatures with a contribution of at least 20% are shown, including MMR/MSI signatures 6, 15, 20 and 26; signature 1, which is associated with aging; and signatures 12 and 28.16,17 The oncoprint (left) shows genomic alterations in MMR genes, and immunohistochemical analysis (IHC) for MMR proteins (MSH2, MSH6, MLH1, and PMS2).
An additional 28 patients (2.7%) had tumors with indeterminate MSIsensor scores (eFigure 1A in the Supplement). To further characterize these tumors, we reviewed each case for deleterious alterations in MMR genes, and in tumors for which adequate tissue was available, we performed IHC to assess for MMR protein loss (Figure 2). Within this subset, tumors were classified as MSI-H/dMMR if they were found to harbor a deleterious germline or somatic alteration in an MMR gene or had MMR protein loss on IHC findings. Of the 28 patients with MSIsensor-indeterminate scores, 9 met these criteria and were considered to have MSI-H/dMMR prostate cancer. These patients typically had high TMBs and evidence of a predominant MSI/MMR signature on mutational signature decomposition (Figures 1B and 2). All patients who were identified as having MSI-H/dMMR tumors based on a deleterious MMR gene alteration were confirmed to have evidence of biallelic loss (eTable 1 in the Supplement). In total, 32 of 1033 patients (3.1%) met the criteria defined above for MSI-H/dMMR prostate cancer (eFigures 1A and 2 in the Supplement). The disease state at the time of tumor collection and the frequencies of MSI-H/dMMR in disease subsets are shown in eFigure 3 in the Supplement. Of note, 7 of the 1033 patients (0.7%) had a deleterious MMR gene alteration (1 germline and 6 somatic) without evidence of MSI or hypermutation (eTables 1 and 3 in the Supplement).
As outlined above, 384 prostate tumors (24.8%) with quality scores sufficiently high for mutational calling had insufficient sequence coverage or tumor purity for MSIsensor analysis. We reviewed these tumors to assess whether orthogonal measures, such as TMB, IHC, or mutational signature decomposition, could help guide clinical decision making in these cases. In total, 9 of 313 patients (2.9%) with inadequate tissue quality for MSIsensor analysis had high TMB and/or a somatic mutation in an MMR-associated gene. Two patients (P-00021600 and P-0024488) had TMBs greater than 20 mut/Mb and were found to have loss of MMR proteins on IHC analysis, consistent with dMMR (eTable 1 in the Supplement). Based on these results, in cases where tumor purity limits MSIsensor assessment, we now recommend confirmatory IHC for tumors with a high TMB or a deleterious alteration in an MMR gene (eFigure 4 in the Supplement).
Germline Alterations in MMR Genes and MSI-H/dMMR Status
Mismatch repair–deficient prostate cancer has been reported to occur at increased frequency in Lynch syndrome mutation carriers.21 We sought to determine the rate of germline alterations in MMR genes in patients with MSI-H/dMMR prostate cancer who were identified through tumor profiling. Seven of the 32 patients with MSI-H/dMMR disease (21.9%) had a pathogenic or likely pathogenic germline mutation in an MMR-associated gene (eFigure 1B in the Supplement), including 5 in MSH2, 1 in MSH6, and 1 in PMS2. One additional patient had a pathogenic germline mutation in MSH6 but no evidence of MSI or hypermutation in his tumor, suggesting that the pathogenic MSH6 mutation did not contribute to the pathogenesis of his prostate cancer (eTable 3 in the Supplement).
Analysis of Longitudinally Profiled Patients
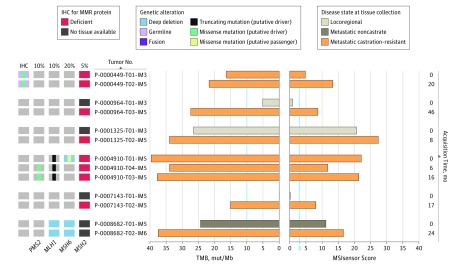
As outlined above, 21.9% of patients with MSI-H/dMMR prostate cancer had a germline MMR gene mutation that presumably played a causative role in the development of their malignant disease. However, most patients with MSI-H/dMMR prostate cancer were not germline mutation carriers (eFigure 1B in the Supplement). Six of the 32 patients with MSI-H/dMMR disease underwent sequencing of 2 or more tumors acquired during their disease course (Figure 3). Two of these demonstrated somatic acquisition of MSI-H/dMMR status in mCRPC tumors obtained later in their disease course, whereas their earlier tumors showed no evidence of MSI. The earlier tumors had adequate tumor content as quantitated by histopathologic review (50%-70%) and bioinformatically (24%-60%) (eMethods in the Supplement). The remaining 4 patients had evidence of MSI-H/dMMR in all profiled tumors, including 1 patient with a germline PMS2 mutation.
Figure 3. Longitudinal Assessment of Microsatellite Instability–High and Mismatch Repair–Deficient Status (MSI-H/dMMR) in Matched Tumors.
Six patients with MSI-H/dMMR prostate cancer had more than 1 tumor profiled. The time of acquisition of these tumors is indicated relative to the time of acquisition of the first profiled tumor (month 0). Two of the 6 patients (P-0000964 and P-0007143) had evidence of somatically acquired MSI in the latter metastatic castration–resistant tumors. For patient P-0007143, the earlier tumor was a prostate sample acquired at his diagnosis, at which time he had de novo metastatic noncastrate disease. This tumor had a very low MSIsensor score and tumor mutation burden (TMB). One patient (P-0000449) with a germline PMS2 mutation displayed MSI in both matched tumors. IHC indicates immunohistochemical analysis.
Clinical Characteristics of Patients With MSI-H/dMMR Prostate Cancer
The clinical characteristics of the 32 patients with MSI-H/dMMR cancer are summarized in eTable 2 in the Supplement. Median age at diagnosis was 64.5 years (range, 39-85 years). One patient had pure small-cell histologic findings. Among the 31 patients with prostate adenocarcinoma, 21 (67.7%) had mCRPC as their last disease state. For those patients, the median time to castration resistance was 8.6 months (range, 1.2-54.2 months) and the median duration of treatment with first-line abiraterone acetate or enzalutamide for mCRPC was 9.9 months (range, 3.0-34.5 months).
Responses to Anti–PD-1/PD-L1 Therapy in MSI-H/dMMR mCRPC
As of May 2018, 11 of the 32 patients with MSI-H/dMMR had received an anti–PD-1 or anti–PD-L1 agent for mCRPC as monotherapy or in combination with another immunomodulatory agent (Figure 4). The remaining 21 patients had not received immune checkpoint blockade owing to death before pembrolizumab approval or had not yet experienced disease progression during standard therapies.
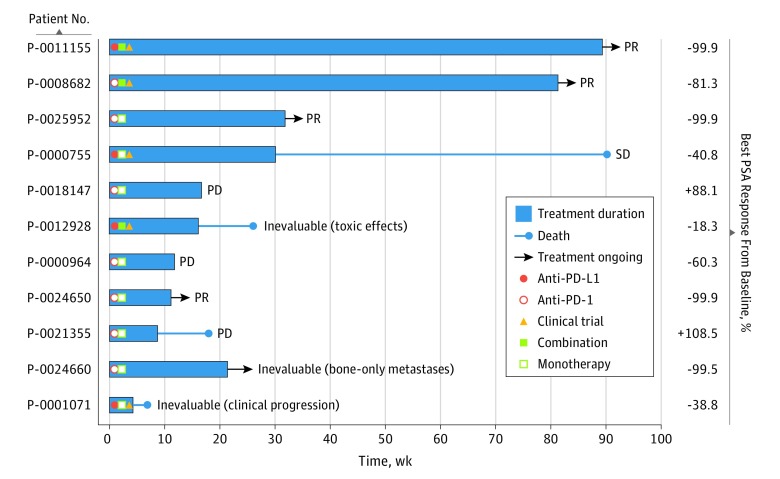
Figure 4. Responses to Immune Checkpoint Blockade in Microsatellite Instability–High and Mismatch Repair Deficient (MSI-H/dMMR) Prostate Cancer.
Eleven patients with MSI-H/dMMR castration-resistant prostate cancer received an anti–programmed cell death protein 1 (PD-1)/programmed cell death 1 ligand 1 (PD-L1) agent. Best prostate-specific antigen (PSA) response from baseline to date, and radiographic responses (partial response [PR], stable disease [SD], progressive disease [PD]) are noted. As of May 31, 2018, 5 patients continued to receive treatment with greater than 50% declines in PSA levels, 4 of whom had objective radiographic responses (eFigure 5 in the Supplement). One patient (eFigure 5B in the Supplement) had an initial radiographic PR, progressed in the prostate and by PSA, underwent palliative radiotherapy to the prostate, and continued to receive immune therapy with further clinical benefit. One patient had stable disease (SD) for approximately 6 months. Five patients showed no benefit, one of whom died, possibly owing to drug-related toxic effects.
For the 11 patients with mCRPC treated with anti–PD-1/PD-L1 therapy, duration of therapy ranged from 4.6 to 89 weeks or longer (Figure 4). Six (54.5%) achieved a decline in prostate-specific antigen (PSA) levels greater than 50%, including 4 with a greater than 99% decline. Eight patients were evaluable for radiographic response; of these, 4 achieved objective responses (eFigure 5 in the Supplement), 1 had stable disease for approximately 6 months, and 3 had radiographic progression as best response. Disease in the remaining 3 patients was inevaluable for radiographic response, owing to early clinical progression or toxic effects in 2 and bone-only disease in 1, although the latter had a durable decline in PSA levels greater than 99%. In total, 5 of 11 patients with MSI-H/dMMR mCRPC (45.5%) continued to receive therapy with durable clinical benefit, 5 (45.5%) had no benefit, and 1 (9.1%) had stable disease for approximately 6 months.
Discussion
The National Comprehensive Cancer Network guidelines for prostate cancer were recently amended to include consideration of MSI-H/dMMR testing and pembrolizumab treatment for MSI-H/dMMR mCRPC in the second-line setting or beyond.7 However, in its approval of pembrolizumab, the US Food and Drug Administration did not define how MSI-H/dMMR status should be evaluated, and detailed guidance is not provided as part of national guidelines. Some next-generation sequencing assays can assess for MSI-H/dMMR status by interrogating microsatellite loci for evidence of MSI, by identifying mutations and copy number alterations in MMR-associated genes, and by inferring TMB, a phenotypic hallmark of MSI-H/dMMR tumors.10,11,13 Next-generation sequencing–based tumor genomic profiling may therefore represent a robust and efficient strategy to identify the subset of patients with prostate cancer who may benefit from anti–PD-1/PD-L1 therapy.
Herein, we report the largest experience to date of MSI-H/dMMR characterization in 1346 patients with prostate cancer. Thirty-two (3.1%) of the 1033 patients with evaluable disease had phenotypic evidence of MSI-H/dMMR. We also found that 21.9% of patients with MSI-H/dMMR prostate cancer had a germline mutation in an MMR gene, suggesting that germline testing should be considered for all patients with MSI-H/dMMR prostate cancer. We acknowledge that the strict criteria used to define MSI-H/dMMR in our study, including the tumor purity threshold required to report MSIsensor scores, may have resulted in false-negative results in a small number of cases. Our experience suggests that, in clinical practice, it will be of benefit to incorporate information on mutational burden and mutational status of MMR genes in cases in which MSIsensor scores are indeterminate or tumor tissue is of low quality or purity. Sequencing can also identify those patients for whom IHC may be of incremental utility.
A notable finding from our study was that the MSI-H/dMMR phenotype was acquired somatically later during disease evolution in 2 of 6 patients who underwent longitudinal tumor profiling. Although MSI may have been subclonal in the earlier samples and thus missed owing to tumor heterogeneity, this result indicates that it was not a truncal event and would have been missed if only an older archival sample had been profiled. These data suggest that metastatic tissue may represent the optimal material for assessment of MSI status.
Because the MSI-H/dMMR phenotype is uncommon in prostate cancer, data describing responses to immune checkpoint blockade in this disease subset remain limited. Herein, we report clinical outcomes for the largest group of patients with MSI-H/dMMR mCRPC treated with anti–PD-1/PD-L1 therapy. Overall, 45.5% of patients with MSI-H/dMMR mCRPC derived durable clinical benefit, in line with other MSI-H/dMMR malignant neoplasms.5 Because approximately half of patients with MSI-H/dMMR had no response to immunotherapy, future studies should explore mechanisms of resistance in this population, which may involve alterations in the tumor antigen–presenting machinery and tumor-extrinsic factors, including inadequate T-cell activation.
Limitations
Our study has several limitations. It is a single-center retrospective analysis, and the number of patients with MSI-H who were treated with anti–PD-1/PD-L1 therapy is limited. Immunohistochemical analysis was not possible in all patients because of limited tumor tissue. Not all patients had tumors of sufficient quality for MSIsensor analysis, which could be addressed in the future with emerging cell-free DNA platforms.
Conclusions
In conclusion, we propose a comprehensive sequencing-based approach to identify patients with MSI-H/dMMR prostate cancer. Our results demonstrate that anti–PD-1/PD-L1 therapy is associated with meaningful clinical benefit in nearly half of MSI-H/dMMR mCRPC.
eMethods. Mutational Signature Analysis and Tumor Purity Assessment
eTable 1.
eTable 2. Summary Clinical Characteristics of MSI-H/dMMR Patients (n = 32)
eTable 3. Patients With Deleterious MMR Gene Alterations and no Evidence of MSI/Hypermutation
eFigure 1. Frequency of MSI-H/dMMR Prostate Cancer and Germline MMR Gene Alterations Among MSI-H/dMMR Patients
eFigure 2. Sample Adequacy for MSIsensor Assessment and Frequencies of MSI-H/dMMR Prostate Cancer
eFigure 3. Clinical Characteristics at the Time of Tumor Acquisition and MSI-H/dMMR Frequency in Advanced Tumors
eFigure 4. Proposed Guideline for the Identification of MSI-H/dMMR Prostate Cancer
eFigure 5. PSA and Radiographic Responses in 4 Patients With MSI-H/dMMR mCRPC Who Received an Anti–PD-1/PD-L1 Agent
References
- 1.Beer TM, Kwon ED, Drake CG, et al. Randomized, double-blind, phase III trial of ipilimumab versus placebo in asymptomatic or minimally symptomatic patients with metastatic chemotherapy-naive castration-resistant prostate cancer. J Clin Oncol. 2017;35(1):40-47. doi: 10.1200/JCO.2016.69.1584 [DOI] [PubMed] [Google Scholar]
- 2.Kwon ED, Drake CG, Scher HI, et al. ; CA184-043 Investigators . Ipilimumab versus placebo after radiotherapy in patients with metastatic castration-resistant prostate cancer that had progressed after docetaxel chemotherapy (CA184-043): a multicentre, randomised, double-blind, phase 3 trial. Lancet Oncol. 2014;15(7):700-712. doi: 10.1016/S1470-2045(14)70189-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Topalian SL, Hodi FS, Brahmer JR, et al. Safety, activity, and immune correlates of anti–PD-1 antibody in cancer. N Engl J Med. 2012;366(26):2443-2454. doi: 10.1056/NEJMoa1200690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Graff JN, Alumkal JJ, Drake CG, et al. Early evidence of anti–PD-1 activity in enzalutamide-resistant prostate cancer. Oncotarget. 2016;7(33):52810-52817. doi: 10.18632/oncotarget.10547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Le DT, Durham JN, Smith KN, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357(6349):409-413. doi: 10.1126/science.aan6733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hansen AR, Massard C, Ott PA, et al. Pembrolizumab for advanced prostate adenocarcinoma: findings of the KEYNOTE-028 study. Ann Oncol. 2018;29(8):1807-1813. doi: 10.1093/annonc/mdy232 [DOI] [PubMed] [Google Scholar]
- 7.National Comprehensive Cancer Network. Prostate cancer (version 3). https://www.nccn.org/professionals/physician_gls/pdf/prostate.pdf. Accessed June 25, 2018.
- 8.Guedes LB, Antonarakis ES, Schweizer MT, et al. MSH2 loss in primary prostate cancer. Clin Cancer Res. 2017;23(22):6863-6874. doi: 10.1158/1078-0432.CCR-17-0955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pritchard CC, Morrissey C, Kumar A, et al. Complex MSH2 and MSH6 mutations in hypermutated microsatellite unstable advanced prostate cancer. Nat Commun. 2014;5:4988. doi: 10.1038/ncomms5988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abida W, Armenia J, Gopalan A, et al. Prospective genomic profiling of prostate cancer across disease states reveals germline and somatic alterations that may affect clinical decision making [published online May 31, 2017]. JCO Precis Oncol. 2017;2017. doi: 10.1200/PO.17.00029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robinson D, Van Allen EM, Wu YM, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161(5):1215-1228. doi: 10.1016/j.cell.2015.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering–Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn. 2015;17(3):251-264. doi: 10.1016/j.jmoldx.2014.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zehir A, Benayed R, Shah RH, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10 000 patients. Nat Med. 2017;23(6):703-713. doi: 10.1038/nm.4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Middha S, Zhang L, Nafa K, et al. Reliable pan-cancer microsatellite instability assessment by using targeted next-generation sequencing data. JCO Precis Oncol. 2017;2017. doi: 10.1200/PO.17.00084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Niu B, Ye K, Zhang Q, et al. MSIsensor: microsatellite instability detection using paired tumor-normal sequence data. Bioinformatics. 2014;30(7):1015-1016. doi: 10.1093/bioinformatics/btt755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alexandrov LB, Nik-Zainal S, Wedge DC, et al. ; Australian Pancreatic Cancer Genome Initiative; ICGC Breast Cancer Consortium; ICGC MMML-Seq Consortium; ICGC PedBrain . Signatures of mutational processes in human cancer [published correction appears in Nature. 2013;502(7470):258]. Nature. 2013;500(7463):415-421. doi: 10.1038/nature12477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alexandrov LB, Nik-Zainal S, Wedge DC, Campbell PJ, Stratton MR. Deciphering signatures of mutational processes operative in human cancer. Cell Rep. 2013;3(1):246-259. doi: 10.1016/j.celrep.2012.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mandelker D, Zhang L, Kemel Y, et al. Mutation detection in patients with advanced cancer by universal sequencing of cancer-related genes in tumor and normal DNA vs guideline-based germline testing. JAMA. 2017;318(9):825-835. doi: 10.1001/jama.2017.11137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pritchard CC, Mateo J, Walsh MF, et al. Inherited DNA-repair gene mutations in men with metastatic prostate cancer. N Engl J Med. 2016;375(5):443-453. doi: 10.1056/NEJMoa1603144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chakravarty D, Gao J, Phillips SM, et al. OncoKB: a precision oncology knowledge base [published online May 16, 2017]. JCO Precis Oncol. 2017;2017. doi: 10.1200/PO.17.00011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dominguez-Valentin M, Joost P, Therkildsen C, Jonsson M, Rambech E, Nilbert M. Frequent mismatch-repair defects link prostate cancer to Lynch syndrome. BMC Urol. 2016;16:15. doi: 10.1186/s12894-016-0130-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eMethods. Mutational Signature Analysis and Tumor Purity Assessment
eTable 1.
eTable 2. Summary Clinical Characteristics of MSI-H/dMMR Patients (n = 32)
eTable 3. Patients With Deleterious MMR Gene Alterations and no Evidence of MSI/Hypermutation
eFigure 1. Frequency of MSI-H/dMMR Prostate Cancer and Germline MMR Gene Alterations Among MSI-H/dMMR Patients
eFigure 2. Sample Adequacy for MSIsensor Assessment and Frequencies of MSI-H/dMMR Prostate Cancer
eFigure 3. Clinical Characteristics at the Time of Tumor Acquisition and MSI-H/dMMR Frequency in Advanced Tumors
eFigure 4. Proposed Guideline for the Identification of MSI-H/dMMR Prostate Cancer
eFigure 5. PSA and Radiographic Responses in 4 Patients With MSI-H/dMMR mCRPC Who Received an Anti–PD-1/PD-L1 Agent