Primary mediastinal nonseminomatous (PMN) germ cell tumor are rare neoplasms arising from the anterior mediastinum in young adults. This article analyzes a large series of patients with germ cell tumors, including PMN germ cell tumors, and aims to identify precision treatments for this aggressive disease.
Abstract
Primary mediastinal nonseminomatous germ cell tumors (PMNSGCT) frequently become refractory to chemotherapy, and no effective salvage therapy exists. We performed genomic profiling on a series of 44 PMNSGCT and compared the results with those from chemorefractory, metastatic pure seminomatous (Sem, n = 22) and nonseminomatous (NS, n = 86) testicular germ cell tumors. Archival tissues were sequenced by a hybrid capture‐based technology (FoundationONE; Foundation Medicine, Inc., Cambridge, MA). Microsatellite instability (MSI) and tumor mutational burden (TMB, mutations [mut]/Mb) were determined.
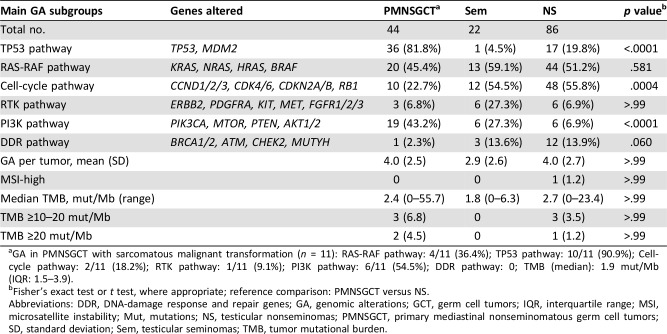
Statistically significant differences in genomic alterations (GA) of PMNSGCT versus NS included higher TP53 pathway GA (p < .0001), PIK3CA pathway GA (p < .0001), and lower cell‐cycle pathway GA (p = .0004). There were no MSI‐high PMNSGCT cases. Mean TMB was similar between the groups, but there were more ≥10 mut/Mb in the PMNSGCT group versus NS (11.4% vs. 4.6%).
The GA identified in PMNSGCT were similar to the findings from NS, with differential opportunities for targeted therapies and immunotherapies. Further study of precision treatments appears warranted.
Introduction
Primary mediastinal nonseminomatous germ cell tumors (PMNSGCT) are rare neoplasms arising from the anterior mediastinum in young adults. Their clinical behavior is usually more aggressive than that associated with their primary gonadal counterpart. PMNSGCT carry a relatively poor prognosis, with only 40%–50% of patients cured with first‐line chemotherapy compared with the 90% or greater cure for testicular germ cell tumors (GCT) treated with similar regimens. Those patients who relapse have only a limited likelihood of being rescued by second‐line chemotherapy, including high‐dose chemotherapy or aggressive surgery [1]. For this reason, clinical investigation of new therapeutic options is warranted for these patients. However, clinical research on GCT has historically suffered from huge limitations owing to the low frequency of the disease and to the lack of known druggable molecular drivers. In the following study, we used genomic profiling to search for new routes to targeted therapy and immunotherapy for this uncommon and challenging disease, and we aimed at comparing the frequency of the genomic alterations identified in refractory GCT of different tumor origin.
Materials and Methods
Archival tissues from 152 patients with chemotherapy‐treated and refractory GCT were sequenced using a U.S. Food and Drug Administration‐approved hybrid capture‐based genomic profiling assay (FoundationONE; Foundation Medicine, Inc., Cambridge, MA) in a Clinical Laboratory Improvement Amendments‐certified, College of American Pathologists‐accredited laboratory (Foundation Medicine, Inc.). The pathologic diagnosis of each case was centrally reviewed by an experienced pathologist (J.S.R.).
In brief, ≥50 ng DNA was extracted from formalin‐fixed, paraffin‐embedded tissue blocks. The samples were assayed using adaptor‐ligation, and hybrid capture for all coding exons from 287 (version 1) to 315 (version 2) cancer‐related genes plus select introns from 19 (version 1) to 28 (version 2) genes frequently rearranged in cancer was performed. Sequencing of captured libraries was performed using the Illumina HiSeq technology (Illumina, San Diego, CA) to a mean exon coverage depth of >500×. Resultant sequences were analyzed for all classes of genomic alterations, including short variant alterations (base substitutions, insertions, and deletions), copy number alterations (focal amplifications [<20 Mb], nonfocal amplifications [≥20 Mb], and homozygous deletions), and selected gene fusions or rearrangements, as previously described [2]. Microsatellite instability (MSI) was determined on 114 loci. Tumor mutational burden (TMB, reported as mutations [mut]/Mb) was determined on 0.8 Mb (version 1) or 1.1 Mb (version 2) of sequenced DNA for each sample based on the number of somatic base substitution or indel alterations per Mb after filtering to remove known functionally oncogenic somatic mutation. TMB definitions followed the cutoffs originally established in melanoma, non‐small cell lung cancer, and urothelial bladder cancer: low (0–5 mut/Mb), intermediate (6–19 mut/Mb), and high (>20 mut/Mb). The analyzed patients should have experienced a relapse after at least one cisplatin‐based chemotherapy regimen. Among these patients, we identified 44 PMNSGCT, 22 testicular seminomas, and 86 testicular nonseminomas or mixed GCT. The characteristics and molecular findings from patients with testicular GCT have been already presented [3].
Results and Discussion
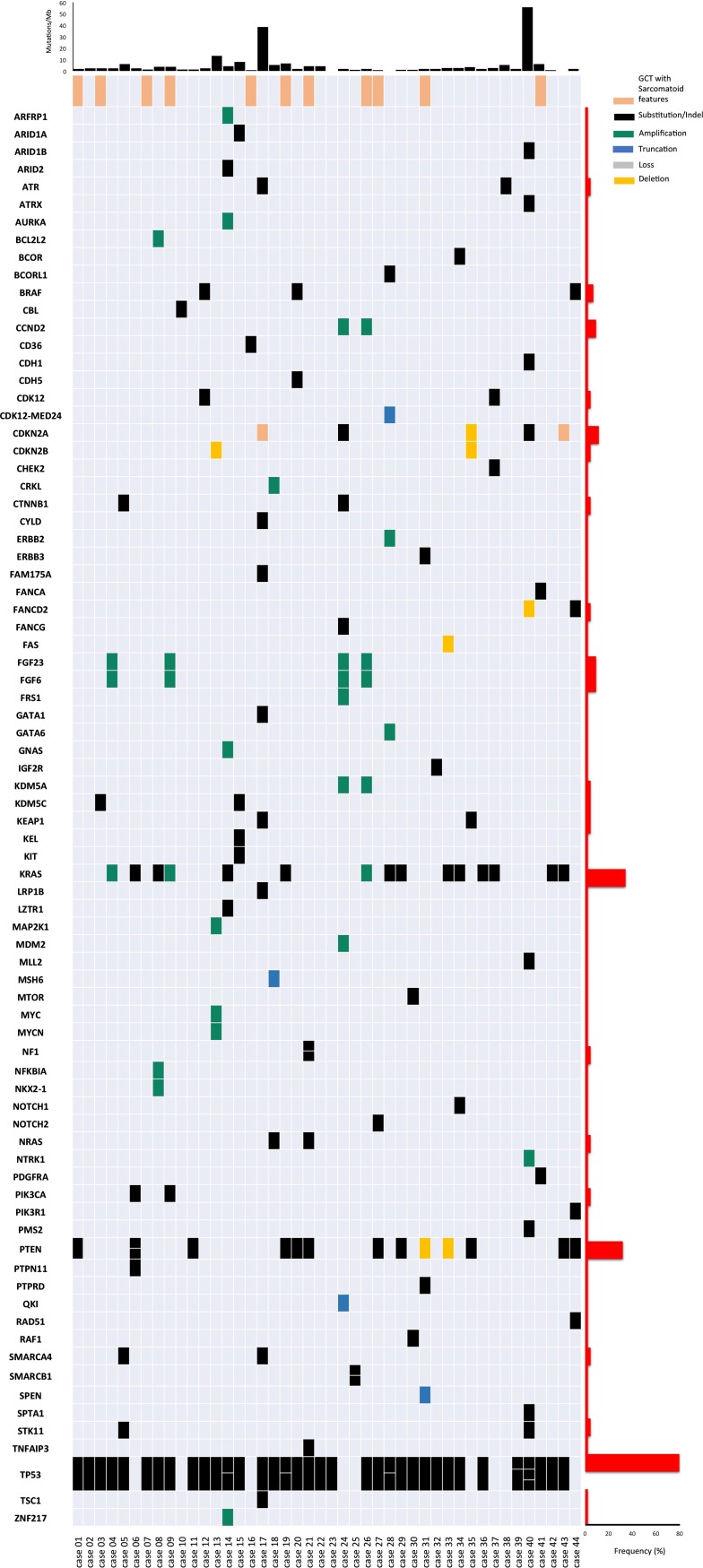
Among PMNSGCT, there were 43 males and 1 female, median age was 28 years (interquartile range: 20–36.7), 11 (25%) showed sarcomatous features suggestive of malignant transformation, 2 (4.5%) had unresectable mature teratoma, and 15 patients (34.1%) showed a yolk sac tumor component. The source of tumor tissue was the primary mediastinal mass, visceral metastases, and lymph node metastases in 23 (52.3%), 19 (43.2%), and 2 (4.5%) cases, respectively. Prechemotherapy tumor tissue was analyzed in 41 patients (93.2%). The comparison of genomic profiling between PMNSGCT and testicular GCT is presented in Table 1. Statistically significant differences in the distribution of genomic alterations consisted of more frequent alterations related to the TP53 and PI3K pathway and less frequent alterations in cell‐cycle pathway. No significant differences were found in the distribution of DNA damage response and repair gene alterations, that were found in 2.3% of PMNSGCT. No cases of MSI‐high tumors were observed among PMNSGCT. Although the median TMB was equally low (2.4 mut/Mb in PMNSGCT, 2.7 mut/Mb in nonseminoma, and 1.8 mut/Mb in seminoma), there were several cases with intermediate or high TMB in PMNSGCT, resembling testicular nonseminomas, in contrast with testicular seminomas. Sarcomatous features were identified in 3 (3.5%) of 86 nonseminomatous testicular GCT. Findings obtained in the population of patients with sarcomatous components resembled those from the overall PMNSGCT, except for a lower median TMB (1.9 mut/Mb). The overall spectrum of the single genomic alterations identified in PMNSGCT is shown in Figure 1. Targetable genomic alterations impacting downstream pathway activations included BRAF (n = 3, 6.8%), ERBB2, NTRK1, MTOR, and TSC1 genes (n = 1, 2.3% in all cases). When the PMNSGCT in which the primary tumor was sequenced is compared with cases where a recurrent or metastatic tumor site was sequenced, the KRAS and PTEN gene alteration frequencies were similar, BRAF alterations were identified in both groups, and ERBB2, NTRK1, KIT, and TSC1 alterations were identified in the recurrent/metastatic tumors only.
Table 1. Genomic alterations observed in PMNSGCT versus features from gonadal primary GCT.
GA in PMNSGCT with sarcomatous malignant transformation (n = 11): RAS‐RAF pathway: 4/11 (36.4%); TP53 pathway: 10/11 (90.9%); Cell‐cycle pathway: 2/11 (18.2%); RTK pathway: 1/11 (9.1%); PI3K pathway: 6/11 (54.5%); DDR pathway: 0; TMB (median): 1.9 mut/Mb (IQR: 1.5–3.9).
Fisher's exact test or t test, where appropriate; reference comparison: PMNSGCT versus NS.
Abbreviations: DDR, DNA‐damage response and repair genes; GA, genomic alterations; GCT, germ cell tumors; IQR, interquartile range; MSI, microsatellite instability; Mut, mutations; NS, testicular nonseminomas; PMNSGCT, primary mediastinal nonseminomatous germ cell tumors; SD, standard deviation; Sem, testicular seminomas; TMB, tumor mutational burden.
Figure 1.
Oncoprint displaying all the genomic alterations observed in the population of 44 patients with primary mediastinal nonseminomatous germ cell tumors. Genes are ordered alphabetically. At the top of the figure, the plot of tumor mutational burden (mutations/Mb) for each single patient is reported. On the right, there is a plot indicating, for each single gene, the frequency of observed genomic alterations.
Abbreviation: GCT, germ cell tumor.
The present study analyzed one of the largest series of patients with refractory GCT, including PMNSGCT, reported thus far; it also included the molecular characterization of sarcomatous malignant transformation arising from PMNSGCT, for which no similar data are available when looking at the most relevant studies [4]. The study is aimed at providing clinical researchers with new opportunities for investigation in the very uncommon PMNSGCT, which is considered an orphan disease. The FoundationONE assay was primarily developed to obtain a comprehensive molecular characterization of tumor samples for clinical use, mainly in those patients who develop resistance to conventional therapies, as well as to give patients the opportunity of being included in clinical trials of targeted drugs based on their molecular profile. As reported in a previous study, TP53 alterations were notably prevalent in PMNSGCT [4]. Therefore, this is the second large study corroborating the negative prognostic significance of TP53 alterations in PMNSGCT, and integrating this feature into clinical prognostic models is warranted.
Similar to the findings in the testicular GCT study, although the genomic alterations found in PMNSGCT did not reveal a high frequency of potential genomic targets, there were intriguing options for both targeted therapies and immunotherapies in the PMNSGCT group in selected patients. Based on our findings, there may be opportunities for targeted therapies in PMNSGCT with several agents targeting the PI3K pathway, added to a few more opportunities for agents targeting BRAF, ERBB2, and NTRK1 in the few genomically altered patients. Given the absence of MSI‐high tumors and the relatively low TMB associated with these neoplasms, we confirmed that there are huge limitations, at the molecular level, for the rational development of immunotherapy in GCT, including PMNSGCT. These findings add to the ultimate literature reporting discouraging preliminary findings from trials of immune checkpoint inhibitors in refractory GCT [5], [6] and enforce the assumption that therapeutic opportunities may come through appropriate patient selection. Limitations in our study include the lack of annotated clinical data, which prevented us from analyzing the effect of genomic profiling on the efficacy of specific therapeutic regimens and from conducting survival analyses based on the genomic findings.
Conclusion
Genomic alterations identified in this study were similar to those identified in primary testicular NS, with a higher frequency of yolk sac differentiation and TP53 alterations and slightly increased opportunities for targeted therapies (BRAF, ERBB2, and NTRK1) and immunotherapies (5% with TMB ≥20 mut/Mb). Further study of precision treatments for this orphan disease appears warranted.
Acknowledgments
This work was supported by Foundation Medicine, Inc., Cambridge, Massachusetts.
Disclosures
Jon Chung: Foundation Medicine, Inc. (E); Sherri Millis: Foundation Medicine, Inc. (E); Laurie M. Gay: Foundation Medicine, Inc. (E, OI); Siraj M. Ali: Foundation Medicine, Inc. (E, OI, IP), Incysus (SAB); Jeffrey S. Ross: Foundation Medicine, Inc. (E, OI). The other authors indicated no financial relationships.
(C/A) Consulting/advisory relationship; (RF) Research funding; (E) Employment; (ET) Expert testimony; (H) Honoraria received; (OI) Ownership interests; (IP) Intellectual property rights/inventor/patent holder; (SAB) Scientific advisory board
References
- 1.International Prognostic Factors Study Group , Lorch A, Beyer J et al. Prognostic factors in patients with metastatic germ cell tumors who experienced treatment failure with cisplatin‐based first‐line chemotherapy. J Clin Oncol 2010;28:4906–4911. [DOI] [PubMed] [Google Scholar]
- 2.Chalmers ZR, Connelly CF, Fabrizio D et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med 2017;9:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Necchi A, Bratslavsky G, Corona RJ et al. Comprehensive genomic characterization of chemotherapy‐resistant testicular germ cell tumors (TGCT). J Clin Oncol 2018;36(suppl 15):4555a. [Google Scholar]
- 4.Bagrodia A, Lee BH, Lee W et al. Genetic determinants of cisplatin resistance in patients with advanced germ cell tumors. J Clin Oncol 2016;34:4000–4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adra N, Einhorn LH, Althouse SK et al. Phase II trial of pembrolizumab in patients with platinum refractory germ‐cell tumors: A Hoosier Cancer Research Network Study GU14‐206. Ann Oncol 2018;29:209–214. [DOI] [PubMed] [Google Scholar]
- 6.Raggi D, Giannatempo P, Mariani L et al. Apache: An open label, randomized, phase 2 study of durvalumab (Durva), alone or in combination with tremelimumab (Treme), in patients (pts) with advanced germ cell tumors (GCT): Results at the end of first stage. J Clin Oncol 2018;36(suppl 15):4547a. [Google Scholar]