Abstract
Accurate and informative microbiological testing is essential for guiding diagnosis and management of pneumonia in patients who are critically ill. Sampling of tracheal aspirate (TA) is less invasive compared with mini-bronchoalveolar lavage (mBAL) and is now recommended as a frontline diagnostic approach in patients who are mechanically ventilated, despite the historical belief that TA was suboptimal due to contamination from oral microbes. Advancements in metagenomic next-generation sequencing (mNGS) now permit assessment of airway microbiota without a need for culture and, as such, provide an opportunity to examine differences between mBAL and TA at a resolution previously unachievable. Here, we engaged shotgun mNGS to assess quantitatively the airway microbiome in matched mBAL and TA specimens from a prospective cohort of critically ill adults. We observed moderate differences between sample types across all subjects; however, we found significant compositional similarity in subjects with bacterial pneumonia, whose microbial communities were characterized by dominant pathogens. In contrast, in patients with noninfectious acute respiratory illnesses, significant differences were observed between sample types. Our findings suggest that TA sampling provides a similar assessment of airway microbiota as more invasive testing by mBAL in patients with pneumonia.
Keywords: lower respiratory tract infection, mini-bronchial alveolar lavage, next-generation sequencing, pneumonia, tracheal aspirate
INTRODUCTION
Pneumonia causes more deaths each year in the United States than any other type of infectious disease (7). The ability to accurately detect etiological pathogens and distinguish them from background commensal microbiota is essential for guiding optimal antimicrobial treatment. In patients requiring mechanical ventilation, less invasive tracheal aspirate (TA) sampling has historically been considered inferior to specimen collection by mini-bronchoalveolar lavage/telescoping catheter (mBAL) due to the potential for oropharyngeal microbiota contamination (3, 5). This idea has been challenged, however, by studies demonstrating a lack of clinically significant differences between sample types (2–5), and a greater acceptance of TA sampling is now reflected in recent updates to clinical practice guidelines (10). Despite the broad potential implications of this shift in diagnostic sampling approach, relatively little information exists regarding microbial composition differences between mBAL and TA specimens and the potential implications of such differences for both clinical diagnostic testing and airway microbiome studies. To address this gap in knowledge, we employed shotgun metagenomic next-generation sequencing (mNGS) to evaluate the microbial compositions of matched mBAL and TA specimens.
METHODS
We enrolled 52 adults who were intubated within 72 h of intensive care unit admission with acute respiratory failure according to University of California, San Francisco institutional review board-approved protocol 10-02701. Demographic and clinical characteristics of the 52 study subjects are summarized in (Table 1). Two-physician adjudication based on retrospective medical record review (blinded to mNGS results) and the US Centers for Disease Control and Prevention surveillance case definition of pneumonia were used to identify 15 subjects with culture-confirmed bacterial pneumonia (PNA-pos; Ref. 18). Adjudication also identified 12 subjects with a clear alternative noninfectious etiology of acute respiratory failure (PNA-neg) and 25 subjects with acute respiratory illnesses of indeterminate etiology (PNA-ind), which included those with negative bacterial cultures but suspected pneumonia based on clinical criteria alone. Subjects with PCR-confirmed viral etiologies were also included in the PNA-ind group because occult bacterial coinfection could not be excluded.
Table 1.
Demographics and clinical characteristics of study cohort
| Total | PNA-pos | PNA-neg | PNA-ind | |
|---|---|---|---|---|
| Total enrolled, n | 52 | 15 | 12 | 25 |
| Age, average yr | 63 | 60 | 64 | 64 |
| Female sex, n | 17 (33%) | 3 (20%) | 7 (58%) | 7 (28%) |
| Race | ||||
| Black, n | 3 (6%) | 1 (7%) | 0 (0%) | 2 (8%) |
| Asian American, n | 14 (27%) | 4 (27%) | 2 (15%) | 8 (32%) |
| White, n | 27 (52%) | 8 (53%) | 8 (67%) | 11 (44%) |
| Other, n | 4 (8%) | 0 (0%) | 2 (17%) | 2 (8%) |
| Hispanic ethnicity, n | 4 (8%) | 2 (13%) | 0 (0%) | 2 (8%) |
| Suspected pneumonia type | ||||
| Community-acquired pneumonia, n | 22 (42%) | 8 (53%) | 0 (0%) | 14 (56%) |
| Hospital-acquired pneumonia, n | 8 (15%) | 5 (33%) | 0 (0%) | 3 (12%) |
| Ventilator-associated pneumonia, n | 2 (4%) | 2 (13%) | 0 (0%) | 0 (0%) |
| Indeterminate pneumonia status, n | 8 (15%) | 0 (0%) | 0 (0%) | 8 (32%) |
| Immunosuppression, n | 18 (35%) | 6 (40%) | 4 (33%) | 8 (32%) |
| Prior antibiotic use, n | 43 (83%) | 11 (73%) | 12 (100%) | 20 (80%) |
| Bacteremia, n | 12 (23%) | 4 (27%) | 2 (17%) | 6 (25%) |
| 30-Day mortality, n | 14 (27%) | 4 (27%) | 2 (17%) | 8 (32%) |
PNA-ind, pneumonia-indeterminate; PNA-neg, pneumonia-negative; PNA-pos, pneumonia-positive.
Excess mBAL and TA specimens collected on the same day and within 72 h of patient intensive care unit admission underwent DNA extraction and sequencing library preparation according to previously described methods (14, 19). After paired-end Illumina sequencing, we employed a previously reported bioinformatics pipeline to detect and profile the airway microbiome. Briefly, this incorporated subtractive alignment of the human genome [National Center for Biotechnology Information Genome Reference Consortium human build 38 (GRCh38)] using Spliced Transcripts Alignment to a Reference (STAR; Ref. 6) followed by quality filtering using PriceSeqFilter (17). Additional filtering to remove Pan troglodytes (University of California, Santa Cruz panTro4) and nonfungal eukaryotes, cloning vectors, and phiX phage was performed using Bowtie 2 (15). The identities of the remaining microbial reads were determined by querying the National Center for Biotechnology Information nucleotide database using Genomic Short-read Nucleotide Alignment Program (GSNAP) -l (14, 19). We sequenced no-template water control samples and restricted analyses to taxa present at >1% of the microbial population by abundance, as previously described (5, 14). No microbe was universally present in every sample, suggesting that systematic contamination across TA or mBAL sampling methods was unlikely. Microbial community composition metrics were calculated using the vegan R package version 2.5.2 (16). P values were computed using Wilcoxon rank sums. When evaluating community richness, one outlier (>3 standard deviations above the mean) was identified and removed before computing significance.
RESULTS
Comparison of mBAL with TA across all patient groups.
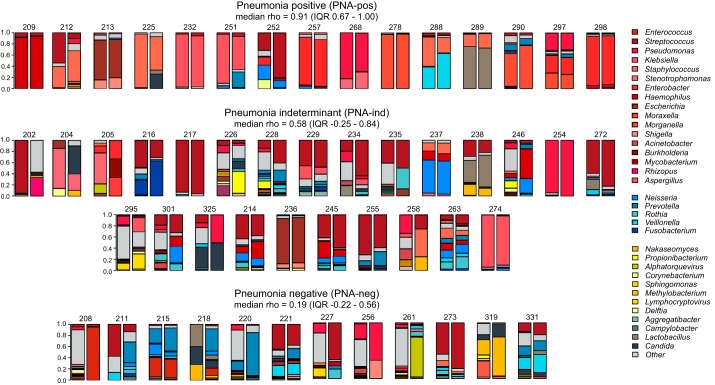
To compare the microbial community compositions of matched mBAL and TA specimens across all patients in the cohort, we first calculated the Bray-Curtis dissimilarity index, which revealed no significant differences (P = 0.31 by permutational multivariate ANOVA). We next asked whether within-subject diversity of the respiratory microbial communities differed by specimen type and did not observe a significant difference in the Shannon diversity index [SDI; 1.05 (0.71–1.55) vs. 1.45 (0.74–2.05) for TA and mBAL, respectively; P = 0.057; Table 2], although the P value approached significance. Community richness (total number of different genera identified in each sample), however, was higher in mBAL samples than in TA (P = 0.046; Table 2). Calculation of Spearman correlation between matched mBAL and TA specimens across all subjects revealed moderate differences, with a mean correlation of 0.41 (interquartile range: 0.03–0.87; Fig. 1).
Table 2.
Microbial community metrics in TA and mBAL samples, including differences between subjects with pneumonia and those with respiratory failure due to etiologies other than respiratory infection, and abundance differences for common oropharyngeal microbes by genus rpM between all TA and mBAL samples, irrespective of PNA group, calculated using Wilcoxon rank sum
| TA |
mBAL |
||||
|---|---|---|---|---|---|
| Metric | Median (IQR) | PNA-pos vs. PNA-neg | Median (IQR) | PNA-pos vs. PNA-neg | mBAL vs. TA (All Samples) |
| Richness | 6.00 (4.00–9.00) | P = 6.5 × 10−2 | 8.00 (4.00–13.50) | P = 1.2 × 10−3 | P = 4.6 × 10−2 |
| Shannon diversity | 1.05 (0.71–1.55) | P = 4.7 × 10−2 | 1.45 (0.74–2.05) | P = 5.2 × 10−6 | P = 5.7 × 10−2 |
| TA | mBAL | ||
|---|---|---|---|
| Genus | Mean rpM (IQR) | Mean rpM (IQR) | mBAL vs. TA |
| Prevotella | 0.07 (0.00–0.06) | 0.02 (0.00–0.03) | P = 0.15 |
| Veillonella | 0.03 (0.00–0.02) | 0.03 (0.00–0.02) | P = 0.99 |
| Streptococcus | 0.20 (0.00–0.33) | 0.18 (0.00–0.34) | P = 0.88 |
| Fusobacterium | 0.02 (0.00–0.00) | 0.01 (0.00–0.00) | P = 0.53 |
| Rothia | 0.03 (0.00–0.03) | 0.01 (0.00–0.00) | P = 0.31 |
| Neisseria | 0.04 (0.00–0.02) | 0.03 (0.00–0.02) | P = 0.72 |
IQR, interquartile range; mBAL, mini-bronchial alveolar lavage; PNA-neg, pneumonia-negative; PNA-pos, pneumonia-positive; rpM, reads per million reads mapped; TA, tracheal aspirate.
Fig. 1.
Fraction of total microbial sequencing reads represented by each genus in matched mini-bronchial alveolar lavage (left column) and tracheal aspirate (right column) specimens. Summary of the Spearman correlation ρ-values for pairwise comparisons in each PNA group is listed under each header. Legend colors correspond to the topmost abundant microbes across all samples, with red shading indicating microbes with established respiratory pathogenicity as recently defined (13), blue indicating genera known to be common oropharyngeal microbiota, and yellow/gray indicating other microbial genera. Category Other refers to all other microbes identified and those identified at abundance <1%. IQR, interquartile range.
Because oropharyngeal microbiota have historically been suspected to compromise TA specimens, we next evaluated for differences in the abundance of common oropharyngeal microbiota (5). Surprisingly, we found no statistically significant differences between mBAL and TA specimens with respect to Prevotella, Veillonella, Streptococcus, Fusobacterium, Rothia, or Neisseria abundance (Table 2). Assessment of microbial relative abundance (total genus alignments per million reads sequenced) also revealed no significant interspecimen type differences [median = 43.30 (interquartile range: 6.69–327.93) vs. 26.89 (5.66–167.63); P = 0.66].
Comparison of mBAL and TA as a function of pneumonia status.
We reasoned that differences in microbial composition between mBAL and TA specimens would be most clinically significant if they impacted diagnostic accuracy in patients with pneumonia and thus assessed taxonomic similarity between the PNA-pos and PNA-neg groups. We found significantly greater correlation in subjects who were PNA-pos compared with subjects who were PNA-neg [pairwise Spearman correlation of 0.75 (0.67–1.00) vs. 0.19 (−0.22 to 0.55); P = 1.62 × 10−3], suggesting that pathogen dominance of the lung microbiome during infection may drive compositional similarity (1, 14).
For both sample types, a culture-confirmed pathogen was the most abundant microbe detected by mNGS in 14 (93% of) subjects who were PNA-pos and the second most abundant in the remaining subject [Table 3; Supplemental Table S1 (https://doi.org/10.17504/protocols.io.wqnfdve)]. Gram-negative pathogens were cultured from a relatively high percentage of subjects who were PNA-pos (65%) compared with prior surveillance studies (8). mNGS of mBAL specimens detected all 23 culture-identified microbes, whereas mNGS of TA samples identified 22. The discrepant microbe was from a polymicrobial culture and was detected by mNGS in the TA specimen but present at <1% relative abundance and, therefore, indistinguishable from background using our bioinformatic approach [Table 3; Supplemental Table S1 (https://doi.org/10.17504/protocols.io.wqnfdve)].
Table 3.
Patient diagnoses and results of clinical microbiology testing for bacterial and viral pathogens
| No. | PNA Group | Diagnosis | Single vs. Polymicrobial | Microbe(s) Detected by Culture | Rank, TA | Rank, mBAL |
|---|---|---|---|---|---|---|
| 209 | PNA-pos | Community-acquired pneumonia | Polymicrobial | Haemophilus | 1 | 1 |
| Streptococcus | 2 | 2 | ||||
| 212 | PNA-pos | Hospital-acquired pneumonia | Polymicrobial | Staphylococcus | 2 | 1 |
| 213 | PNA-pos | Ventilator-associated pneumonia | Polymicrobial | Escherichia | 1 | 1 |
| Klebsiella | 4 | 5 | ||||
| 225 | PNA-pos | Community-acquired pneumonia | Single microbe | Staphylococcus | 1 | 1 |
| 232 | PNA-pos | Community-acquired pneumonia | Single microbe | Klebsiella | 1 | 1 |
| 251 | PNA-pos | Community-acquired pneumonia | Single microbe | Klebsiella | 1 | 1 |
| 252 | PNA-pos | Community-acquired pneumonia | Polymicrobial | Streptococcus | 1 | 1 |
| Enterobacter | 4 | 3 | ||||
| Staphylococcus | 7 | * | ||||
| 257 | PNA-pos | Hospital-acquired pneumonia | Single microbe | Enterobacter | 1 | 1 |
| 268 | PNA-pos | Community-acquired pneumonia | Polymicrobial | Pseudomonas | 1 | 1 |
| Stenotrophomonas | 2 | 2 | ||||
| 278 | PNA-pos | Hospital-acquired pneumonia | Single microbe | Moraxella | 1 | 1 |
| 288 | PNA-pos | Community-acquired pneumonia | Single microbe | Staphylococcus | 1 | 2 |
| 289 | PNA-pos | Community-acquired pneumonia | Single microbe | Staphylococcus | 2 | 2 |
| 290 | PNA-pos | Community-acquired pneumonia | Single microbe | Moraxella | 1 | 1 |
| 297 | PNA-pos | Hospital-acquired pneumonia | Polymicrobial | Enterobacter | 1 | 1 |
| Morganella | 2 | 2 | ||||
| Klebsiella | 3 | 3 | ||||
| Pseudomonas | 5 | 5 | ||||
| 298 | PNA-pos | Community-acquired pneumonia | Single microbe | Enterobacter | 1 | 1 |
| No. | PNA Group | Diagnosis | Microbe(s) Detected by Culture or PCR | |||
| 208 | PNA-neg | Septic shock due to Enterococcus bacteremia | ||||
| 211 | PNA-neg | Acute myocardial infarction | ||||
| 215 | PNA-neg | Craniotomy for resection of arteriovenous malformation | ||||
| 218 | PNA-neg | Small bowel obstruction, pancreatitis, and hypoxic respiratory failure | ||||
| 220 | PNA-neg | Elective craniotomy for aneurysm bypass surgery | ||||
| 221 | PNA-neg | Seizure | ||||
| 227 | PNA-neg | Hemorrhagic shock and PEA cardiac arrest | ||||
| 256 | PNA-neg | Status after heart transplant with suspected drug-related fever | ||||
| 261 | PNA-neg | Volume overload secondary to constrictive pericarditis | ||||
| 273 | PNA-neg | Hemorrhagic stroke | ||||
| 319 | PNA-neg | Status after balloon angioplasty of hepatic artery | ||||
| 331 | PNA-neg | Acute liver failure secondary to portal vein thrombosis | ||||
| 202 | PNA-ind | MSSA bacteremia, septic shock, and suspected pneumonia | ||||
| 204 | PNA-ind | Osteomyelitis and hypercarbic respiratory failure | ||||
| 205 | PNA-ind | Community-acquired pneumonia | Rhinovirus | |||
| 214 | PNA-ind | Intracranial hemorrhage | ||||
| 216 | PNA-ind | COPD exacerbation with hypercarbic respiratory failure and suspected pneumonia | ||||
| 217 | PNA-ind | Hypoxic respiratory failure and suspected pneumonia | ||||
| 226 | PNA-ind | Hypoxic respiratory failure and suspected pneumonia | ||||
| 228 | PNA-ind | Community-acquired pneumonia | RSV | |||
| 229 | PNA-ind | Altered mental status, septic shock, and suspected pneumonia | ||||
| 234 | PNA-ind | Severe sepsis and hypoxia with urinary tract infection | ||||
| 235 | PNA-ind | Acute renal failure, shock, hypoxic respiratory failure, and suspected pneumonia | ||||
| 236 | PNA-ind | Aspiration secondary to altered mental status | ||||
| 237 | PNA-ind | Seizure, hypernatremia, and suspected pneumonia | ||||
| 238 | PNA-ind | Hypoxemic and hypercarbic respiratory failure following PEA arrest | ||||
| 245 | PNA-ind | Seizure, altered mental status, and suspected aspiration | ||||
| 246 | PNA-ind | Hypoxic respiratory failure and suspected pneumonia | ||||
| 254 | PNA-ind | Respiratory failure, shock, and suspected pneumonia | ||||
| 255 | PNA-ind | Status after aortic valve repair | ||||
| 258 | PNA-ind | Subarachnoid hemorrhage | ||||
| 263 | PNA-ind | Altered mental status and hypoxic respiratory failure | ||||
| 272 | PNA-ind | Sepsis with possible aspiration pneumonia | ||||
| 274 | PNA-ind | Altered mental status following perioperative intubation | ||||
| 295 | PNA-ind | Community-acquired pneumonia | RSV | |||
| 301 | PNA-ind | Acute respiratory distress syndrome | ||||
| 325 | PNA-ind | Hypoxic respiratory failure |
For patients with positive clinical microbiology results, the rank of each bacterial genus based on abundance (reads per million reads mapped, rpM) of sequencing reads in matched mini-bronchial alveolar lavage or tracheal aspirate specimens is listed. Samples for which clinical diagnostics returned polymicrobial cultures contain multiple lines and are noted as polymicrobial. COPD, chronic obstructive pulmonary disease; MSSA, methicillin-sensitive Staphylococcus aureus; PEA, pulseless electrical activity; PNA-ind, acute respiratory illnesses of indeterminate etiology including patients with negative bacterial cultures but suspected bacterial pneumonia based on clinical criteria alone; PNA-neg, subjects with a clear alternative noninfectious etiology of acute respiratory failure; PNA-pos, clinically identified pneumonia with a positive bacterial culture; RSV, respiratory syncytial virus.
Present at <1% relative abundance.
Reduced α-diversity of the human respiratory microbiome has been described as an ecological marker of infection (1, 14), and thus we next asked whether SDI differed by specimen type. We found that SDI differed significantly between subjects who were PNA-pos and subjects who were PNA-neg within mBAL samples, and a similar, albeit less significant, difference was also observed within TA samples (P = 5.2 × 10−6 and 4.7 × 10−2, respectively; Fig. 2; Table 2). Community richness (total number of different genera identified in each sample) was decreased in subjects who were PNA-pos compared with subjects who were PNA-neg when assessed by mBAL and trended toward significance when assessed by TA (P = 1.2 × 10−3 and 6.5 × 10−2, respectively; Table 2). In addition, we calculated SDI and richness for patients in the PNA-ind group with culture-negative suspected pneumonia. Unlike the subjects who were PNA-pos, we did not observe significant differences in terms of SDI and richness compared with the subjects who were PNA-neg for either fluid type (P = 0.211 and P = 0.679 for SDI of mBAL and TA, respectively; P = 0.156 and P = 0.756 for richness of mBAL and TA, respectively).
Fig. 2.

Shannon diversity index in subjects who were pneumonia-positive (PNA-pos; △) vs. subjects who were pneumonia-negative (PNA-neg; ▲) by specimen type [mini-bronchial alveolar lavage (mBAL), left; tracheal aspirate (TA), right]. Subjects who were PNA-pos had lower diversity compared with subjects who were PNA-neg when assessed by either specimen type.
To explore differences in sample type as a function of pneumonia status further, we compared mBAL versus TA across each patient subgroup (PNA-pos, PNA-ind, and PNA-neg) with respect to Shannon diversity and richness [Supplemental Fig. S1 (https://doi.org/10.17504/protocols.io.wqnfdve)]. Whereas overall, mBAL samples trended toward increased community richness, we observed that this was driven largely by differences in the PNA-neg group. In subjects who were PNA-neg, significant differences between sample types were observed with respect to SDI and richness (P = 5.0 × 10−3 and 4.0 × 10−2, respectively) as well as by Spearman correlation [0.19 (−0.22 to 0.55)]. In contrast, in subjects who were PNA-pos, no differences were observed between fluid types based on SDI or richness (P = 0.46 and 0.88, respectively).
DISCUSSION
Advances in genome sequencing have revealed that the lung, previously considered sterile, supports diverse microbial communities that play a role in both health and disease (5). With the use of shotgun mNGS, we compared the microbial compositions of matched mBAL and TA samples from critically ill adults. Across all patient groups, moderate differences were observed based on Spearman correlation, differences approached significance with respect to α-diversity (SDI; P = 0.057), and richness was significantly higher in mBAL samples (P = 0.046). In contrast, we did not find systematic differences in the abundance of oropharyngeal microbes or in β-diversity, measured by Bray-Curtis index. Notably, however, we found that fluid type differences became inconsequential in the setting of clinically identified pneumonia and became more pronounced in patients with noninfectious acute respiratory illnesses.
Prior studies using 16S rRNA amplicon sequencing have observed differences in community richness in the setting of pneumonia (11, 12), and although we only found significant differences by mBAL, those for TA trended toward significance and may have demonstrated an association with a larger sample size. Finally, despite the historical assumption that TA specimens are compromised by oropharyngeal contamination, we found that abundance of oropharyngeal microbiota did not significantly differ by sample type.
Reflective of current practices in the intensive care unit, the majority of patients in this study received broad-spectrum antibiotics before sample collection. As such, the possibility that antibiotic exposure may have driven compositional similarity between fluid types must be considered. The observation that a greater fraction of subjects who were PNA-neg versus subjects who were PNA-pos received antibiotics, however, suggests this may be less likely.
Together, our data indicate that from a metagenomic perspective, TA sampling is an effective alternative to more invasive mBAL testing for patients with pneumonia, a conclusion consistent with findings of prior clinical studies (2, 4) and the clinical practice guidelines from the Infectious Diseases Society of America and the American Thoracic Society (10). Future studies with a larger sample size may clarify trends in diversity differences that approached, but did not reach, significance. These results may help inform both culture-independent clinical microbiological testing and research on the lung microbiome.
GRANTS
This study was supported by National Heart, Lung, and Blood Institute Grants K23-HL-138461-01A1 (to C. Langelier), K23-HL-123778 (to S. C. Christenson), and R01-HL-110969 and R01-HL-140026 (to C. S. Calfee) and Chan Zuckerberg Biohub (to J. L. DeRisi).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
K.L.K. and C.L. conceived and designed research; K.L.K., T.D., S.C., S.B., and C.L. performed experiments; K.L.K. analyzed data; K.L.K., F.M., C.S.C., and C.L. interpreted results of experiments; K.L.K. and C.L. prepared figures; K.L.K. and C.L. drafted manuscript; K.L.K., F.M., S.C.C., J.W., T.D., A.B., K.V., S.C., A.J., S.B., J.L.D., C.S.C., and C.L. edited and revised manuscript; K.L.K., F.M., S.C.C., J.L.D., C.S.C., and C.L. approved final version of manuscript.
ACKNOWLEDGMENTS
Data are available publicly via BioProject accession identifier PRJNA445982. Supplemental Table S1 and Fig. S1 and analyses of primary sequencing data including preprocessed data files and an R markdown file with documentation are available at dx.doi.org/10.17504/protocols.io.yktfuwn.
Preprint is available at https://doi.org/10.1101/375360.
REFERENCES
- 1.Abreu NA, Nagalingam NA, Song Y, Roediger FC, Pletcher SD, Goldberg AN, Lynch SV. Sinus microbiome diversity depletion and Corynebacterium tuberculostearicum enrichment mediates rhinosinusitis. Sci Transl Med 4: 151ra124, 2012. doi: 10.1126/scitranslmed.3003783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berton DC, Kalil AC, Teixeira PJ. Quantitative versus qualitative cultures of respiratory secretions for clinical outcomes in patients with ventilator-associated pneumonia. Cochrane Database Syst Rev 10: CD006482, 2014. doi: 10.1002/14651858.CD006482.pub4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boersma WG, Erjavec Z, van der Werf TS, de Vries-Hosper HG, Gouw AS, Manson WL. Bronchoscopic diagnosis of pulmonary infiltrates in granulocytopenic patients with hematologic malignancies: BAL versus PSB and PBAL. Respir Med 101: 317–325, 2007. doi: 10.1016/j.rmed.2006.04.021. [DOI] [PubMed] [Google Scholar]
- 4.Canadian Critical Care Trials Group A randomized trial of diagnostic techniques for ventilator-associated pneumonia. N Engl J Med 355: 2619–2630, 2006. doi: 10.1056/NEJMoa052904. [DOI] [PubMed] [Google Scholar]
- 5.Dickson RP, Erb-Downward JR, Freeman CM, McCloskey L, Falkowski NR, Huffnagle GB, Curtis JL. Bacterial topography of the healthy human lower respiratory tract. MBio 8: e02287-16, 2017. doi: 10.1128/mBio.02287-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21, 2013. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hansen V, Oren E, Dennis LK, Brown HE. Infectious disease mortality trends in the United States, 1980–2014. JAMA 316: 2149–2151, 2016. doi: 10.1001/jama.2016.12423. [DOI] [PubMed] [Google Scholar]
- 8.Jain S, Self WH, Wunderink RG, Fakhran S, Balk R, Bramley AM, Reed C, Grijalva CG, Anderson EJ, Courtney DM, Chappell JD, Qi C, Hart EM, Carroll F, Trabue C, Donnelly HK, Williams DJ, Zhu Y, Arnold SR, Ampofo K, Waterer GW, Levine M, Lindstrom S, Winchell JM, Katz JM, Erdman D, Schneider E, Hicks LA, McCullers JA, Pavia AT, Edwards KM, Finelli L; CDC EPIC Study Team . Community-acquired pneumonia requiring hospitalization among U.S. adults. N Engl J Med 373: 415–427, 2015. doi: 10.1056/NEJMoa1500245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalil AC, Metersky ML, Klompas M, Muscedere J, Sweeney DA, Palmer LB, Napolitano LM, O’Grady NP, Bartlett JG, Carratalà J, El Solh AA, Ewig S, Fey PD, File TM Jr, Restrepo MI, Roberts JA, Waterer GW, Cruse P, Knight SL, Brozek JL. Management of adults with hospital-acquired and ventilator-associated pneumonia: 2016 clinical practice guidelines by the Infectious Diseases Society of America and the American Thoracic Society. Clin Infect Dis 63: e61–e111, 2016. doi: 10.1093/cid/ciw353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kelly BJ, Imai I, Bittinger K, Laughlin A, Fuchs BD, Bushman FD, Collman RG. Composition and dynamics of the respiratory tract microbiome in intubated patients. Microbiome 4: 7, 2016. doi: 10.1186/s40168-016-0151-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kitsios GD, Fitch A, Manatakis DV, Rapport SF, Li K, Qin S, Huwe J, Zhang Y, Doi Y, Evankovich J, Bain W, Lee JS, Methé B, Benos PV, Morris A, McVerry BJ. Respiratory microbiome profiling for etiologic diagnosis of pneumonia in mechanically ventilated patients. Front Microbiol 9: 1413, 2018. doi: 10.3389/fmicb.2018.01413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Langelier C, Kalantar KL, Moazed F, Wilson MR, Crawford ED, Deiss T, Belzer A, Bolourchi S, Caldera S, Fung M, Jauregui A, Malcolm K, Lyden A, Khan L, Vessel K, Quan J, Zinter M, Chiu CY, Chow ED, Wilson J, Miller S, Matthay MA, Pollard KS, Christenson S, Calfee CS, DeRisi JL. Integrating host response and unbiased microbe detection for lower respiratory tract infection diagnosis in critically ill adults. Proc Natl Acad Sci USA 115: E12353–E12362, 2018. doi: 10.1073/pnas.1809700115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Langelier C, Zinter MS, Kalantar K, Yanik GA, Christenson S, O’Donovan B, White C, Wilson M, Sapru A, Dvorak CC, Miller S, Chiu CY, DeRisi JL. Metagenomic sequencing detects respiratory pathogens in hematopoietic cellular transplant patients. Am J Respir Crit Care Med 197: 524–528, 2018. doi: 10.1164/rccm.201706-1097LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359, 2012. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MH, Wagner H. vegan: Community Ecology Package version 2.3-5 from R-Forge (Online), 2017. https://rdrr.io/rforge/vegan/.
- 17.Ruby JG, Bellare P, Derisi JL. PRICE: software for the targeted assembly of components of (meta) genomic sequence data. G3 (Bethesda) 3: 865–880, 2013. doi: 10.1534/g3.113.005967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.United States Centers for Disease Control and Prevention CDC/NHSN Surveillance Definitions for Specific Types of Infections (Online), 2019. https://www.cdc.gov/nhsn/pdfs/pscmanual/17pscnosinfdef_current.pdf.
- 19.Wilson MR, O’Donovan BD, Gelfand JM, Sample HA, Chow FC, Betjemann JP, Shah MP, Richie MB, Gorman MP, Hajj-Ali RA, Calabrese LH, Zorn KC, Chow ED, Greenlee JE, Blum JH, Green G, Khan LM, Banerji D, Langelier C, Bryson-Cahn C, Harrington W, Lingappa JR, Shanbhag NM, Green AJ, Brew BJ, Soldatos A, Strnad L, Doernberg SB, Jay CA, Douglas V, Josephson SA, DeRisi JL. Chronic meningitis investigated via metagenomic next-generation sequencing. JAMA Neurol 75: 947–955, 2018. doi: 10.1001/jamaneurol.2018.0463. [DOI] [PMC free article] [PubMed] [Google Scholar]