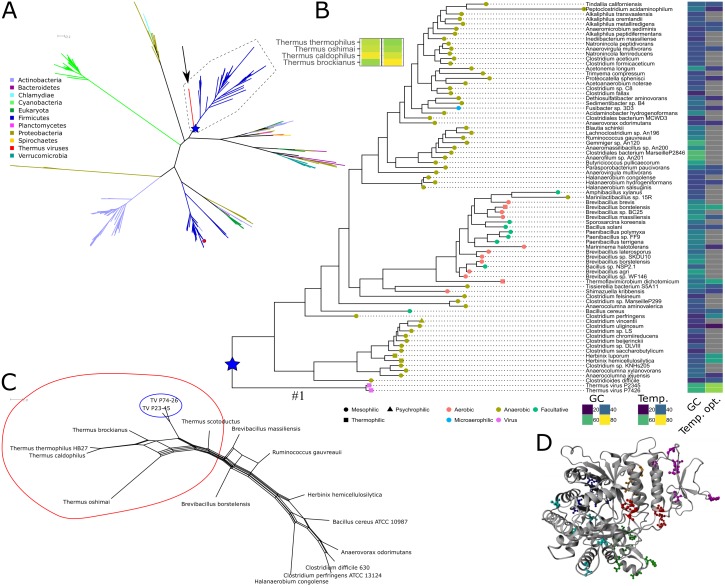
Figure 1. Phylogeny, sequence characteristics and growth optima.
(A) Unrooted maximum likelihood phylogeny of representative NrdJm amino acid sequences. The two virus sequences are indicated with an arrow. Phyla, with viruses assumed from their names to infect organisms from the same phylum, are indicated with coloured branches. The only solved structure from Lactobacillus leichmannii (PDB: 1L1L) is indicated with a small red circle. The root of the TV and Firmicutes clan is marked with a blue star. Full tree file available at Figshare (https://doi.org/10.17045/sthlmuni.7117430.v1). (B) Detailed view of the clan containing the Thermus virus sequences, surrounded by a dashed line in A, with GC contents, optimum growth temperatures (when available; viruses set to typical host 70 °C), indications of temperature ranges (leaf node shape) and oxygen requirements (node colour). (C) NeighborNet depicting patterns of codon usage between Thermus thermophilus, Thermus viruses and selected Firmicutes measured in euclidean distances. A split containing Thermus spp. and TVs is encircled in red; the TVs are encircled in blue. (D) Positions of mutations with a posterior probability ≥ 95% to be positively selected, mapped on a homology model of TVNrdJm: red: specificity site, magenta: dimer mimicking domain, orange: proximity of the specificity site, green: B12 binding site. Blue: surface patch, cyan: others.