Abstract
Model membranes in the form of freely suspended liposomes are widely used as biophysical tools for studying lipid-lipid and protein-lipid interactions. Liposomes prepared by conventional methods have bilayer leaflets that are chemically identical. In contrast, living cells actively maintain a different lipid composition in the two leaflets of the plasma membrane, resulting in asymmetric membrane properties that are crucial for normal cell function. Here, we present a protocol for the preparation of unilamellar asymmetric phospholipid vesicles that better mimic biological membranes. Asymmetry is generated by methyl-β-cyclodextrin catalyzed exchange of the outer leaflet lipids between vesicle pools of differing lipid composition, followed by separation of these pools by centrifugation. We further present two complementary assays for the quantification of each leaflet’s lipid composition: the overall lipid composition is determined by chromatography, while the lipid distribution between the two leaflets is determined by NMR using the lanthanide shift reagent Pr3+. The preparation protocol and the chromatographic assay can be applied to any type of phospholipid bilayer, while the NMR assay is specific to lipids with choline headgroups, such as phosphatidylcholine and sphingomyelin. In approximately 12 hours, the protocol can produce a large yield of asymmetric vesicles (up to 20 mg) suitable for a wide range of biophysical studies. We discuss possible extensions of the protocol for the preparation of asymmetric proteoliposomes and cholesterol-containing vesicles, as well as alternative assays for quantifying the transbilayer lipid distribution of lipids that do not possess a choline headgroup.
Keywords: membrane asymmetry, leaflet composition, cyclodextrin, model membrane, liposome
Introduction
The plasma membrane is a marvel of evolutionary nanoengineering. The physical properties of this remarkable organelle are optimized for its dual roles as a cellular barrier and gateway. In a sense, these roles are contradictory: the plasma membrane must be sturdy enough to provide protection against an often harsh external environment, yet malleable enough to allow for cell growth, division and motility, and the passage of water and nutrients. Nature solved this problem with a quasi- two-dimensional fluid formed from a complex mixture of lipid, protein, and carbohydrate. Although its basic architecture is well established, there is an emerging consensus that some critical processes occurring at and within the plasma membrane cannot be adequately explained without invoking nanoscopic membrane structure 1–3. This realization has generated renewed interest in biophysical characterization of membranes at the level of molecular interactions. However, teasing apart these interactions is hardly feasible in a natural plasma membrane that might contain several thousand chemically distinct lipids, and several hundred unique proteins. Instead, uncovering the specific interactions that are responsible for functional phenotypes in vivo, requires the study of simplified models in vitro, where their composition can be finely controlled.
Lipid bilayer vesicles are among the most widely used model systems for biophysical studies of lipid-lipid and protein-lipid interactions. Conventional liposome preparations by hydration of a dry lipid film (typically followed by sonication or extrusion to generate unilamellar vesicles) produce bilayers in which the two leaflets have an identical composition. Cellular plasma membranes, on the other hand, actively maintain a different lipid composition in the exofacial (outer) and cytofacial (inner) leaflets 4,5. Phosphatidylserine (PS), for instance, is located exclusively in the inner leaflet of the plasma membrane (PM), and its exposure on the outer leaflet serves as a marker of impending cell death in eukaryotes 6. Mounting evidence suggests that the lipid compositional asymmetry of the PM not only confines certain lipids to their respective leaflets in order to facilitate their direct interaction with other molecules, but also results in unique membrane properties that, although currently poorly understood, are likely to be crucial for normal cellular function 7. It follows that symmetric model membranes lack at least some key structural characteristics of natural cell membranes that may critically affect membrane interactions with proteins and small molecules. There is a widely recognized need to expand the biophysical toolkit with model systems that more closely mimic the asymmetric cell membrane environment, while still allowing for easily controlled variation of the inner and outer leaflet lipid compositions.
Preparation of asymmetric vesicles
Several techniques exist for preparing asymmetric bilayers 8. In particular, asymmetry in large unilamellar vesicles (LUVs) is desirable because their size is well-defined, and their bilayer properties are not greatly affected by the high curvature found in small unilamellar vesicles (SUVs), or by interactions with a substrate found in supported lipid bilayers (SLBs). These problems are also absent in asymmetric giant unilamellar vesicles (GUVs) of micron size that can be prepared by e.g. passing an inverted emulsion droplet through a lipid lined oil-water interface 9. However, the size of the asymmetric GUVs is harder to control precisely and, in the case of the inverted emulsion technique, the vesicles contain residual organic solvent that could affect bilayer properties. The generation of asymmetric LUVs (aLUVs) has been achieved by three different methods: (1) application of a pH differential to vesicles that contain anionic lipids; (2) external addition of enzymes that selectively modify outer leaflet lipid headgroups; and (3) external addition of lipid carrier molecules that catalyze intervesicular exchange of outer leaflet lipids. The pH adjustment method exploits the fact that certain anionic phospholipids (namely, phosphatidylglycerol and phosphatidic acid, PG and PA respectively) can rapidly diffuse between bilayer leaflets in their uncharged, protonated form 10. Applying a pH differential across the bilayer thus induces a redistribution of these lipids with an accumulation on the more alkaline side 11. Enzymatic headgroup modification has been used to convert outer leaflet phosphatidylserine (PS) to phosphatidylethanolamine (PE) using PS-decarboxylase 12, or outer leaflet phosphatidylcholine (PC) to either PE or PS using phospholipase D 13. In contrast to these methods—each of which acts by chemically modifying a single population of initially symmetric vesicles—catalyzed lipid exchange requires mixing together two symmetric vesicle populations of different composition in the presence of a lipid carrier such as, phospholipid exchange protein 14, bovine serum albumin 15, or certain classes of cyclodextrins 16. The primary disadvantage of catalyzed exchange is that the two vesicle populations must eventually be separated to recover the asymmetric liposomes; below, we describe two different strategies to address this problem. Its primary advantage, however, is its versatility. While pH adjustment and enzymatic modification are each specific for a subset of phospholipids, catalyzed exchange is compatible with a wide variety of phospholipids, and can be used to create asymmetric vesicles with diverse inner and outer leaflet compositions.
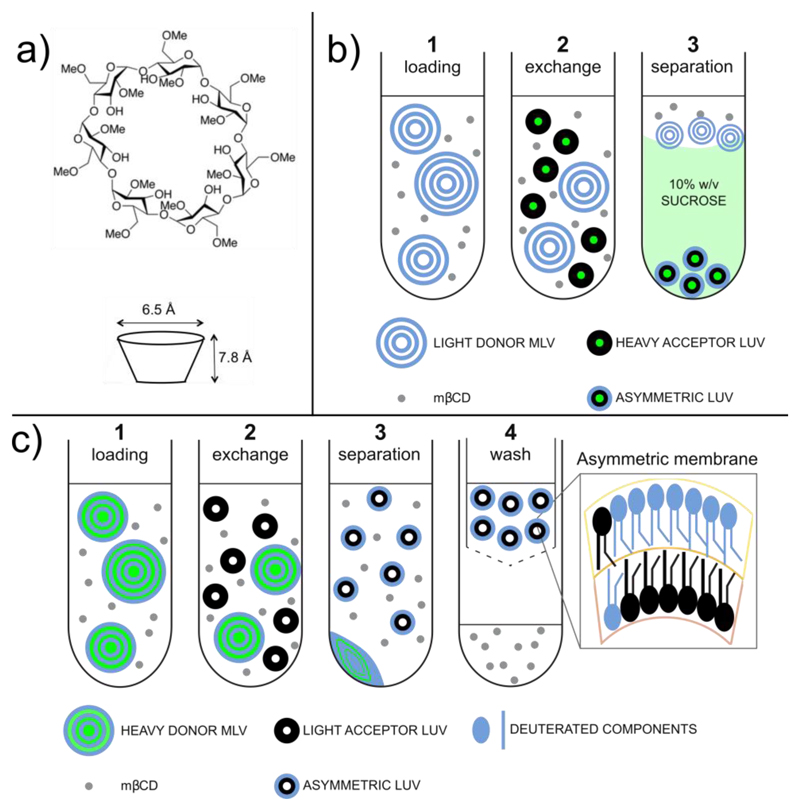
Here, we focus on the preparation of aLUVs using catalyzed exchange, with methyl-β-cyclodextrin (mβCD) as the lipid carrier. mβCD is a water-soluble, ring-shaped oligosaccharide possessing a hydrophilic outer surface and a central hydrophobic cavity that is large enough to accommodate a lipid chain (Fig. 1a). The reversible formation of an mβCD/lipid complex effectively displaces water from the cavity, replacing unfavorable interactions with favorable ones 17. At high concentrations, mβCD will completely dissolve a lipid vesicle suspension, but at lower concentrations, a dynamic equilibrium exists between intact vesicles, mβCD/lipid complexes, and free mβCD 18. This “exchange-competent” solution of mβCD and donor lipid is the starting point for preparing aLUVs, as shown in Step 1 of Fig. 1b and c. Addition of acceptor LUVs to the mβCD/donor solution results in mβCD-catalyzed exchange of the acceptor vesicles’ exposed outer leaflet lipids with the donor lipid pool, while leaving the inner leaflet of the acceptor vesicles relatively unperturbed, as shown in Step 2 16.
Figure 1.
Illustration of the different aLUV preparation protocols. (a) Chemical structure of methyl-β-cyclodextrin (mβCD), and the size and shape of its hydrophobic pocket. (b) Heavy acceptor strategy: (1) mβCD is incubated with donor lipid MLVs suspended in buffer; (2) mβCD facilitates the exchange of the outer leaflet of acceptor LUVs (entrapped with sucrose) with donor lipid; (3) the desired aLUVs are recovered from the pellet after ultracentrifugation through a sucrose cushion. (c) Heavy donor strategy: (1) mβCD is incubated with donor lipid MLVs entrapped with sucrose. The donor lipid is composed of the desired outer leaflet lipid; (2) mβCD facilitates the exchange of the outer leaflet of acceptor LUVs with donor lipid; (3) remaining sucrose loaded donor MLVs are removed by centrifugation; (4) the aLUV sample is further purified through the removal of mβCD and mβCD-lipid complexes with a centrifugal concentrator. The desired aLUVs are then recovered from the retentate (figure adapted from ref. 19).
Two strategies have been used to separate the original acceptor pool (which contains the aLUVs) from the donor/acceptor/mβCD exchange slurry; each exploits density and/or size differences between the donor and acceptor vesicle pools. The heavy acceptor strategy (Fig. 1b) makes use of a dense sucrose solution (typically 25% w/v) trapped in the acceptor vesicle lumen. Following exchange, separation is achieved by layering the exchange slurry onto a sucrose solution of intermediate density (typically 10% w/v) followed by ultracentrifugation at 190,000 × g. The lighter solution composed primarily of undissolved donor multilamellar vesicles (MLVs) and mβCD remains at the top of the centrifuge tube, while the heavy acceptor LUVs sink through the sucrose cushion and are recovered in the pellet, as shown in Fig. 1b Step 3. The heavy donor protocol (Fig. 1c) essentially reverses this strategy: the donor lipid film is hydrated in a sucrose solution (typically 20% w/w) to create large, dense MLVs, while the acceptor lipid is hydrated in pure water or buffer. Following exchange, the donor MLVs are separated from the lighter, smaller acceptor LUVs by low-speed centrifugation (20,000 × g), with the now-asymmetric acceptors recovered in the supernatant, as shown in Fig. 1c Step 3. Residual mβCD is then removed with a centrifugal ultrafiltration device (Fig. 1c Step 4).
The dense sucrose core of aLUVs prepared using the heavy acceptor strategy, though acceptable for many applications, may present problems for some experiments. One potential consequence of entrapped sucrose is the introduction of an osmotic stress that can cause bilayer thinning and lipid area expansion 19. Extruded vesicles are often non-spherical and can easily accommodate small osmotic gradients without accumulating stress by “rounding up”, effectively diluting the internal solution by increasing their volume 20. Even accounting for such volume changes, typical experimental conditions—for example, vesicles with a 25% (w/v) sucrose core, suspended in PBS buffer—result in a residual osmotic differential of ~ 300 mOsm/kg in aLUVs prepared using the heavy acceptor strategy. For 100 nm diameter vesicles, the corresponding tension calculated from Laplace’s law is ~ 20 mN/m, a value that can exceed the rupture tension of some lipid bilayers 21. In cases where it is desirable to minimize or eliminate this tension, the buffer osmolality can be adjusted, but the osmolyte should be carefully chosen to maintain compatibility with the chosen experimental techniques, as well as to avoid direct interactions with lipids that might alter membrane properties. The heavy donor strategy—described in detail in the PROCEDURE below— circumvents these issues and eliminates the need for ultracentrifugation.
Quantifying leaflet composition
Both exchange efficiency and spontaneous lipid flip-flop rate depend on lipid structure and possibly other factors 22. It is therefore critical to determine the composition of each bilayer leaflet after the asymmetric liposomes are prepared; it is not safe to assume that the outer leaflet contains only donor lipid, or that the inner leaflet contains only acceptor lipid. Quantifying the leaflet composition requires a separate determination of the overall vesicle composition and the asymmetric distribution of each lipid species. The former is generally accomplished with chromatography, for example thin layer chromatography (TLC) 16, gas chromatography (GC) 19, or ultra-high performance liquid chromatography (UHPLC) 23. Asymmetry assays generally involve the selective modification, extraction, or interaction of outer leaflet lipids with reagents added externally to the aLUV suspension, followed by detection and quantification; these assays are usually specific for lipid headgroups. Among the methods that have been used to quantify asymmetry are: labeling of exposed aminophospholipids (i.e., PS or PE) with trinitrobenzenesulfonic acid (TNBS), followed by thin layer chromatography 24; periodate oxidation of exposed PG, followed by detection of oxidation byproducts 10; selective outer leaflet extraction of radiolabeled lipids with bovine serum albumin or phospholipid exchange protein 10; binding of a cationic peptide to exposed negatively charged lipids (e.g., PS or PG) 25; zeta potential measurements of asymmetric bilayers containing charged lipids 26; and hydrolysis of PC by phospholipase D, followed by detection of free choline 13.
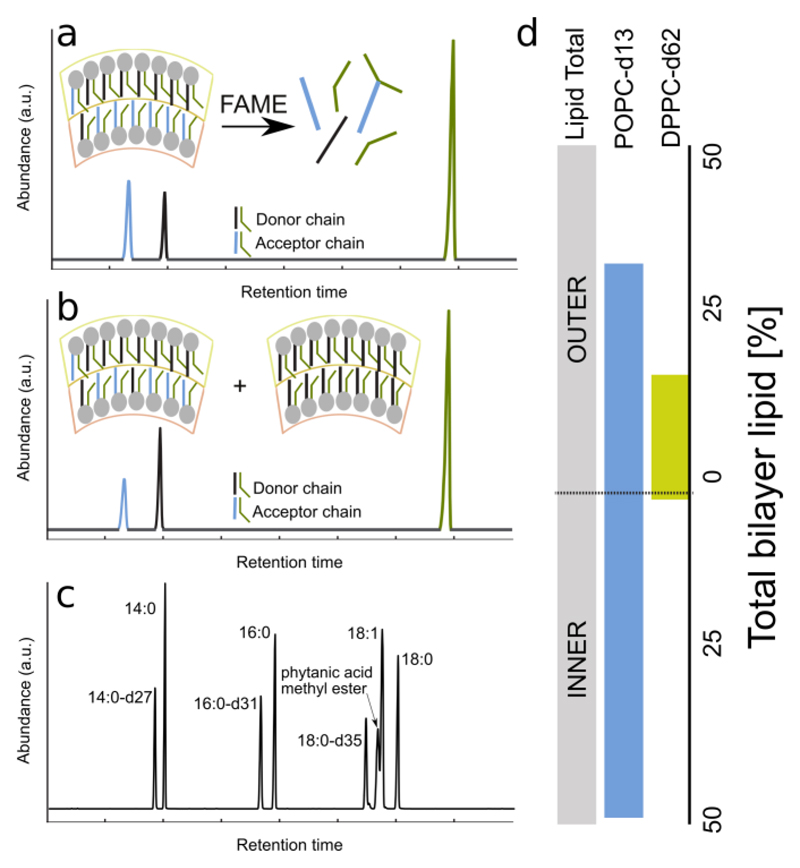
In the PROCEDURE section, we describe a two-part assay for determining leaflet composition that follows the general outline given above. In the first step, GC coupled to mass spectrometry is used to quantify the total mole fraction of each lipid species. As shown in Fig. 2c, GC is extremely sensitive to even small differences in fatty acid composition (i.e., length and degree of unsaturation) or isotopic content (i.e., protiated vs. deuterated chains), and is easily quantified. As mentioned below, an alternative technique, UHPLC, can be used instead to determine the overall composition of lipids with the same acyl chains but different headgroups 23. In the second step, 1H NMR coupled with a shift reagent is used to determine the asymmetric distribution of lipids with protiated choline headgroups (Fig. 3). This assay can distinguish choline headgroups located in the inner and outer leaflets because the shift reagent (e.g., a lanthanide ion such as Pr3+) is added externally and selectively interacts with outer leaflet lipids 19. Compared to the other asymmetry assays mentioned above, NMR is the most straightforward to implement because the signals from the inner and outer leaflet populations are resolved in a single measurement. The primary limitations of NMR are: (1) it requires that at least one of the lipid components of the aLUV has a choline headgroup (i.e., PC or sphingomyelin (SM)); and (2) it often fails when aLUVs contain charged lipid in the outer leaflet (in our hands, more than 10 mol% PG, or 2 mol% PS). The latter limitation results from interactions between Pr3+ and exposed anionic lipid, which can cause liposome aggregation that degrades the NMR signal.
Figure 2.
Gas chromatography (GC) assay for quantifying vesicle composition. To analyze the lipid composition, the phospholipid must be derivatized into fatty acid methyl esters (FAMEs). (a) An illustration of a GC chromatograph from a typical aLUV preparation; (b) An illustration of a GC chromatograph where more donor than acceptor is present, an example of donor contamination. (c) A GC chromatograph of different fatty acids and their deuterated variants. (d) Individual leaflet compositions, determined by combining results from GC and 1H NMR assays, of an aLUV prepared from DPPC-d62 donor exchanged into POPC-d13 acceptor.
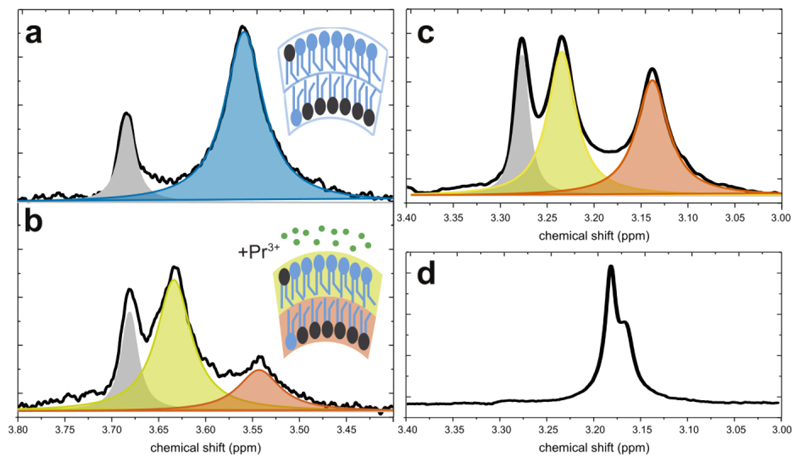
Figure 3.
1H NMR assay for quantifying lipid asymmetry. (a) 1H NMR of aLUVs using a deuterated-choline acceptor lipid. The blue area represents the signal from the inner and outer leaflet protiated cholines in the sample (the gray peak is residual mβCD). (b) After external addition of Pr3+, inner leaflet (red) and outer leaflet (yellow) choline resonances are resolved, and the area under each peak is proportional to the amount of protiated choline present in each leaflet. (c) 1H NMR of scrambled aLUVs shows inner and outer leaflet peaks of approximately equal area. (d) 1H NMR of SUVs shows distinct inner and outer peaks in the absence of shift reagent. SUVs can be generated by an imbalance in the mβCD:lipid ratio. Figure adapted from refs. 19 and 28.
Experimental design
In the PROCEDURE section below we describe the preparation of aLUVs composed of a DPPC-enriched outer leaflet and a POPC-enriched inner leaflet 19,23. In principle, the variety of compositions that can be prepared is limited only by the nature of the interaction between mβCD and the different types of lipids. In our own work (summarized in Table 1), we have used the protocol to prepare chemically asymmetric vesicles composed of POPC and POPE 27, DPPC and POPE, egg sphingomyelin and POPE, and DMPC and POPC, as well as isotopically labelled aLUVs composed of deuterated variants of POPC 19,27 or DPPC 28.
Table 1.
Two-component aLUVs prepared using the heavy donor strategy described in the PROCEDURE section. Shown are: the donor lipid; the acceptor lipid; their ratio; the temperature during mβCD/donor and mβCD/donor/acceptor incubation (Steps 13 and 14 in the PROCEDURE); the mole fractions of donor lipid in the inner (χD,innerner) and outer (χD,outer) leaflets of the aLUVs; the observed exchange efficiency (Eobs); the fraction of donor lipid in the outer leaftlet (Dasym); and the respective references.
| Donor (D) | Acceptor (A) | D:A ratio | Exchange temp. [C] | χD,inner | χD,outer | Eobs | Dasym | Reference |
|---|---|---|---|---|---|---|---|---|
| POPC-d44 | POPC | 2 | 22 | 0.1 | 0.62 | 0.72 | 0.86 | 19 |
| POPC | POPC-d44 | 2 | 22 | 0.16 | 0.6 | 0.74 | 0.79 | 19 |
| DMPC-d54 | POPC-d13 | 2 | 22 | 0.05 | 0.51 | 0.55 | 0.91 | this work |
| DPPC-d62 | POPC-d13 | 3 | 22 | 0.02 | 0.34 | 0.36 | 0.95 | 19 |
| POPE | POPC | 2 | 22 | 0 | 0.6 | 0.64 | 1.00 | 27 |
| POPE | POPC | 3 | 22 | 0.19 | 0.76 | 1.04 | 0.8 | 27 |
| DPPC-d62 | DPPC-d13 | 3 | 22 | 0.3 | 0.59 | 0.89 | 0.66 | 28 |
| POPC | POPE | 2 | 35 | 0.06 | 0.54 | 0.58 | 0.90 | 27 |
| POPC | POPE | 3 | 35 | 0.11 | 0.64 | 0.72 | 0.85 | 27 |
| eSM | POPE | 3 | 35 | 0.09 | 0.44 | 0.54 | 0.83 | this work |
| DPPC | POPE | 3 | 35 | 0.08 | 0.8 | 0.88 | 0.91 | this work |
Some basic sample characterization is an integral part of the protocol. Essential analyses include measuring the vesicle size distribution with dynamic light scattering (DLS), and quantifying the inner and outer leaflet lipid composition (the example described here uses GC and NMR for this purpose, although other assays may be necessary depending on the choice of lipids). There are a few important considerations when modifying the described protocol for a different lipid composition:
-
1)
Exchange conditions. The mβCD concentration is carefully chosen to ensure membrane integrity at each step of the protocol and should not be changed without performing appropriate control experiments. In order to ensure that acceptor vesicles are not osmotically stressed during the exchange of lipid between the donors and acceptors, the acceptor LUVs are prepared in a dilute salt solution (e.g., 20-30 mM NaCl) to balance the osmolarity of the mβCD solution. If it is necessary to prepare aLUVs in a higher molarity buffer, appropriate tests should be performed to ensure vesicle integrity.
-
2)
Overall composition assay. The GC assay for quantifying liposome composition is sensitive to differences in the acyl chains (including isotopic differences) of the acceptor and donor lipids, as shown in Fig. 2c. If lipids with identical acyl chains are desired (e.g, POPC and POPE), TLC 16 or UHPLC 23 can be used instead of GC, as these techniques separate lipids based on chemical differences in their headgroups rather than their chains.
-
3)
Asymmetry assay. Asymmetry assays are specific for lipid headgroups, and the 1H NMR assay described below quantifies the inner/outer distribution of lipids with protiated choline headgroups (i.e., PC and SM). If an asymmetric bilayer composed of two different PC lipids is desired, it is possible to determine the transbilayer distribution of the lipids using NMR, provided that one of the lipids has a deuterated choline (as in the example used in the PROCEDURE below). Other assays have been described to quantify the asymmetry of mixtures that do not contain any PC lipids or that have a high concentration of charged lipids, as mentioned previously.
-
4)
Preparation of asymmetric proteoliposomes. If incorporation of a transmembrane protein into the asymmetric liposomes is desired, the protein should be reconstituted into the acceptor vesicles. Additional control experiments should be performed to ensure that the presence of the protein does not interfere with the composition assays.
-
5)
Preparation of cholesterol-containing aLUVs. If cholesterol-containing aLUVs are desired, the protocol should be modified by either: (1) adding cholesterol to both the donor and acceptor vesicles; (2) introducing cholesterol after preparation of phospholipid-only aLUVs, using cholesterol-loaded mβCD 29; or (3) adding cholesterol only to the acceptor vesicles and optimizing the exchange conditions for the use of either hydroxypropyl-α-cyclodextrin 30 or methyl-α-cyclodextrin 31 (α-CDs can transport phospholipids but not cholesterol due to their smaller hydrophobic cavity). We note that determining the transbilayer distribution of cholesterol is not straightforward.
Materials
Reagents
1,2-Dipalmitoyl-d62-sn-glycero-3-phosphocholine [di-16:0 PC-d62, DPPC-d62, chain perdeuterated DPPC, 796.4 g/mol] (Avanti Polar Lipids cat. no. 860355)
1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine-1,1,2,2-d4-N,N,N-trimethyl-d9 [16:0/18:1 PC(d13), POPC-d13, headgroup deuterated POPC, 773.08 g/mol] (Avanti Polar Lipids cat. no. 790433)
1-Palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (sodium salt) [16:0/18:1 PG, POPG, 771.0 g/mol] (Avanti Polar Lipids cat. no. 840457)
Methyl-β-cyclodextrin (mβCD, 1310 g/mol) (Fisher Scientific cat. no. AC377110000)
Sucrose (342.3 g/mol) (Fisher Scientific cat. no. BP2201)
HPLC-grade chloroform (Fisher Scientific cat. no. AA43685)
HPLC-grade methanol (Fisher Scientific cat. no. AA22909)
-
CAUTION Hydrochloric acid HCl (Fisher Scientific cat. no. SA48).
Harmful if inhaled, ingested, or comes in contact with the skin. Proper goggles and gloves should be worn during handling and manipulation.
Praseodymium(III) nitrate hexahydrate {Pr(NO3)3 6H2O} (Pr3+, 435.0 g/mol) (Fisher Scientific cat. no. AA11240)
Ultrapure H2O
99.9% D2O (Cambridge Isotopes cat. no. DLM-4).
Equipment
Avance III 400 MHz spectrometer (Bruker, Billerica, MA) using Bruker TopSpin acquisition software, and analyzed with TopSpin 3.2.
Wilmad LabGlass 5 mm NMR tubes (Fisher Scientific cat. no. 1680008).
Agilent 5890A gas chromatograph (Santa Clara, CA) with a 5975C mass-sensitive detector operating in electron-impact mode.
An HP-5MS capillary column (30 m × 0.25 mm, 0.25 μm film thickness) (Agilent cat. no. 19091S-433E)
Centrifugal filter devices: Amicon Ultra-15, 100,000 Da molecular weight cutoff (Fisher Scientific cat. no. UFC910024)
Sorvall LYNX 4000 Superspeed Centrifuge (Thermo Fisher Scientific cat. no. 75006580)
BI-200SM Research Goniometer and Laser Light Scattering system (Brookhaven Instruments cat. no. BI-200SM).
Hand-held miniextruder with a 100 nm pore-diameter polycarbonate filter (Avanti Polar Lipids cat. no. 610026)
250 μL glass syringe (Hamilton cat. no. 81175)
Reagent Setup
Solutions: Prepare the following solutions using H2O unless specified: sucrose (20% w/w, 0.632 M, 1.08 g/mL), NaCl (25 mM), Pr(NO3)3 (20 mM in D2O). Prepare the following lipid stock solutions in glass vials with Teflon-lined caps: DPPC-d62 (30 mM in CHCl3); POPC-d13 (30 mM in CHCl3); POPG (2 mM in CHCl3). In a fume hood, prepare methanolic HCl (1 M) by adding 3.04 mL concentrated HCl to 30 mL methanol in a beaker, then stir to mix. Transfer the solution to a graduated cylinder, then add methanol to a total volume to 37 mL. Store the solution in a glass vial with a Teflon-lined cap.
Prepare methyl-β-cyclodextrin (35 mM) as follows. Weigh the cyclodextrin powder in a volumetric flask (for 25 mL of solution, 1.1463 g mβCD is needed). Add water until the flask is about half full, then cover the opening with foil and incubate at room temperature for 1-2 hours, or until the powder is fully dissolved. Then bring the solution to the appropriate volume with water and mix by inverting the flask 20 times. The density of the solution should be 1.0114 g/cm3. The solution can be safely kept for a few days on the bench, but freezing at -20 °C is recommended for longer term storage to prevent bacterial growth 23. Allow frozen solution to warm to room temperature, and gently mix before use.
Centrifugal filters: Amicon filters contain trace amounts of glycerol that can interfere with NMR analysis. Prior to use, rinse the filters 7 times by filling with 10 mL ultrapure water and centrifuging at 5000 × g for 2-5 min. Fill pre-rinsed filters with water and store at 4 °C until use.
Procedure
Preparation of donor and acceptor films
TIMING ~14 h
1. Transfer 647 μL (15.46 mg, 19.4 μmol) of the DPPC-d62 stock solution to a glass scintillation vial (“donor”) using a glass syringe.
2. Transfer 205 μL (4.75 mg, 6.15 μmol) of the POPC-d13 stock solution and 162 μL (0.25 mg, 0.324 μmol) of the POPG stock solution to a glass scintillation vial (“acceptor”) using a glass syringe.
CRITICAL STEP Acceptor films are doped with 5 mol % POPG (negatively charged lipid) to prevent formation of paucilamellar vesicles.
3. Remove the organic solvent from the scintillation vials from Steps 1 and 2 with an inert gas stream and gentle heating, followed by overnight drying under high vacuum (~12 h).
PAUSE POINT Lipid films can be stored frozen for several months prior to hydration.
Preparation of acceptor unilamellar vesicles
TIMING ~2.5 h
4. Preheat the acceptor film from Step 2 to 40 °C and hydrate it with 500 μL of preheated 25 mM NaCl solution to a lipid concentration of 10 mg/mL, followed by vigorous vortexing to disperse the lipid and create an MLV suspension.
CRITICAL STEP The temperature for hydration should always be above the melting temperature of the lipids.
5. Incubate the acceptor MLV suspension at 40 °C for 1 h with occasional vortexing.
6. Transfer the acceptor MLV suspension to a -80 °C freezer for ~ 10 min or until completely frozen. Thaw at 40 °C for 5-10 min and then vortex. Repeat the freeze-thaw cycle five times.
PAUSE POINT MLV suspensions can be stored frozen for several months. Prior to use a few freeze/thaw cycles should be performed.
7. To obtain unilamellar vesicles, pass the acceptor MLV suspension 31 times through a miniextruder assembled with a 100 nm pore-diameter polycarbonate filter.
CRITICAL STEP The extrusion should always be carried out at a temperature higher than the melting temperature of the lipids. Use a hot plate if needed, and preheat the extruder before loading the lipid suspension. Then incubate for at least 10 min on the hot plate to equilibrate the temperature before extrusion.
CRITICAL STEP Measure the size distribution of the acceptor LUVs with DLS. Checking the size of the acceptors at this point is useful for diagnosing problems in the final aLUVs.
?Troubleshooting
Preparation of donor multilamellar vesicles
TIMING ~3 h
8. Preheat the donor film from Step 1 to 50 °C and hydrate it with 775 μL of preheated 20% (w/w) sucrose to a lipid concentration of 20 mg/mL, followed by vortexing to disperse the lipid and create an MLV suspension.
CRITICAL STEP The temperature for hydration should always be above the melting temperature of the lipids.
9. Incubate the donor MLV suspension at 50 °C for 1 h with occasional vortexing.
10. Transfer the donor MLV suspension to a -80 °C freezer for ~ 10 min or until completely frozen. Thaw at 50 °C for 5-10 min and then vortex. Repeat the freeze-thaw cycle five times.
11. Dilute the donor suspension 20-fold by adding 14.7 mL H2O, and immediately centrifuge at 20,000 × g for 30 min at 20 °C. Discard the supernatant by tipping the centrifuge tube.
CRITICAL STEP Perform this step immediately before proceeding to the next step.
?Troubleshooting
Incubation of donor vesicles with mβCD
TIMING ~2 h
12. Suspend the donor pellet with 4.43 mL of the 35 mM mβCD solution for a mβCD:lipid molar ratio of ~ 8:1. Transfer the suspension to a 20 mL glass scintillation vial with a magnetic stir bar.
CRITICAL STEP Calculate the mβCD:lipid ratio assuming that no lipid is lost during centrifugation in Step 11.
CRITICAL STEP To minimize the formation of bubbles and foam, gently pipet the mβCD solution onto the pellet, then triturate slowly until the pellet is dissolved.
CRITICAL STEP To ensure efficient mixing during the next step, check that the level of the mβCD/donor solution is not much higher than the level of the stir bar. If the level is much higher, a beaker or other flat bottom glass container can be used instead of the scintillation vial.
13. Incubate the mβCD/donor solution at room temperature with gentle stirring (~ 250 rpm) for 2 h.
CRITICAL STEP Vigorous stirring can create foam, which may reduce the exchange efficiency.
CRITICAL STEP For optimal exchange efficiency, the incubation should be performed at a temperature higher than the melting temperature of the acceptor lipids.
CRITICAL STEP Perform this step immediately before proceeding to the next step.
Incubation of acceptor vesicles with mβCD/donor solution
TIMING ~0.5h
14. Add the acceptor LUVs from Step 7 to the mβCD/donor solution, and then incubate at room temperature with gentle stirring (~ 250 rpm) for 30 min.
CRITICAL STEP We find little gain in exchange efficiency beyond 30 min for this lipid composition (unpublished).
CRITICAL STEP For optimal exchange efficiency, the incubation should be performed at a temperature higher than the melting temperature of the acceptor lipids.
Purification of aLUVs
TIMING ~4 h
15. Measure the volume of the mβCD/donor/acceptor slurry and dilute it 8-fold by adding ~ 36 mL of ultrapure H2O. Transfer the mixture to a 50 mL conical tube and centrifuge at 20,000 × g for 30 min at 20 °C.
16. Without disturbing the pellet, carefully remove the supernatant with a glass Pasteur pipet and transfer to a clean 50 mL container. Discard the pellet.
CRITICAL STEP When a fixed angle centrifuge rotor is used in Step 15, the pellet will form on the side wall of the centrifuge tube. Do not be overly aggressive when removing the supernatant. Keep the centrifuge tube immobilized on the bench, and stop when ~ 1 mL of supernatant remains above the topmost point of the pellet.
17. Transfer the supernatant to two 15 mL centrifugal filter devices. Centrifuge at 5000 × g until the volume has been reduced below 500 μL in each filter (~ 30 min).
CRITICAL STEP This step proceeds more quickly when using a swinging bucket rotor even at a lower speed (e.g., 2500 × g).
CRITICAL STEP For larger scale preps, use additional centrifugal filters to reduce the time required to concentrate the supernatant.
18. Discard the filtrate (which contains mβCD and mβCD/lipid complexes) from each centrifugal filter device, and then add D2O to the maximum volume (~ 15 mL). Centrifuge at 5000 × g until the volume has again been reduced below 500 μL in each filter (~ 30 min). Repeat this step three times to remove mβCD, and to replace H2O with D2O.
CRITICAL STEP Washing with D2O instead of H2O enables the use of 1H NMR without the need for solvent suppression, as described in Steps 28-31.
19. Using a mechanical pipet with a 200 μL tip, transfer the retentate into a 1.5 mL snap-top centrifuge tube.
CRITICAL STEP Measure the size distribution of the aLUVs with DLS and compare to the acceptor LUV size measured in Step 7, to check vesicle integrity and test for the presence of donor MLVs or other contaminants.
?Troubleshooting
Assessing exchange efficiency with gas chromatography (GC)
TIMING ~3 h
20. Transfer 5-10 μL of the aLUV suspension (50-100 μg of lipid) into a 13×100 mm screw top glass culture tube for conversion to fatty acid methyl esters (FAMEs) via acid catalyzed methanolysis.
CRITICAL STEP The DLS count rates measured in Steps 7 and 19 can be used to estimate the aLUV concentration, which is helpful for determining the amount of sample required for the GC assay.
21. Add 1 mL methanolic HCl to the culture tube, vortex, flush with Ar, and seal tightly with a Teflon-lined cap. Incubate in a dry block heater at 85 °C for 1 h, and then allow the tube to cool for 5 min.
22. Add 1 mL H2O and vortex to mix. Then, add 1 mL hexane and vortex vigorously to extract the FAMEs.
23. Centrifuge at low speed (~ 400 × g) for 5 min to break the emulsion, then remove 500-800 μL of the upper (hexane) phase and transfer to a GC autosampler vial. Bring the total volume to 1 mL with hexane.
24. Load the autosampler vial into the GC automatic liquid sampler. Using an inlet temperature of 270 °C and helium carrier gas at a 1 mL/min flow rate, initiate the following column temperature program for a 1 μL sample aliquot in splitless injection mode: 2 min at 60°C; 20 °C/min to 170 °C; 5 °C/min to 240 °C; 30 °C/min to 300 °C; 2 min at 300°C, for a total run time of 25.5 min.
25. GC/MSD ChemStation Enhanced Data Analysis software (Agilent Technologies, Santa Clara, CA) will assign and integrate the total ion chromatogram peaks.
26.Calculate the donor mole fraction χD in the aLUVs:
where Ad16:0, A16:0, and A18:1 are the integrated areas of the peaks corresponding to methyl palmitate-d31, methyl palmitate, and methyl oleate, respectively.
Estimate the fraction of outer leaflet exchange (i.e., the exchange efficiency):
Note that the previous equation assumes negligible donor flip-flop and equal lipid packing density within the two leaflets.
CRITICAL STEP The accuracy of the determined composition can be increased with the use of a standard curve for the specific lipid mixture as the area fractions may not vary linearly with the component mole fractions.
27. Compare Eobs calculated in Step 26 with the maximum theoretical exchange efficiency Emax = ρ/(ρ + 0.5) where ρ is the donor-to-acceptor ratio; for the 3:1 ratio used in this protocol, Emax = 0.86.
CRITICAL STEP The equation for Emax assumes that all donor lipid is available for exchange with the outer leaflet acceptor lipid, and that mβCD does not preferentially interact with either the donor or acceptor lipid. Emax therefore represents an upper limit of the exchange efficiency. Assuming negligible donor flip-flop during the exchange step, Eobs > Emax may indicate the presence of donor contamination.
?Troubleshooting
Assessing donor asymmetry with 1H NMR
TIMING ~1 h
28. In a plastic snap-cap centrifuge tube, bring ~ 50-100 μL of the aLUV suspension to 500 μL with D2O for a final lipid concentration of ~ 0.5 mg/mL, then transfer to a 5 mm NMR tube.
29. Load the sample into the NMR spectrometer and collect a standard 1H pulse sequence with a 30° flip angle and 1 s delay time at 50 °C. Collect 32−256 transients, depending on signal-to-noise. Locate the singlet resonance at ~ 3.1-3.6 ppm corresponding to the 9 equivalent protons of the donor choline moiety (Fig. 3a).
CRITICAL STEP An asymmetric choline resonance may indicate contamination from small unilamellar vesicles (SUVs), as shown in Fig. 3d.
?Troubleshooting
30. Dispense 2 μL of the 20 mM Pr3+/D2O solution directly into the NMR tube. Cap the tube and invert it a minimum of three times to mix the contents. Reload the sample in the NMR and collect 32−256 transients. All or part of the choline resonance will shift downfield (i.e., to higher ppm) and broaden, as in Fig. 3b; the shifted portion corresponds to protiated choline lipid (here, the donor lipid) located in the outer leaflet. Repeat this procedure a minimum of 2-3 times, or until a shift of at least 0.05 ppm is achieved.
31. Find the integrated areas R of the shifted and unshifted (if present) parts of the choline resonance, and then determine the fraction of donor lipid located in the outer leaflet:
CRITICAL STEP This analysis can be easily performed using the researcher’s preferred data analysis software by first exporting the NMR spectrum as an ASCII file, and then fitting the resonances to Lorentz functions (Fig. 3). Two Lorentzians are used to model NMR data in the absence of Pr3+: one for the choline and one for the CD (if multiple CD peaks are present, each can be fit with a separate Lorentzian). In the presence of Pr3+ a third Lorentzian is applied to account for the second (outer leaflet) choline resonance. To assist with the fitting routine, the peak area ratio between the choline and CD peaks in the absence of Pr3+ is used to constrain the area ratio of the total choline (i.e. the sum of the shifted and unshifted choline peaks) and CD in the presence of Pr3+ since neither choline nor CD are being lost from or added to the system.
CRITICAL STEP Values of fDout close to 0.5 indicate a symmetric donor distribution, as shown in Fig. 3c. While a gradual loss of asymmetry over several days will occur due to spontaneous lipid flip-flop, this process is slow for the specific sample described here when stored at room temperature (elevated temperatures will accelerate flip-flop 28). An initial absence of asymmetry, though not expected here, may occur for different choices of donor and acceptor lipids. The explanation may be trivial (i.e., an intrinsically fast flip-flop rate for the chosen composition), or it may indicate experimental conditions that are incompatible with aLUV preparation (i.e., an mβCD concentration that is too high for the chosen donor and acceptor lipids).
Determining the composition of the inner and outer leaflets
TIMING ~ 1 min
32. Using the experimentally determined quantities of from NMR (Step 31) and χD from GC/MS (Step 26), calculate the donor mole fraction in each leaflet:
The acceptor mole fraction in the outer and inner leaflets is then equal to and respectively.
?Troubleshooting
Acceptor LUVs size distribution is too large or too small (Step 7)
Vesicles prepared by extrusion through 100 nm pores typically have size distributions centered at a 100-150 nm hydrodynamic diameter, as measured by DLS. If the acceptor vesicles are larger than 150 nm, repeat Step 7. Diameters much smaller than 100 nm may indicate that the extruder was accidentally assembled with a smaller pore size filter.
Donor pellet appears loose (Step 11)
A loose pellet is an indication that the donor MLVs are significantly heterogeneous in size and/or density, which may result in donor contamination in the final aLUV sample. Remove the diffuse parts of the pellet along with the supernatant, and retain only that part of the pellet that adheres firmly to the wall of the centrifuge tube.
Recovered aLUV sample has visible aggregates and/or a large increase in average size (Step 19)
Visible aggregates and/or a significant increase in the average vesicle size (i.e., more than 10% larger than the acceptor LUVs) is an indication of donor MLV contamination. Dilute the sample 20-fold with H2O, vortex briefly and then spin at 20,000 × g for 10-20 min. Carefully recover the supernatant with a glass transfer pipet, leaving the last ~ 5 mL even if there is no visible pellet. Repeat Steps 17-19.
Recovered aLUV sample contains more donor lipid than acceptor lipid (Step 27)
More donor than acceptor lipid (as measured by GC) is an indication of donor MLV contamination. Dilute the sample 20-fold with H2O, vortex briefly and then spin at 20,000 × g for 10-20 min. Carefully recover the supernatant with a glass transfer pipet, leaving the last ~ 5 mL even if there is no visible pellet. Repeat Steps 17-27.
NMR shows an asymmetric choline resonance before the addition of shift reagent (Step 29)
Fig. 3d demonstrates how the NMR line shape can reveal the presence of contaminating SUVs that can be generated during incubation of mβCD and lipid. SUVs are diagnosed by an asymmetric choline resonance (often appearing as two distinct peaks) in the absence of a shift reagent. The two peaks are the result of different packing constraints in the outer and inner leaflets. If SUVs are present, it may be necessary to reduce the mβCD concentration.
Anticipated Results
Beginners will need to perform several practice runs to obtain consistent results. Of note, factors that influence mixing of the exchange slurry (e.g., temperature and stirring speed) in Steps 13-14 may influence the exchange efficiency, and careful removal of the supernatant in Step 16 is critical for avoiding contamination. With time and experience, aLUV compositional variation of < 10% and a yield of > 60% of the initial acceptor LUV mass can be achieved. If contamination of donor vesicles is present and additional dilution-concentration-wash steps are needed as outlined in the Troubleshooting section, the yield may be smaller.
For the specific lipid composition described above, the exchange efficiency Eobs varies between 0.35 and 0.45 in our hands. If greater outer leaflet replacement is desired, the donor:acceptor ratio can be increased or multiple rounds of exchange can be performed (keeping in mind that additional exchange steps will reduce the yield). Unsurprisingly, we find that exchange efficiency depends on the identity of both the donor and acceptor lipids, with some systems approaching the maximum theoretical efficiency 19,23,28. This is likely due to preferential interactions between mβCD and certain types of lipids, that depend on both chain and headgroup structure 18,32. We find nearly complete donor asymmetry for this composition, with ~ 95% of DPPC-d62 residing in the outer leaflet, as determined by 1H NMR. For other choices of donor and acceptor lipids, donor asymmetry measured immediately post-preparation ranges from 65-95% 19,23,28. Table 1 summarizes the aLUVs prepared in our laboratories.
Much can be learned about the fundamental physical and chemical properties of cell membranes using aLUVs. For example, it is possible to systematically study the nature and mechanisms of interleaflet coupling in vesicles that mimic the asymmetric lipid distribution found in the plasma membrane, including whether phase separation in one leaflet induces demixing of lipid components in the other leaflet. Such basic insights are necessary for understanding cellular processes such as transmembrane signaling. Using the procedure described above, we prepared aLUVs having an outer leaflet composed of DPPC-d62/POPC-d13 in a 34/66 molar ratio, and an inner leaflet composed of POPC-d13 19. Using small-angle neutron scattering, we determined structural parameters for the inner and outer leaflets, including thickness and area per lipid. At room temperature, phase coexistence was observed in the outer leaflet of these aLUVs. Interestingly, we found a relatively low packing density for DPPC-d62 located in the outer leaflet compared to the typical tight packing of gel phases, suggesting a disordering effect from interactions with the fluid inner leaflet. This apparent strong interleaflet coupling was abolished at higher temperatures, where both leaflets were in the disordered fluid phase 23.
Without question, asymmetric vesicles are better biological mimetics when compared to their symmetric counterparts that have dominated membrane biophysical studies for nearly 50 years. The tradeoff is ease-of-preparation, although we expect significant improvements will come with more widespread use of the procedures presented here and elsewhere 16. Studies using aLUVs are still in their relative infancy, but it is already clear that asymmetry can significantly alter lipid lateral diffusion 33, packing density 19 and phase behavior 34, and the conformation 25 and partitioning 7 of transmembrane proteins. Continued investigation will provide a deeper understanding of the properties of asymmetric bilayers, and shed new light on the functional significance of membrane asymmetry in living cells.
Acknowledgements
This work acknowledges support from the Austrian Science Fund (FWF): project number P27083. J.K. and F.A.H. are supported through the Scientific User Facilities Division of the U.S. Department of Energy (DOE), Office of Basic Energy Sciences (BES), under contract no. DE-AC05 00OR2275. DM is supported by University of Windsor startup funds.
Footnotes
Author contributions statement
M.D., F.A.H. and D.M. wrote the manuscript; M.D., F.A.H., B.E., G.P, E.L, R.F.S. and J.K. provided input and edited the manuscript; M.D., F.A.H., B.E. and D.M. conducted the experiments.
References
- 1.Lorent JH, et al. Structural determinants and functional consequences of protein affinity for membrane rafts. Nature Communications. 2017;8 doi: 10.1038/s41467-017-01328-3. 1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lingwood D, Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 3.Doktorova M, et al. Cholesterol Promotes Protein Binding by Affecting Membrane Electrostatics and Solvation Properties. Biophysical Journal. 2017;113:2004–2015. doi: 10.1016/j.bpj.2017.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Op den Kamp JAF. Lipid Asymmetry in Membranes. Ann Rev Biochem. 1979;48:47–41. doi: 10.1146/annurev.bi.48.070179.000403. [DOI] [PubMed] [Google Scholar]
- 5.Verkleij AJ, et al. The asymmetric distribution of phospholipids in the human red cell membrane. A combined study using phospholipases and freeze-etch electron microscopy. Biochimica et Biophysica Acta. 1973;323:178–193. doi: 10.1016/0005-2736(73)90143-0. [DOI] [PubMed] [Google Scholar]
- 6.Fadok VA, et al. Exposure of phosphatidylserine on the surface of apoptotic lymphocytes triggers specific recognition and removal by macrophages. Journal of Immunology. 1992;148:2207–2216. [PubMed] [Google Scholar]
- 7.Perillo VL, Penalva DA, Vitale AJ, Barrantes FJ, Antollini SS. Transbilayer asymmetry and sphingomyelin composition modulate the preferential membrane partitioning of the nicotinic acetylcholine receptor in Lo domains. Archives of Biochemistry and Biophysics. 2016;591:76–86. doi: 10.1016/j.abb.2015.12.003. [DOI] [PubMed] [Google Scholar]
- 8.Clair JR, Wang Q, Li G, London E. In: The Biophysics of Cell Membranes. Epand R, Ruysschaert JM, editors. Vol. 19. Springer; 2017. pp. 1–27. [Google Scholar]
- 9.Pautot S, Frisken BJ, Weitz DA. Engineering asymmetric vesicles. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:10718–10721. doi: 10.1073/pnas.1931005100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Redelmeier TE, Hope MJ, Cullis PR. On the mechanism of transbilayer transport of phosphatidylglycerol in response to transmembrane pH gradients. Biochemistry. 1990;29:3046–3053. doi: 10.1021/bi00464a022. [DOI] [PubMed] [Google Scholar]
- 11.Hope MJ, Redelmeier TE, Wong KF, Rodrigueza W, Cullis PR. Phospholipid asymmetry in large unilamellar vesicles induced by transmembrane pH gradients. Biochemistry. 1989;28:4181–4187. doi: 10.1021/bi00436a009. [DOI] [PubMed] [Google Scholar]
- 12.Mui BL, Dobereiner HG, Madden TD, Cullis PR. Influence of transbilayer area asymmetry on the morphology of large unilamellar vesicles. BiophysicalJjournal. 1995;69:930–941. doi: 10.1016/S0006-3495(95)79967-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takaoka R, Kurosaki H, Nakao H, Ikeda K, Nakano M. Formation of asymmetric vesicles via phospholipase D-mediated Transphosphatidylation. Biochimica et Biophysica Acta. 2017 doi: 10.1016/j.bbamem.2017.10.011. [DOI] [PubMed] [Google Scholar]
- 14.Bloj B, Zilversmit DB. Asymmetry and transposition rates of phosphatidylcholine in rat erythrocyte ghosts. Biochemistry. 1976;15:1277–1283. doi: 10.1021/bi00651a017. [DOI] [PubMed] [Google Scholar]
- 15.Herrmann A, Zachowski A, Devaux PF. Protein-mediated phospholipid translocation in the endoplasmic reticulum with a low lipid specificity. Biochemistry. 1990;29:2023–2027. doi: 10.1021/bi00460a010. [DOI] [PubMed] [Google Scholar]
- 16.Cheng HT, London E. Preparation and properties of asymmetric large unilamellar vesicles: interleaflet coupling in asymmetric vesicles is dependent on temperature but not curvature. Biophysical Journal. 2011;100:2671–2678. doi: 10.1016/j.bpj.2011.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szejtli J. Introduction and General Overview of Cyclodextrin Chemistry. Chemical Reviews. 1998;98:1743–1754. doi: 10.1021/cr970022c. [DOI] [PubMed] [Google Scholar]
- 18.Bozelli JC, Jr, Hou YH, Epand RM. Thermodynamics of Methyl-beta-cyclodextrin-Induced Lipid Vesicle Solubilization: Effect of Lipid Headgroup and Backbone. Langmuir. 2017;33:13882–13891. doi: 10.1021/acs.langmuir.7b03447. [DOI] [PubMed] [Google Scholar]
- 19.Heberle FA, et al. Subnanometer Structure of an Asymmetric Model Membrane: Interleaflet Coupling Influences Domain Properties. Langmuir. 2016;32:5195–5200. doi: 10.1021/acs.langmuir.5b04562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mui BL, Cullis PR, Evans EA, Madden TD. Osmotic properties of large unilamellar vesicles prepared by extrusion. Biophysical Journal. 1993;64:443–453. doi: 10.1016/S0006-3495(93)81385-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Evans E, Heinrich V, Ludwig F, Rawicz W. Dynamic tension spectroscopy and strength of biomembranes. Biophysical Journal. 2003;85:2342–2350. doi: 10.1016/s0006-3495(03)74658-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Homan R, Pownall HJ. Transbilayer diffusion of phospholipids: dependence on headgroup structure and acyl chain length. Biochimica et Biophysica Acta. 1988;938:155–166. doi: 10.1016/0005-2736(88)90155-1. [DOI] [PubMed] [Google Scholar]
- 23.Eicher B, et al. Joint small-angle X-ray and neutron scattering data analysis of asymmetric lipid vesicles. Journal of Applied Crystallography. 2017;50:419–429. doi: 10.1107/S1600576717000656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Denkins YM, Schroit AJ. Phosphatidylserine decarboxylase: generation of asymmetric vesicles and determination of the transbilayer distribution of fluorescent phosphatidylserine in model membrane systems. Biochimica et Biophysica Acta. 1986;862:343–351. doi: 10.1016/0005-2736(86)90237-3. [DOI] [PubMed] [Google Scholar]
- 25.Lin Q, London E. The influence of natural lipid asymmetry upon the conformation of a membrane-inserted protein (perfringolysin O) The Journal of Biological Chemistry. 2014;289:5467–5478. doi: 10.1074/jbc.M113.533943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Markones M, et al. Engineering asymmetric lipid vesicles: accurate and convenient control of the outer leaflet lipid composition. Langmuir. 2018 doi: 10.1021/acs.langmuir.7b03189. [DOI] [PubMed] [Google Scholar]
- 27.Eicher B, et al. Intrinsic Curvature-Mediated Transbilayer Coupling in Asymmetric Lipid Vesicles. Biophysical Journal. 2018;114:146–157. doi: 10.1016/j.bpj.2017.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marquardt D, et al. 1H NMR Shows Slow Phospholipid Flip-Flop in Gel and Fluid Bilayers. Langmuir. 2017;33:3731–3741. doi: 10.1021/acs.langmuir.6b04485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng HT, Megha, London E. Preparation and properties of asymmetric vesicles that mimic cell membranes: effect upon lipid raft formation and transmembrane helix orientation. The Journal of Biological Chemistry. 2009;284:6079–6092. doi: 10.1074/jbc.M806077200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin Q, London E. Preparation of artificial plasma membrane mimicking vesicles with lipid asymmetry. PloS One. 2014;9:e87903. doi: 10.1371/journal.pone.0087903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li G, et al. Efficient replacement of plasma membrane outer leaflet phospholipids and sphingolipids in cells with exogenous lipids. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:14025–14030. doi: 10.1073/pnas.1610705113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang Z, London E. Effect of cyclodextrin and membrane lipid structure upon cyclodextrin-lipid interaction. Langmuir. 2013;29:14631–14638. doi: 10.1021/la4031427. [DOI] [PubMed] [Google Scholar]
- 33.Chiantia S, Schwille P, Klymchenko AS, London E. Asymmetric GUVs prepared by MbetaCD-mediated lipid exchange: an FCS study. Biophysical Journal. 2011;100:L1–3. doi: 10.1016/j.bpj.2010.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin Q, London E. Ordered raft domains induced by outer leaflet sphingomyelin in cholesterol-rich asymmetric vesicles. Biophysical Journal. 2015;108:2212–2222. doi: 10.1016/j.bpj.2015.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]