Abstract
Naphthalene (NA) is a ubiquitous environmental pollutant and possible human carcinogen that forms tumors in rodents with tissue/regional and species selectivity. This study seeks to determine whether NA is able to directly adduct DNA in an ex vivo culture system. Metabolically active lung tissue was isolated and incubated in explant culture with carbon-14 labeled NA (0, 25, 250μM) or 1,2-naphthoquinone (NQ), followed by AMS analyses of metabolite binding to DNA. Despite relatively low metabolic bioactivation in the primate airway, dose-dependent NA-DNA adduct formation was detected. More airway adducts were detected in female mice (4.7-fold) and primates (2.1-fold) than in males of the same species. Few adducts were detected in rat airway or nasal epithelium. NQ, which is a metabolic product of NA, proved to be even more potent, with levels of adduct formation 70 to 80-fold higher than seen when tissues were incubated with the parent compound NA. This is the first study to demonstrate NA-DNA adduct formation at a site of carcinogenesis, the mouse lung. Adducts were also detected in non-human primate lung and with a NQ metabolite of NA. Taken together, this suggests that NA may contribute to in vivo carcinogenesis through a genotoxic mechanism.
Keywords: naphthalene, naphthoquinone, DNA adducts, respiratory toxicity
1. Introduction
Naphthalene (NA) is a possible human carcinogen that causes respiratory tract lesions in two mammalian species (Abdo et al., 1992; Abdo et al., 2001; National Toxicology Program, 1992). The respiratory tract toxicity of NA has been studied extensively in rodents, though the mechanism of NA carcinogenicity is a topic of debate. Recurrent cytotoxicity and proliferation is thought to be the driving force behind NA exposure-related formation of mouse lung tumors (Van Winkle et al., 1995; West et al., 2001) and rat nasal tumors (Long et al., 2003; National Toxicology Program, 2000), but the formation of DNA adducts has not been measured. Determining whether NA can cause stable DNA adducts in tissues identified as likely targets of NA carcinogenesis, or not, is highly relevant to ongoing risk assessment.
Near lifetime exposure to NA vapor causes tumors to form in the respiratory tissues of rodents, with tissue and species selectivity (Abdo et al., 1992; Abdo et al., 2001; National Toxicology Program, 1992). It has been suggested that species differences in metabolism and protein adduction are indicative of different modes of action in target tumor tissues (Piccirillo et al., 2012). NA respiratory toxicity is dependent on bioactivation by cytochrome P450 monooxygenase (CYP) enzymes, which are elevated in respiratory tract target tissues identified by the NTP chronic bioassay (female mouse conducting airways of the lung, male/female rat nasal respiratory epithelium, and female rat olfactory epithelium). Enzyme kinetics for NA metabolism in microsomes obtained from microdissected airway and nasal samples found that the steady state kinetic parameters for NA disappearance support the involvement of enzymes in toxicity and tumorigenicity (rat olfactory epithelium > mouse olfactory epithelium > mouse airways ≫ rat airways) (Buckpitt et al., 2013).
There is a correlation between the severity of NA toxicity and the amount of protein adducts (Cho et al., 1994; DeStefano-Shields et al., 2010). Known metabolites capable of forming protein adducts include the 1,2-NA epoxide, NA-diol epoxide, 1,4-NQ and 1,2-NQ (Boland et al., 2004; Cho et al., 1994; DeStefano-Shields et al., 2010; Lin et al., 2006; Pham et al., 2012a; Pham et al., 2012b; Waidyanatha et al., 2002). In the sole study of rodent DNA adducts, depurinated and stable NA DNA adducts were found in mouse skin (female SENCAR mice; 500 or 1200 nmol on shaved skin for 4h) (Saeed et al., 2009). The relevance of that study to lung carcinogenesis is uncertain because skin is not a known target tissue for inhaled NA cytotoxicity. Further, the results are difficult to interpret because authors reported similar levels of NA- and 1,2-NQ-adduct formation despite the lack of NA metabolic activation in the mouse skin.
Understanding the mechanism of NA carcinogenesis, and the relevance of this mechanism to humans, is necessary for the protection of human health. NA-derived DNA adducts have not been demonstrated in vivo or in vitro in the respiratory system of any species. If NA-derived DNA adducts form in the same rodent respiratory tissues that are susceptible to tumor formation in the chronic bioassay, this may indicate a possible genotoxic mode of action for NA lung carcinogenesis. The argument has been made that NA does not pose a cancer risk to humans because NA is bioactivated by cytochrome P450 monooxygenase enzymes that have very high efficiency in rodent lung tissue and low efficiency in human lung tissue. However, despite low CYP activity in the primate, which results in reduced metabolism to reactive metabolites, NA DNA adducts form in primate airway. NA DNA adducts also form in mouse airway but not in rat lung.
2. Materials and Methods
2.1. Rodent tissue
Adult male and female B6C3F1 mice and male Sprague Dawley rats were purchased from Harlan Labs. The Sprague Dawley strain was selected because NA has been demonstrated to be highly cytotoxic in the Sprague Dawley rat nose (Buckpitt et al., 2013; Lee et al., 2005; Plopper et al., 1992) and NA was known to form protein adducts in Sprague Dawley nasal epithelium (DeStefano-Shields et al., 2010). In one study, the Sprague Dawley rat was reported to be more sensitive to NA than F344 rats, with nasal olfactory epithelial necrosis at lower doses (0.1–30 ppm, 6h) in Sprague Dawley than in F344 rats (1–30 ppm, 6h)(Dodd et al., 2010).
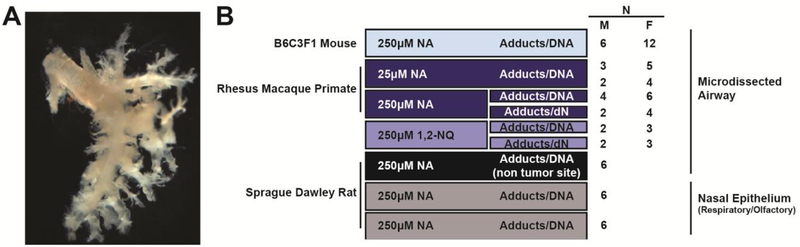
All rodents were maintained in a barrier facility with filtered air in AAALAC approved conditions on a 12-hour light/dark cycle with food and water ad libitum. All animal experiments were performed under protocols approved by the University of California Davis or Lawrence Livermore National Laboratory (LLNL) IACUC in accordance with National Institutes of Health guidelines. Rodents were euthanized with an intraperitoneal overdose of Fatal-Plus® C IIN (Vortech Pharmaceuticals, pentobarbital sodium). Lungs were removed en bloc and inflated via tracheal cannula with a 50:50 solution of deficient Waymouth’s media (prepared without cysteine, cystine, methionine, reduced or oxidized glutathione, (Duan et al., 1996)) and 2% agarose (2-hydroxyethyl agarose, low gelling temperature; Sigma, St. Louis, MO). Airways from mice and rats were microdissected as previously described to isolate the metabolically active region of the lung (Plopper et al., 1991; Van Winkle et al., 1996). Previous studies using this approach have demonstrated that tissues isolated in this manner retain active cytochrome P450 monooxygenase activity (Buckpitt et al., 1995; Van Winkle et al., 1996) and that these airway explants demonstrate cytotoxicity in vitro (West et al., 2003). One half of a microdissected mouse lung was used per exposure group (10–20 mg tissue); exposure endpoints were randomized between donor animals. Figure 1A shows an example of an agarose-inflated, microdissected airway. For rats, the microdissected left lung lobe was split into three parts, which yielded a comparable volume of tissue per exposure group to the mouse airway). Exposure endpoints were randomized between two pieces such that each animal received two distinct treatments (250μM or control). Respiratory and olfactory epithelium were harvested from rat nose using methods previously described (Cichocki et al., 2014). Incubation concentrations were based on estimated nasal tissue doses following a 30 ppm exposure in mice (20–200μM) in (Morris, 2013).
Figure 1. Methods and study design.
(A) An example of metabolically active microdissected airway tissue from a B6C3F1 mouse. (B) Study design: metabolically active tissue from male and female adult animals of three species was isolated and incubated in explant culture with 14C labeled NA or 14C labeled1,2-NQ for 1 hour, followed by 14C-AMS analyses of metabolite binding to DNA or deoxynucleosides (dN) that include dA, dG, dC, and dT. Each 14C exposure was run in parallel with non-radiolabeled controls. Abbreviations: N, number of samples per endpoint; F, female; M, male.
2.2. Primate tissue
Lung lobes were harvested from adult male and female rhesus macaques from the California National Primate Research Center. All rhesus macaque tissues were from adult animals euthanized for normal colony maintenance at the center over a three-year period from 2015–2017. Tissues from animals with respiratory infections, or from juvenile or geriatric animals were omitted from the study. Primate characteristics are reported in Supplementary Table 1. Fresh tissue was kept on ice in Nutrient Mixture F-12 Hams media (Sigma, St. Louis, MO), and microdissected on ice within 2 hours of tissue harvest. Distal airway pieces approximately ≤2 mm thick were removed from the surrounding parenchyma. Approximately 50 mg of tissue was used per exposure group (0, 25 or 250 μM). Sample volumes for ex vivo exposures were determined through method optimization, which aimed to reduce animal usage. Volume of tissue needed was determined to obtain robust yields of purified DNA (> 5 μg) for sample processing, carbon-14 (14C) accelerator mass spectrometry (AMS) analyses, and data normalization. Airways sampled across all three species were from the same airway level and approximately the same volume. Sample sizes and tissues utilized are reported in Figure 1B.
2.3. Radiochemicals
14C-NA (≥ 98% pure) was diluted to a specific activity of 4.8 mCi/mmol (low dose, 2.5mM) or 0.46 mCi/mmol (high dose, 25mM) with non-radiolabeled NA in acetonitrile from a stock solution of 14C-NA (1,4,5,8-14C; ≥ 98% pure) in methanol (MC 2147; 500 μCi, 58 mCi/mmol; Moravek Biochemicals, Brea, CA). 14C-1,2-NQ was diluted to a specific activity of 0.51 mCi/mmol (25mM) with non-radiolabeled 1,2-NQ in acetonitrile from a stock solution of 14C-1,2-NQ (1,4-14C; ≥ 98% pure) in acetonitrile (100 μCi, 55 mCi/mmol; American Radiolabeled Chemicals, St. Louis, MO).
2.4. Tissue incubation
Metabolically active tissue was placed in a 20-ml scintillation vial with 990 μl of media and kept on ice until NA, NQ or sham was added. Vials of tissue were randomly assigned to a treatment group and given a sample number that lacked descriptive identifying information. 14C labeled NA or 1,2-NQ NA (10 μl was added to each vial to yield final concentrations of 25 or 250 μM). Vials were sealed with electrical tape to prevent volatilized compounds from escaping, and incubated in a water bath for 1 hour at 37°C. Parallel negative control incubations were conducted with tissue in 990 μl media with 10 μl acetonitrile, and used as an indicator of background radiation level. At the end of the treatment, all vials were placed on ice for 10 minutes and then samples were transferred to microfuge tubes. Tissue was washed 15–25 times, each time with 1–2 ml of 200 proof ethanol (the microfuge tubes were changed 5–8 times) to remove unbound NA and 1,2-NQ. Washes were checked using a liquid scintillation counter and considered clean when the ethanol exhibited background levels of 14C (approximately 10–15 CPM at UC Davis, 29–32 CPM at LLNL). Tissues were stored in −80°C freezer until DNA isolation. As an internal check that tissues were metabolically active and toxicity was induced, several samples were incubated with NA and then with Ethidium homodimer-1 to label permeable cells. More permeable cells were present following incubation with naphthalene indicating that metabolism was occurring (data not shown).
2.5. DNA isolation and quantification
Washed, dry, frozen tissue was placed in 2-ml FastPrep Lysing Matrix Z tubes (2-mm zirconia beads; MP Biomedicals, Santa Ana, CA) with 500 μl ATL buffer (Qiagen, Hilden, Germany). Tissue was homogenized using a mini bead beater mill (Cole-Parmer; Vernon Hills, IL) for 40-second intervals (x8–10), placed on ice between bead beating steps. DNA was isolated from homogenized tissue using DNeasy Blood & Tissue kits (Cat No./ID: 69504; Qiagen, Hilden, Germany) according to manufacturer instructions with the following modifications. Because NA metabolites are known to adduct protein, an additional proteinase K lysis step was added to the Base Qiagen protocol (Purification of Total DNA from Animal Tissues (Spin-Column Protocol)) so that the final proteinase K incubation time was 4 hours, with 25 μl of proteinase K added at the beginning and 25 μl additional proteinase K added at 2 hours. 4 μl RNase A (100 mg/ml) was added to each sample after proteinase K lysis step, and samples were incubated with RNase A for 2 minutes at room temperature. Each sample was divided and purified through two DNAeasy columns to maximize yield, and then recombined after elution. DNA was eluted twice, first with 200 μl and then with 100 μl of Buffer AE. DNA concentration and purity was monitored using a NanoDrop™ spectrophotometer. An aliquot of DNA from the primate airway samples was saved for enzymatic digestion.
2.6. Enzymatic digestion of DNA and sample processing
Up to 45 μg of primate airway DNA was used for digestion with LCMS grade solvents. The DNA solution was brought to 400 μl with 10 mM Tris-MgCl2 buffer, pH 7.1. Enzymatic digestion of DNA samples began with the addition of 30 μl of 1-mg/ml DNase I (prepared in 0.15M NaCl) to the DNA solution. The DNA solution was incubated using a Thermomixer at 37°C and 500 rpm for 1 hour. Subsequently, 30 μl of 0.5mg/ml nuclease P1 (prepared in 1mM ZnCl2) was added into the DNA solution and incubated as previously described for 2 hours. Afterwards, 30 μl of 1-mg/ml alkaline phosphatase (prepared in 1mM MgCl2) and 30 μl of 50 μg/ml phosphodiesterase I (prepared in 110 mM Tris-HCl, 110 mM NaCl, 15 mM MgCl2 in 50% glycerol, pH 8.9) were added into the DNA solution and incubated as previously described for 18 hours. After completion of the enzymatic digestion a 20 μl aliquot was transferred into 60 μl of water for measurement of deoxyguanosine. The remaining DNA digest was then diluted 1:2 with water and processed using solid phase extraction (SPE), using Strata-X C18 columns (Phenomenex, Torrance, CA). The column was activated with 1 ml of methanol, equilibrated with 1 ml of water, and then the sample was loaded. The column was washed sequentially with 1 ml of water and 1 ml of 25% methanol, followed by elution with 1 ml of methanol. The column eluate was evaporated to dryness at 45°C using a Savant SpeedVac sample concentrator (Thermo Fisher Scientific, Waltham, MA). The samples were reconstituted in 200 μl of water and stored at −80°C until analysis using AMS.
2.7. Quantification of deoxynucleosides using UPLC-UV
Deoxynucleosides (dN) include dA, dG, dC, and dT. In this study, dG was measured as an indication of how much total dN was in each sample. The ratio of dG to dN was constant. Measurement of dN, expressed as adducts/pg deoxyguanosine (dG), was performed using an Agilent 1290 Infinity series ultra-high performance liquid chromatography system coupled to an Agilent 1260 diode array detector (DAD) (Agilent Technologies, Santa Clara, CA). The DAD was set to 260nm for detection of dG. Chromatographic separation was performed using a Kinetex C18 column (1.7μm; 2.1×100mm) (Phenomenex, Torrance, CA). The mobile phases consisted of water containing 0.1% formic acid (mobile phase A) and acetonitrile containing 0.1% formic acid (mobile phase B). The following mobile phase gradient was used: 15%B from 0 to 6min, with a linear increase to 95%B between 6 and 8 min, held at 95%B from 8 to 12 min, returned to 15% B between 12 and 12.5 min, and re-equilibration of the column at 15%B from 12.5 to 16 min. The mobile phase flow rate was 0.2 deoxyguanosine/min. A 10 μl sample injection volume was used. A five-point calibration curve, ranging from 0 to 300 ng dG on column, was created and used for quantification of dG in DNA digest samples.
2.8. AMS sample preparation and analysis
All glass used in AMS sample prep was baked prior to use to remove residual carbon and standard laboratory protocols were followed (Buchholz et al. 2000). Quartz was baked at 900°C for 2 hours and borosilicate was baked at 500 °C for 2 h. Each DNA or dN sample suspended in water was transferred by pipet into a quartz vial (6mm o.d. × 30-mm length) and dried overnight in a vacuum centrifuge. Upon removal from the centrifuge, 1 μl tributyrin (ICN Pharmaceuticals; Costa Mesa, CA) was added as carbon carrier (0.59 mg C) to each vial using a volumetric capillary tube (Drummond Scientific Company, Broomall, PA). Each quartz vial received ~40 mg of CuO as it was transferred to a quartz combustion tube that was evacuated and sealed with a torch. The samples in combustion tubes were combusted at 900°C for 3.5 hours to oxidize all organic carbon to CO2 and then reduced to filamentous carbon as described previously (Ognibene et al., 2003). Carbon samples were packed into aluminium sample holders and carbon isotope ratios were measured on a National Electrostatics Corporation (Middleton, WI) 250-kV single stage AMS spectrometer at LLNL. AMS measurement times were typically 3–10 minutes/sample with a counting precision (relative standard deviation, RSD) of 0.5% to 3% and a standard deviation among 3 to 10 measurements of 1% to 3%. The 14C/13C ratios of the samples were normalized to measurements of four identically prepared isotopic standard samples of known isotope concentration (IAEA C-6 also known as ANU sucrose).
Measured isotope ratios were converted to biologically relevant units using the carbon concentration, isotope ratio and mass of carbon carrier, the specific activity of the labelled compound (NA or 1,2 NQ), and the DNA or dN mass. Details on the procedure can be found in the literature (Kwok et al., 1999; Vogel and Love, 2005; Zimmermann et al., 2017).
2.9. Statistics
GraphPad Prism 7 was used to perform one-way or two-way ANOVA with Tukey’s multiple comparisons post-hoc test, or linear regression analysis (dose-response only, Figure 3). Data are reported as mean ± standard error. Values of p < 0.05 were considered statistically significant (*p<0.05, **p<0.01, ***p<0.001, ****p<0.0001).
3. Results
3.1. NA DNA adducts were detected in lung tissue from B6C3F1 mice and Rhesus macaque primates, and at low levels in rat tissues.
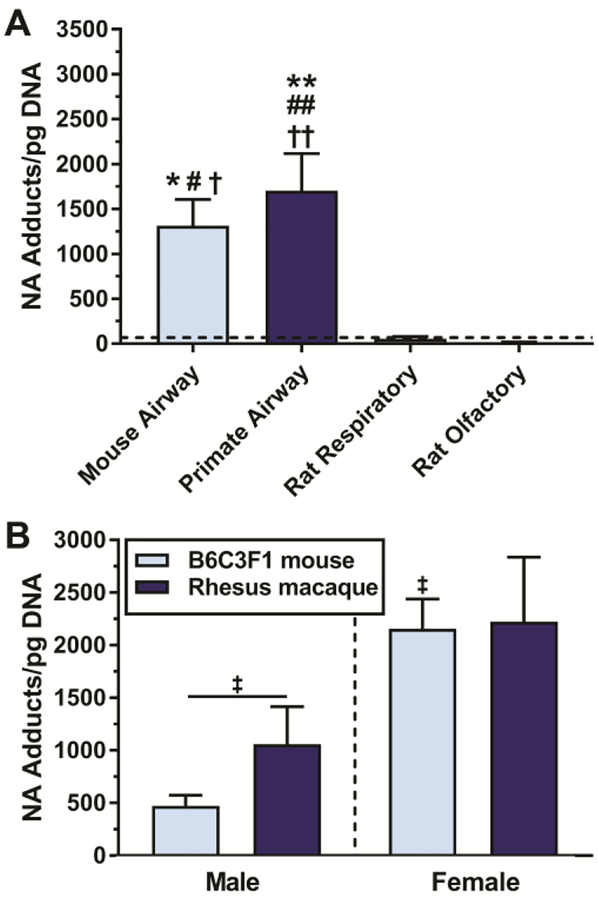
A comparison of DNA adduct levels in the tissues tested is shown in Figure 2. The level of NA DNA adduct formation in mouse airways was 1706 ± 409 adducts/pg DNA (Figure 2A). Neither rat olfactory nor respiratory nasal epithelium had levels of 14C in isolated DNA exceeding levels in rat lung (69.0 ± 24.8 adducts/pg DNA). Nasal DNA adduct levels were 55.4 ± 23.9 adducts/pg DNA for respiratory epithelium, and 13.9 ± 3.7 adducts/pg DNA for olfactory epithelium which was below the limit of quantitation (LOQ) established from measurement of vehicle controls (N=20). Levels of DNA adducts were not different between the two types of nasal epithelium (p>0.9999). Vehicle treated tissues processed under the same conditions had radioactivity levels corresponding to a LOQ = 24 adducts/pg DNA (LOQ = mean of vehicle controls + 2*standard deviation of vehicle controls). For mice and primates, sex is a significant main effect in determining the level of DNA adduct formation (p=0.0011, Figure 2B). Airways of female B6C3F1 mice (N=12) form 4.7-fold more NA DNA adducts than airways from male B6C3F1 mice (N=6, p=0.0139). The level of NA DNA adduct formation in female mouse airways (2160 ± 285 adducts/pg DNA) was statistically similar to female primate airways (2220 ± 612 adducts/pg DNA).
Figure 2. Sex and species differences in NA DNA adducts.
Mouse and primate airway are susceptible to DNA adduct formation after a 250μM NA ex vivo incubation. (A) Pooled B6C3F1 mouse airway (N=12), pooled rhesus macaque primate airway (N=9), and male nasal epithelium (respiratory (N=6) and olfactory (N=6)). The dashed black line reflects the level of radioactivity present in male rat lung, which is not a known site of carcinogenesis. Neither rat olfactory or respiratory nasal epithelium has levels of 14C exceeding levels in rat lung. (B) Adduct formation is elevated in lung tissue from female mice and primates. *=significantly different from rat respiratory, #=significantly different from rat olfactory, †=significantly different from rat lung, ‡ significantly different from male mouse airway.
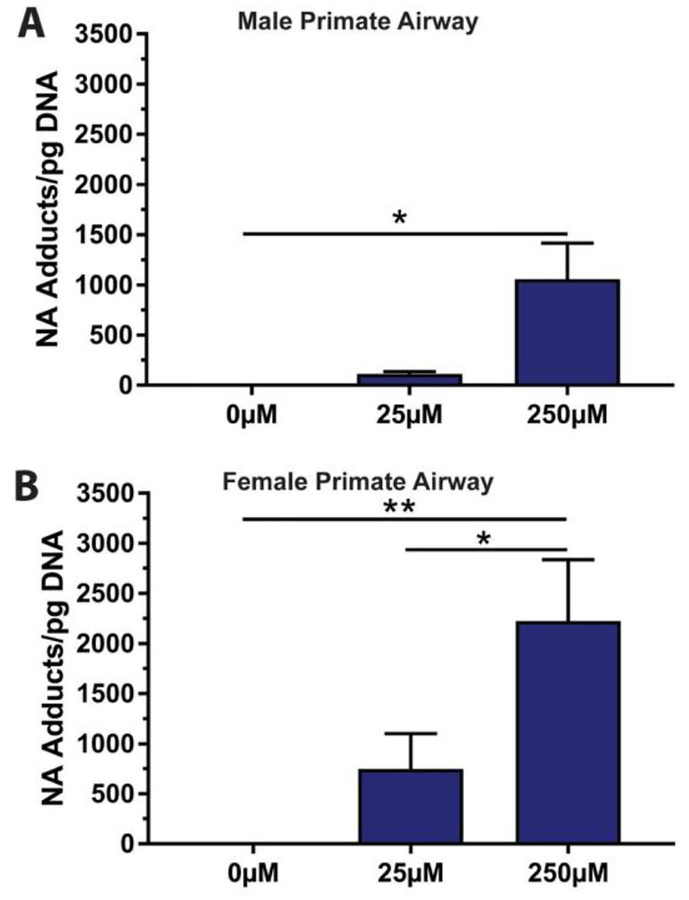
NA DNA adduct dose-response was quantified for male (Figure 3A) and female (Figure 3B) primate airway explants incubated with 0 μM, 25 μM or 250 μM NA. The mean at 250 μM was 1060 ± 357 adducts/pg DNA for males and 2220 ± 612 adducts/pg DNA for females. The number of NA DNA adducts increased with NA concentration in both males (y=4.23x + 1.46, R2=0.631, N=3–4) and females (y=8.09x + 233, R2=0.544, N=5–6).
Figure 3. Dose dependent NA DNA adduct formation.
NA DNA adduct levels in (A) male and (B) female primate airway explants after a 1 hour incubation. Sample size for these comparisons was 3 (25μM) to 4 (250μM) for the male dose response, and 5 (25μM) to 6 (250μM) for female dose response.
3.2. 1,2-NQ was a potent DNA adductor in primate airway.
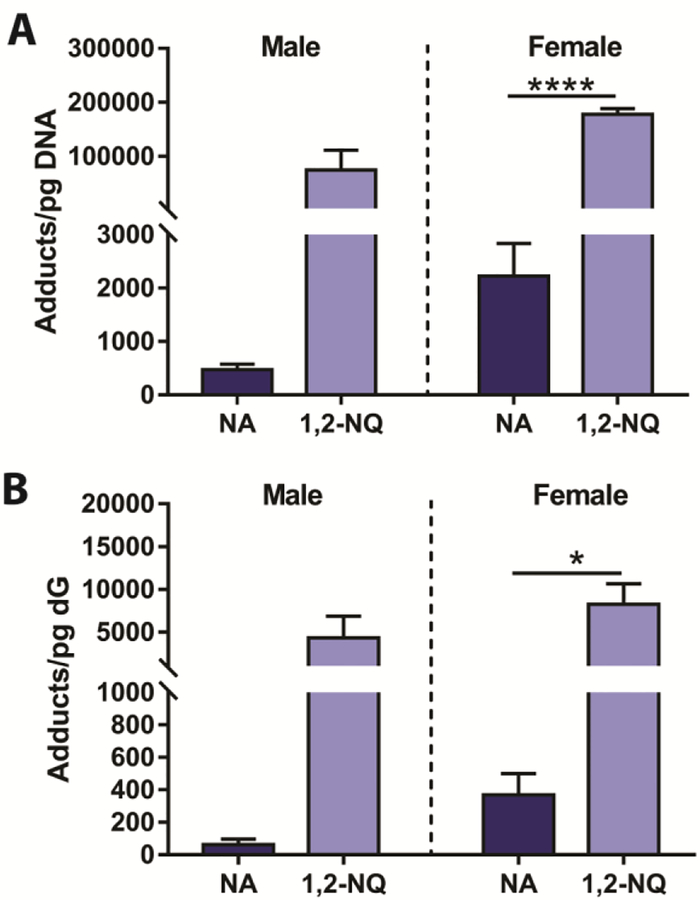
The 1,2-NQ metabolite can be generated by CYP bioactivation of NA and subsequent metabolism, however it does not require bioactivation when the metabolite is directly incubated with explant tissue. Therefore, as a reactive metabolite of NA, 1,2-NQ can presumably adduct DNA independent of CYP bioactivation. In primate airway explants, the ability of 1,2-NQ to form DNA adducts greatly exceeded that of NA (Figure 4A). Interpretation of sex differences is limited by sample size (NMale=2–4, Nfemale=3–6). Incubation with 1,2-NQ resulted in approximately 70-fold greater adduct formation in males, and 80-fold greater adduct formation in females (p<0.0001) than was observed with 250 μM NA incubations.
Figure 4. In primate airway explants, the ability of 1,2-NQ to form DNA adducts, and remain bound to dG, greatly exceeds that of NA.
(A) The 250μM incubation with 1,2-NQ resulted in 70-fold greater adduct formation in males and 80-fold greater adduct formation in females (p<0.0001; N1,2-NQ=3, NNA=6) than was observed with 250μM NA incubations. (B) NA and 1,2-NQ DNA adducts are still detected in enzymatically digested DNA, bound to dN.
3.3. Use of enzymatic digestion to demonstrate covalent binding of NA and NQ to DNA.
NA and 1,2-NQ DNA adducts were still detected in enzymatically digested DNA, bound to isolated dG. As with DNA adducts, higher levels of dN adducts were formed with 1,2-NQ than with NA (Figure 4B). There was only a 22-fold greater 1,2-NQ dG adduct formation in females (p=0.022; N1,2-NQ=3, NNA=4) than was observed with 250μM NA incubations.
4. Discussion
We detected adduct formation in both sexes of B6C3F1 mice following a one hour 250μM NA exposure. However, female mice were nearly five times more sensitive to airway NA DNA adduct formation than were males. The observed sex difference in DNA adduct formation corresponds with existing pathology and metabolism data. Susceptibility to acute cellular damage by NA was increased in adult female mice (Carratt et al., 2016; Van Winkle et al., 2002), and the process of epithelial regeneration was slower in female mice relative to males (Oliver et al., 2009).
There was a clear difference between DNA adduct formation for NA and for the NA-metabolite tested: 1,2-NQ. Our findings that 1,2-NQ is highly reactive with DNA, and is stably adducted to dN, is important for occupational and environmental health. 1,2-NQ bound to DNA in an ex vivo culture system at 70 to 80 times the rate of NA (greater than 22-fold for dN). If people are able to metabolize NA to the 1,2-NQ, this becomes very important. Coke oven workers exposed to a variety of PAHs have 1.7-fold higher levels of 1,2-NQ serum albumin adducts compared to control personnel (76.6 pmol/g vs. 44.9 pmol/g) (Dai et al., 2004). 1,2-NQ and 1,4-NQ serum adducts were also detected in a study of blood donors in Taiwan, with more 1,2-NQ adducts than 1,4-NQ adducts (Lin et al., 2009). It is unclear if levels of 1,2-NQ serum albumin adducts detected in people are a result of NA metabolism or direct exposure to environmental/occupational 1,2-NQ, but human tissues/enzymes are able to metabolize NA to NQs (Cho et al., 2005). Rats given NA intraperitoneally (100–800 mg/kg) have dose dependent 1,2-NQ and 1,4-NQ hemoglobin and albumin adduct formation, with more 1,2-NQ adducts than 1,4-NQ adducts (Cho et al., 2005; Waidyanatha et al., 2002).
The formation of NA DNA adducts and 1,2-NQ DNA adducts in primate tissue, at or above levels of adduct formation in mouse tissue is surprising. Previous work has demonstrated that there is a 100-fold difference in metabolism between primates and mice, favoring the formation of reactive metabolites in mice and likely contributing to GSH depletion (Boland et al., 2004). However, this difference in metabolism does not appear to directly correspond with formation of protein adducts either, as there is a 3-fold difference in protein adduct formation between mice and primates (Cho et al., 1994; DeStefano-Shields et al., 2010). Elevated 14C interpreted as DNA adducts might be artificially high when detected using AMS if there is protein contamination in the DNA or dN. DNA purification included additional proteinase digestion steps and the ratio of absorbance at 260 nm and 280 nm was checked and confirmed to be in the range considered free of protein contamination before AMS analysis. Further, little adduct was detected in DNA samples obtained from rats, which were incubated and processed under identical conditions as were the mouse and primate DNA samples. Thus, we believe that the DNA adducts detected are genuine, and the differences between mice and primates with regards to NA metabolism, and the subsequent adduction of proteins and DNA, indicates a critical knowledge gap with regards to which enzymes metabolize NA in primates and humans.
The near absence of DNA adducts in rat nasal epithelium is supportive of the presence a non-mutagenic threshold for the rat nasal tumors in the chronic bioassay as suggested in Bailey et al. (Bailey et al., 2016). Severe GSH depletion and subsequent cycles of cytotoxicity and repair may be sufficient to predispose epithelial cells to non-mutagenic carcinogenesis.
The current study details a novel method by which volatile, possible human carcinogens can be evaluated in a highly sensitive system without the need to generate a toxic and radioactive atmosphere. Aerosol exposures for a radioactive toxic substance present a number of regulatory and safety concerns, including contamination in the laboratory from escaped vapors. Ex vivo incubations with metabolically active tissue provides a reasonable alternative, and AMS allows us to use a relatively modest amount of 14C and still retain sufficient sensitivity to easily quantify DNA adducts. The ex vivo dosing procedure has been validated in previously published protein adduct studies (DeStefano-Shields et al., 2010), and 250 μM NA is toxicologically equivalent to a 10 ppm aerosol (Morris, 2013).
It has been demonstrated that NA-protein adducts formed in vivo following a 200 mg/kg intraperitoneal NA injection can be detected in respiratory tissue using 14C-NA, even with very low levels of radioactivity (1 pCi/nmol) (Buchholz et al., 2010). In designing this experiment, we opted to use an ex vivo culture system instead of intraperitoneal injection for the following reasons. (1) It was not feasible to perform 14C-NA injections in live, adult primates. (2) Intraperitoneal injection of NA would introduce hepatic first-pass metabolism into the equation and confound the results. Isolating the tissues of interest allowed us to test whether local metabolism and bioactivation was sufficient to cause respiratory DNA adducts to form, and we were able to demonstrate that airway NA-DNA adducts form independent of liver metabolism. (3) Microdissecting the airways from the whole lung allowed us to enrich the portion of the metabolically active injury target cell type (Club cells), in the 14C-NA incubations, increasing our sensitivity to detect toxicologically relevant DNA adducts.
A limitation of this study is that it does not tell us which NA metabolite adducts DNA; but, based on what we know about NA metabolism, it could be 1,2-NQ, 1,4-NQ, or an epoxide/quinone imine metabolite. An additional limitation of this study is that we do not know whether the formed adducts would be efficiently removed/repaired. Future studies should address these issues. Specifically, studies that define which metabolite adducts DNA, either through use of transgenic models that allow manipulation of various biotransformation enzymes or via adduct identification using HPLC in tandem with AMS are needed so that biomarkers of exposure and effect can be developed.
5. Conclusions
This study demonstrates NA-DNA adduct formation at the site of carcinogenesis in chronic bioassays, the lung. Further, NA-DNA adducts were detected in non-human primate airways. NA-DNA adducts are formed in both primate and mouse airway explants following ex vivo treatment with NA at a range of concentrations. Primate tissue incubations with 1,2-NQ produced 70 to 80-fold more DNA adducts than incubation with NA, and greater than 22-fold more stable adducts (quantified as adducts with deoxynucleosides in digested DNA).
Supplementary Material
Acknowledgements
Microdissection of lung tissues was assisted by Patti Edwards, Ryan Mendoza, and Skye Kelty (UC Davis). Alan Buckpitt and Dexter Morin assisted with method development and radioisotope training. Alan Buckpitt also provided valuable feedback during the preparation of this manuscript. AMS work was assisted by Kurt Haack (LLNL).
Funding Information
Funded by R01 ES020867, ES020867S1, P41GM103483, P30 ES023513, DOD LC130820. During the period this research was conducted, Sarah Carratt was supported by Robert Emrie Smith Memorial Research Fellowship and NIEHS T32 Fellowship ES007058.
Disclosures
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors declare that they have no actual or potential competing financial interests. This work was performed in part under the auspices of the US Department of Energy by Lawrence Livermore National Laboratory under contract DE-AC52–07NA27344. Reviewed and released as LLNL-JRNL-739704.
References
- Abdo K, Eustis SL, McDonald M, Jokinen MP, Adkins B, Haseman JK, 1992. Naphthalene: A Respiratory Tract Toxicant and Carcinogen for Mice. Inhalation Toxicology 4, 393–409. [Google Scholar]
- Abdo KM, Grumbein S, Chou BJ, Herbert R, 2001. Toxicity and carcinogenicity study in F344 rats following 2 years of whole-body exposure to naphthalene vapors. Inhalation toxicology 13, 931–950. [DOI] [PubMed] [Google Scholar]
- Bailey LA, Nascarella MA, Kerper LE, Rhomberg LR, 2016. Hypothesis-based weight-of-evidence evaluation and risk assessment for naphthalene carcinogenesis. Critical Reviews in Toxicology 46, 1–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland B, Lin CY, Morin D, Miller L, Plopper C, Buckpitt A, 2004. Site-specific metabolism of naphthalene and 1-nitronaphthalene in dissected airways of rhesus macaques. The Journal of pharmacology and experimental therapeutics 310, 546–554. [DOI] [PubMed] [Google Scholar]
- Buchholz BA, Freeman SP, Haack KW, Vogel JS, 2000. Tips and traps in the C-14 bio-AMS preparation laboratory. Nucl. Instrum. Methods Phys. Res. B 172, 404–408. [Google Scholar]
- Buchholz BA, Haack KW, Sporty JL, Buckpitt AR, Morin D, 2010. Free flow electrophoresis separation and AMS quantitation of C-naphthalene-protein adducts. Nucl Instrum Methods Phys Res B 268, 1324–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckpitt A, Chang A, Weir A, Van Winkle L, Duan X, Philpot R, Plopper C, 1995. Relationship of cytochrome P450 activity to Clara cell cytotoxicity. IV. Metabolism of naphthalene and naphthalene oxide in microdissected airways from mice, rats and hamsters. Mol Pharm 47, 74–81. [PubMed] [Google Scholar]
- Buckpitt A, Morin D, Murphy S, Edwards P, Van Winkle L, 2013. Kinetics of naphthalene metabolism in target and non-target tissues of rodents and in nasal and airway microsomes from the Rhesus monkey. Toxicol Appl Pharmacol 270, 97–105. [DOI] [PubMed] [Google Scholar]
- Carratt SA, Morin D, Buckpitt AR, Edwards PC, Van Winkle LS, 2016. Naphthalene cytotoxicity in microsomal epoxide hydrolase deficient mice. Toxicology letters 246, 35–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho M, Chichester C, Morin D, Plopper C, Buckpitt A, 1994. Covalent interactions of reactive naphthalene metabolites with proteins. J Pharmacol Exp Ther 269, 881–889. [PubMed] [Google Scholar]
- Cho TM, Rose RL, Hodgson E, 2005. IN VITRO METABOLISM OF NAPHTHALENE BY HUMAN LIVER MICROSOMAL CYTOCHROME P450 ENZYMES. Drug Metabolism and Disposition 34, 176. [DOI] [PubMed] [Google Scholar]
- Cichocki JA, Smith GJ, Morris JB, 2014. Tissue sensitivity of the rat upper and lower extrapulmonary airways to the inhaled electrophilic air pollutants diacetyl and acrolein. Toxicological sciences : an official journal of the Society of Toxicology 142, 126–136. [DOI] [PubMed] [Google Scholar]
- Dai YF, Leng SG, Pan ZF, Rappaport SM, Zheng YX, 2004. [Preliminary study on naphthalene-metabolites-albumin adduct as an exposure biomarker for coke oven workers]. Zhonghua Yu Fang Yi Xue Za Zhi 38, 392–395. [PubMed] [Google Scholar]
- DeStefano-Shields C, Morin D, Buckpitt A, 2010. Formation of covalently bound protein adducts from the cytotoxicant naphthalene in nasal epithelium: species comparisons. Environmental health perspectives 118, 647–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd DE, Gross EA, Miller RA, Wong BA, 2010. Nasal olfactory epithelial lesions in F344 and SD rats following 1- and 5-day inhalation exposure to naphthalene vapor. Int J Toxicol 29, 175–184. [DOI] [PubMed] [Google Scholar]
- Duan X, Plopper C, Brennan P, Buckpitt A, 1996. Rates of glutathione synthesis in lung subcompartments of mice and monkeys: possible role in species and site selective injury. The Journal of pharmacology and experimental therapeutics 277, 1402–1409. [PubMed] [Google Scholar]
- Kwok ESC, Buchholz BA, Vogel JS, Turteltaub KW, Eastmond DA, 1999. Dose-Dependent Binding of ortho-Phenylphenol to Protein but Not DNA in the Urinary Bladder of Male F344 Rats. Toxicology and applied pharmacology 159, 18–24. [DOI] [PubMed] [Google Scholar]
- Lee MG, Phimister A, Morin D, Buckpitt A, Plopper C, 2005. In situ naphthalene bioactivation and nasal airflow cause region-specific injury patterns in the nasal mucosa of rats exposed to naphthalene by inhalation. J Pharmacol Exp Ther 314, 103–110. [DOI] [PubMed] [Google Scholar]
- Lin CY, Boland BC, Lee YJ, Salemi MR, Morin D, Miller LA, Plopper CG, Buckpitt AR, 2006. Identification of proteins adducted by reactive metabolites of naphthalene and 1-nitronaphthalene in dissected airways of rhesus macaques. Proteomics 6, 972–982. [DOI] [PubMed] [Google Scholar]
- Lin P-H, Chen D-R, Wang T-W, Lin C-H, Chuang M-C, 2009. Investigation of the cumulative tissue doses of naphthoquinones in human serum using protein adducts as biomarker of exposure. Chemico-biological interactions 181, 107–114. [DOI] [PubMed] [Google Scholar]
- Long PH, Herbert RA, Peckham JC, Grumbein SL, Shackelford CC, Abdo K, 2003. Morphology of nasal lesions in F344/N rats following chronic inhalation exposure to naphthalene vapors. Toxicologic pathology 31, 655–664. [DOI] [PubMed] [Google Scholar]
- Morris JB, 2013. Nasal dosimetry of inspired naphthalene vapor in the male and female B6C3F1 mouse. Toxicology 309, 66–72. [DOI] [PubMed] [Google Scholar]
- National Toxicology Program, 1992. Toxicology and Carcinogenesis Studies of Naphthalene (CAS No. 91-20-3) in B6C3F1 Mice (Inhalation Studies). National Toxicology Program technical report series 410, 1–172. [PubMed] [Google Scholar]
- National Toxicology Program, 2000. Toxicology and carcinogenesis studies of naphthalene (cas no. 91-20-3) in F344/N rats (inhalation studies). National Toxicology Program technical report series, 1–173. [PubMed] [Google Scholar]
- Ognibene TJ, Bench G, Vogel JS, Peaslee GF, Murov S, 2003. A high-throughput method for the conversion of CO2 obtained from biochemical samples to graphite in septa-sealed vials for quantification of 14C via accelerator mass spectrometry. Analytical chemistry 75, 2192–2196. [DOI] [PubMed] [Google Scholar]
- Oliver JR, Kushwah R, Wu J, Cutz E, Yeger H, Waddell TK, Hu J, 2009. Gender differences in pulmonary regenerative response to naphthalene-induced bronchiolar epithelial cell injury. Cell Prolif 42, 672–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham NT, Jewell WT, Morin D, Buckpitt AR, 2012a. Analysis of naphthalene adduct binding sites in model proteins by tandem mass spectrometry. Chemico-biological interactions 199, 120–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham NT, Jewell WT, Morin D, Jones AD, Buckpitt AR, 2012b. Characterization of model peptide adducts with reactive metabolites of naphthalene by mass spectrometry. PLoS One 7, e42053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccirillo VJ, Bird MG, Lewis RJ, Bover WJ, 2012. Preliminary evaluation of the human relevance of respiratory tumors observed in rodents exposed to naphthalene. Regul Toxicol Pharmacol 62, 433–440. [DOI] [PubMed] [Google Scholar]
- Plopper CG, Chang AM, Pang A, Buckpitt AR, 1991. Use of microdissected airways to define metabolism and cytotoxicity in murine bronchiolar epithelium. Experimental lung research 17, 197–212. [DOI] [PubMed] [Google Scholar]
- Plopper CG, Suverkropp C, Morin D, Nishio S, Buckpitt A, 1992. Relationship of cytochrome P-450 activity to Clara cell cytotoxicity. I. Histopathologic comparison of the respiratory tract of mice, rats and hamsters after parenteral administration of naphthalene. J Pharmacol Exp Ther 261, 353–363. [PubMed] [Google Scholar]
- Saeed M, Higginbotham S, Gaikwad N, Chakravarti D, Rogan E, Cavalieri E, 2009. Depurinating naphthalene-DNA adducts in mouse skin related to cancer initiation. Free radical biology & medicine 47, 1075–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Winkle LS, Buckpitt AR, Nishio SJ, Isaac JM, Plopper CG, 1995. Cellular response in naphthalene-induced Clara cell injury and bronchiolar epithelial repair in mice. The American journal of physiology 269, L800–818. [DOI] [PubMed] [Google Scholar]
- Van Winkle LS, Buckpitt AR, Plopper CG, 1996. Maintenance of differentiated murine Clara cells in microdissected airway cultures. American journal of respiratory cell and molecular biology 14, 586–598. [DOI] [PubMed] [Google Scholar]
- Van Winkle LS, Gunderson AD, Shimizu JA, Baker GL, Brown CD, 2002. Gender differences in naphthalene metabolism and naphthalene-induced acute lung injury. American journal of physiology Lung cellular and molecular physiology 282, L1122–1134. [DOI] [PubMed] [Google Scholar]
- Vogel JS, Love AH, 2005. Quantitating isotopic molecular labels with accelerator mass spectrometry. Methods in enzymology 402, 402–422. [DOI] [PubMed] [Google Scholar]
- Waidyanatha S, Troester MA, Lindstrom AB, Rappaport SM, 2002. Measurement of hemoglobin and albumin adducts of naphthalene-1,2-oxide, 1,2-naphthoquinone and 1,4-naphthoquinone after administration of naphthalene to F344 rats. Chemico-biological interactions 141, 189–210. [DOI] [PubMed] [Google Scholar]
- West JA, Pakehham G, Morin D, Fleschner CA, Buckpitt AR, Plopper CG, 2001. Inhaled naphthalene causes dose dependent Clara cell cytotoxicity in mice but not in rats. Toxicology and applied pharmacology 173, 114–119. [DOI] [PubMed] [Google Scholar]
- West JA, Van Winkle LS, Morin D, Fleschner CA, Forman HJ, Plopper CG, 2003. Repeated Inhalation Exposures of the Bioactivated Cytotoxicant Naphthalene (NA) Produce Airway Specific Clara Cell Tolerance in Mice. Toxicological sciences : an official journal of the Society of Toxicology 75, 161–168. [DOI] [PubMed] [Google Scholar]
- Zimmermann M, Wang SS, Zhang H, Lin TY, Malfatti M, Haack K, Ognibene T, Yang H, Airhart S, Turteltaub KW, Cimino GD, Tepper CG, Drakaki A, Chamie K, de Vere White R, Pan CX, Henderson PT, 2017. Microdose-Induced Drug-DNA Adducts as Biomarkers of Chemotherapy Resistance in Humans and Mice. Mol Cancer Ther 16, 376–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.