Abstract
The S. cerevisiae Pop2 protein is an exonuclease in the Ccr4-Not complex that is a conserved regulator of gene expression. Pop2 regulates gene expression post-transcriptionally by shortening the poly(A) tail of mRNA. A previous study has shown that Pop2 is phosphorylated at threonine 97 (T97) by Yak1 protein kinase in response to glucose limitation. However, the physiological importance of Pop2 phosphorylation remains unknown. In this study, we found that Pop2 is phosphorylated at serine 39 (S39) under unstressed conditions. The dephosphorylation of S39 was occurred rapidly after glucose depletion, and the addition of glucose to the glucose-deprived culture recovered this phosphorylation, suggesting that Pop2 phosphorylation at S39 is regulated by glucose. This glucose-regulated phosphorylation of Pop2 at S39 is dependent on Pho85 kinase. We previously reported that Pop2 takes a part in the cell wall integrity pathway by regulating LRG1 mRNA; however, S39 phosphorylation of Pop2 is not involved in LRG1 expression. On the other hand, Pop2 phosphorylation at S39 is involved in the expression of HSP12 and HSP26, which encode a small heat shock protein. In the medium supplemented with glucose, Pop2 might be phosphorylated at S39 by Pho85 kinase, and this phosphorylation contributes to repress the expression of HSP12 and HSP26. Glucose starvation inactivated Pho85, which resulted in the derepression of HSP12 and HSP26, together with other glucose sensing mechanisms. Our results suggest that Pho85-dependent phosphorylation of Pop2 is a part of the glucose sensing system in yeast.
Introduction
Post-transcriptional regulation, including the control of mRNA degradation and translation, plays important roles in regulation of gene expression. mRNA degradation is generally initiated by poly(A) tail shortening referred to as deadenylation, which is catalyzed by the Ccr4-Not and the Pan2-Pan3 complexes [1]. In Saccharomyces cerevisiae, the Ccr4-Not complex consists of 9 core subunits, including Ccr4, Pop2/Caf1, Caf40, Caf130 and Not1-5, all of which have homologs in most other eukaryotes [2]. Ccr4 and Pop2 are two active 3’ to 5’ exonucleases: Ccr4 is the major one; Pop2 may modulate specific mRNA expression in certain conditions [1]. Pop2 is reportedly involved in expression control of glucose-repressed genes and regulation of the cell wall integrity (CWI) pathway in S. cerevisiae [3, 4]. The loss of POP2 results in pleiotropic phenotypes including temperature-sensitive growth, abnormal cell morphology, weak cell lysis and defective glucose repression [3, 5]. Pop2 is phosphorylated by Yak1 kinase at threonine 97 (T97) in response to a glucose-regulating signal [6]. This modification is required for cell growth control and cell cycle arrest at G1 phase after glucose starvation [6]. We have previously shown that Pop2 functions in the CWI pathway by regulating the expression of LRG1, encoding a GTPase-activating protein for Rho1 small GTPase [4]. The LRG1 mRNA level is elevated in pop2Δ and the high temperature-sensitivity phenotype caused by pop2Δ mutation is suppressed by deletion of LRG1 [4].
In the budding yeast, Pho85 is a multifunctional cyclin-dependent protein kinase (CDK), which was originally identified as a negative regulator of PHO5 expression through the control of a transcription factor Pho4 [7, 8]. Pho85 is regulated by the association with ten cyclins, Pho80 and Pcls, and involved in multiple cellular processes beyond the important roles in cell cycle control: for example, metabolism of nutrients such as phosphate and carbon sources [9, 10]. The pho85Δ mutant shows pleiotropic phenotypes, including the slow growth with a G1 delay on rich media, a severe growth defect on non-fermentable carbon sources, abnormal morphology, irregular budding and hyperaccumulation of glycogen [9, 11–13]. Additionally, PHO85 deletion causes the elevated expression not only of phosphate-starvation-dependent genes but also of the stress responsive genes such as HSP12 and UBI4 [14, 15]. Thus, Pho85 is activated in satisfactory environmental conditions in order to ensure the turn-off of stress responses and inappropriate activities [16]. However, few Pho85 targets have been unambiguously identified; therefore, a long-time goal is understanding molecular functions and substrates that provide interpretations for each phenotype caused by the pho85Δ mutation.
In this study, to investigate how Pop2 is regulated upon glucose availability, we examined the phosphorylation of Pop2 and its function. Our results presented here suggest that Pop2 is phosphorylated at S39 in a Pho85-dependent manner upon glucose availability. Furthermore, this post-transcriptional modification of Pop2 specifically contributes to the glucose repression of the stress response genes, HSP12 and HSP26.
Materials and methods
Strains, plasmids and general methods
Escherichia coli DH5α was used for DNA manipulations. The present yeast strains and plasmids are described in S1 and S2 Tables. Cells were grown in yeast extract-peptone dextrose (YPD), yeast extract-peptone glycerol lactate (YPGL), synthetic complete medium (SC), and SC media lacking either amino acids or other nutrients (SC–Ura, SC lacking uracil). General procedures were performed as described previously [17].
Gene deletion and protein tagging
Gene disruption and insertion were performed using PCR-based gene replacement, as described previously [18, 19].
Western Blot analysis
Cells grown to exponential phase were subjected to a mild alkali treatment-based protein extraction method [20]. Samples were loaded onto SDS-PAGE gel and then electroblotted onto Immobilon polyvinylidene difluoridemembranes (MerckMillipore, USA). Phos-tag (Wako, Japan) was added to a mix of SDS-PAGE gel when required. Blots were blocked for 1 h at room temperature with TBS-M buffer containing 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, and 5% non-fat dry milk, and then incubated with 1:1,000-diluted primary antibodies in TBS-M buffer overnight at 4°C. After three final washes with TBS buffer containing 20 mM Tris-HCl (pH 7.5) and 150 mM NaCl, blots were incubated with secondary antibodies, and were developed using enhanced chemiluminescence detection kits (Merck Millipore, USA). Signal intensities were quantified by Image Studio software (LI-COR).
Immunoprecipitation of Pop2Flag
Cells were grown in YPD medium at 30°C to exponential phase and harvested by centrifugation. The cells were then re-suspended in XT buffer containing 50 mM HEPES-KOH (pH 7.3), 20 mM potassium acetate, 2 mM EDTA, 0.1% Triton X-100, 5% glycerol protease inhibitors, phenylmethylsulfonyl fluoride (PMSF), aprotinin, and leupeptin. Glass beads were then added and cells were broken by rigorous vortexing at 4°C (4 times at 3,500 rpm for 30 s). Lysates were then centrifuged for 10 min at 15,000 g and supernatants were collected.
To immunoprecipitate Pop2Flag, extracts were incubated with anti-Flag antibody coupled to protein G-Sepharose beads (Sigma Aldrich, USA) for 30 min at 4°C. Pop2Flag-bound beads were then washed three times in XT buffer, and the bound material was eluted with elution buffer containing 0.1 mg/mL Flag peptide in XT buffer for 10 min at 4°C. Eluted Pop2Flag samples were split and one-half treated with λ-phosphatase (NEB #P0753S, USA) for 2 h at 30°C. Samples were then subjected to SDS-PAGE with or without phos-tag followed by Immunoblotting with anti-Flag antibody.
RNA isolation, RT-qPCR, and microarray analysis
Total RNA was isolated using ISOGEN reagent (Nippon Gene, Japan) and the RNeasy Mini kit (Qiagen, Germany). First strands of cDNA were generated using the PrimeScript RT reagent Kit (Takara, Japan). The cDNAs of LRG1, HSP12, HSP26, and PIR3 were quantitated by a quantitative real-time PCR (qRT-PCR) method using a 7500 fast real-time RT-PCR system (Applied Biosystems) with SYBR Premix Ex Taq (Takara, Japan). A standard curve was generated from diluted cDNA derived from wild-type cells, and levels of gene expression were normalized to ACT1 expression. The microarray analysis was performed by the KURABO Bio-Medical Department (Japan) using the Affymetrix GeneChip Yeast Genome 2.0 Array (Affymetrix, Santa Clara, Califronia, USA). Microarray data sets are available at the Gene Expression Omnibus at http://www.ncbi.nlm.nih.gov/geo (GEO accession number GSE124908).
Statistical analysis
Data are presented as means ± standard deviations (SD) of 3 independent experiments. Statistical analyses were performed using analysis of variance (ANOVA) followed by Tukey’s test or student’s t-test test. Differences were considered significant when p < 0.01 or p < 0.05.
Results
Pop2 is a phosphorylated protein
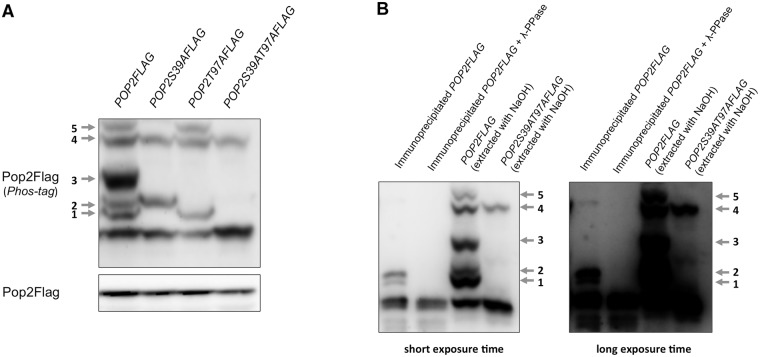
Protein phosphorylation plays an important role in regulating protein function in response to various stimuli. There are few studies investigating post-translational modifications of the Ccr4-Not subunits and the relevance of such modification for their functions. Therefore, we examined the phosphorylation of Pop2, one of the exonucleases in the Ccr4-Not complex, using phosphate affinity (phos-tag) SDS-PAGE. This technique allows for the detection of phosphorylation by the mobility shift [21]. We observed clear electrophoretic shift bands of Pop2Flag under unstressed conditions (Fig 1A). These bands were numbered from 1 to 5 according to their positions. To confirm that the band shifts are caused by Pop2 phosphorylation, immunoprecipitated Pop2Flag were treated with λ-phosphatases. We found that Pop2 mobility shift is abolished by λ-phosphatase treatment (Fig 1B), suggesting that Pop2 is a phosphoprotein. The difference of band patterns between the immunoprecipitated Pop2 and Pop2 in the NaOH-treated crude extract is due to the difference in cell extraction.
Fig 1. Pop2 is phosphorylated at S39 and T97.
(A) pop2Δ mutant cells harboring YCplac33-POP2FLAG (POP2FLAG), YCplac33-POP2S39AFLAG (POP2S39AFLAG) were grown at 30°C until exponential phase. Extracts prepared from each strain were run on phos-tag and conventional gels, then immunoblotted with anti-Flag antibody. Phosphorylated Pop2Flag is indicated with arrows and numbers according to the positions. Band 3 appears as dominant in the pop2Δ mutant harboring the POP2-FLAG plasmid, but less dominant in the WT strain harboring the POP2-FLAG plasmid (see Fig 2B). (B) Wild-type cells harboring YCplac33-POP2FLAG (POP2FLAG) were grown at 30°C until exponential phase. Extracts prepared from this strain were immunoprecipitated using anti-Flag antibody. Immunoprecipitated Pop2Flag was treated with/withoutλphosphatase. The band patterns of immunoprecipitated Pop2 are compared with the band patterns of Pop2 and Pop2S39AT97A extracted with NaOH as analyzed in (A).
Previous global phosphorylation analyses revealed the Serine 39 residue (S39) of Pop2 as a potential phosphorylation site [22–24]. It also has been proposed that Pop2 is phosphorylated at Threonine 97 (T97) [6]. Therefore, we investigated whether Pop2 is phosphorylated at S39 and T97 in a pop2Δ background. We constructed a strain expressing Pop2 with phospho-defective mutations at either S39 (Pop2S39A), T97 (Pop2T97A), or both (Pop2S39AT97A), and then examined the mobility of Pop2. Interestingly, the shifted-bands 1, 3, and 5 of Pop2 were lost by S39A mutation, while shifted-bands 2 and 3 were lost by T97A mutation (Fig 1A). Consistently, S39AT97A caused the disappearance of shifted-band 1, 2, 3, and 5 (Fig 1A). Our data suggest that Pop2 is phosphorylated at both S39 and T97. Furthermore, the shifted-band 3 was vanished by either S39A or T97A mutation, providing strong evidence that this band is a result of S39 and T97 phosphorylation combination.
Pop2 phosphorylation is regulated by glucose availability
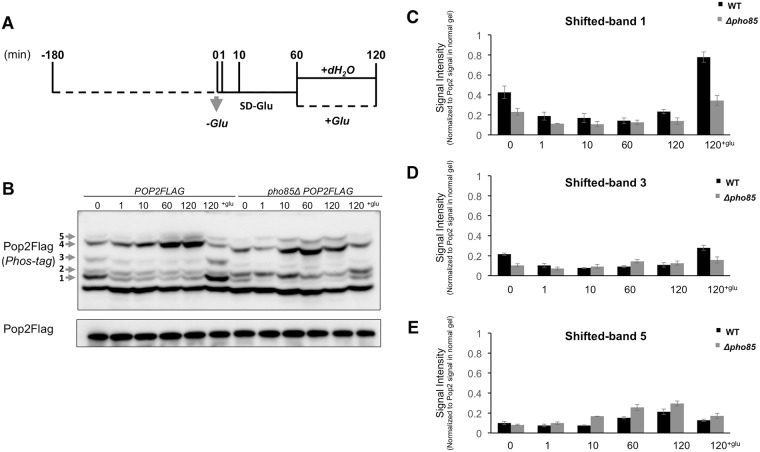
Recent data suggested that the Ccr4-Not complex is involved in glucose-regulating pathways [25]. Therefore, we examined Pop2 phosphorylation in cells cultured transiently in media with and without glucose (Fig 2A). Phosphorylated forms of Pop2 showing in shifted-band 1 and 3 were diminished within 1 min after glucose starvation. Adding glucose to the glucose-deprived culture recovered these bands (Fig 2B, 2C and 2D). On the other hand, the shifted-band 5 was increased upon glucose limitation and decreased after glucose re-addition (Fig 2E). Remarkably, S39A mutation caused the disappearance of shifted-band 1,3, and 5 in the presence of glucose (Fig 1), indicating that phosphorylation of Pop2 at S39 is regulated by glucose availability.
Fig 2. The S39 phosphorylation of Pop2 is regulated by glucose.
(A) Schematic illustration of cell collection. Cells were grown at 30°C until exponential phase, washed once with medium lacking glucose and re-suspended in medium lacking glucose. Glucose was then added to the culture to a final concentration of 2%. Cells were collected at indicated times. (B) Wild-type cells and pho85Δ mutant cells harboring YCplac33-POP2FLAG were collected as in (A). Extracts prepared from each strain were run on phos-tag and conventional gels, then immunoblotted with anti-Flag antibody. Phosphorylated Pop2Flag is indicated with arrows and numbers according to the positions. Representative data are shown. (C, D, E) The intensities of Pop2Flag shifted-band 1, 3, and 5 signals in phos-tag gel were measured and normalized to the Pop2Flag signals in normal gel.
The S39 phosphorylation regulates Pop2 function to repress the expression of HSP12 and HSP26
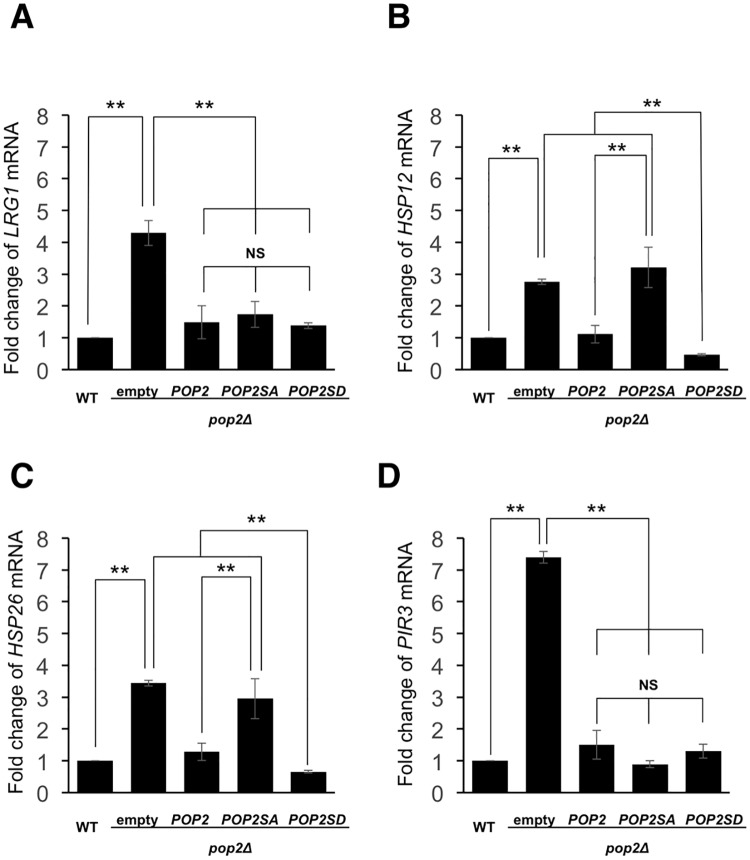
We previously reported that LRG1 mRNA level is upregulated by pop2 deletion [4]. Therefore, we examined the effect of S39 phosphorylation on LRG1 expression. The pop2 deletion caused increased LRG1 expression, which was efficiently suppressed by both wild-type Pop2 and Pop2S39A (Fig 3A). Furthermore, Pop2 with a phospho-mimetic mutation at S39 (S39D) also efficiently suppressed the increased LRG1 expression caused by pop2Δ mutation (Fig 3A). This result suggests that S39 phosphorylation is unrelated to the regulation of LRG1 mRNA levels.
Fig 3. Effect of the S39 phosphorylation on the LRG1, HSP12, HSP26 and PIR3 expression in glucose-containing medium.
(A, B, C, D) Expression of LRG1, HSP12, HSP26, PIR3 mRNAs in WT harboring YCplac33 (empty vector) and pop2Δ harboring YCplac33, YCplac33-POP2FLAG, YCplac33-POP2S39AFLAG, YCplac33-POP2S39DFLAG. The cells were grown at 30°C until exponential phase and collected for RNA isolation. mRNA levels were quantified by qRT-PCR. The relative mRNA levels were calculated using delta delta Ct method normalized to ACT1 reference gene. The data show mean ± the standard deviation (n = 3). NS, not significant, **P < 0.01 as determined by Tukey’s test.
To identify other Pop2 targets, we measured the gene expression levels in wild-type and pop2Δ mutant cells by carrying out a microarray analysis. Our microarray data showed that pop2 deletion upregulated expression of many genes involved in stress responses, such as HSP12, HSP26, and PIR3 (S3 Table). To confirm the microarray data, we examined the mRNA levels of HSP12, HSP26, and PIR3 in wild-type and pop2Δ mutant cells by real time RT-PCR. These mRNA levels were increased by pop2Δ mutation, indicating that Pop2 functions in glucose repression of HSP12, HSP26 and PIR3 (Fig 3B, 3C and 3D). We next asked whether S39 phosphorylation affects the ability of Pop2 to repress the expression of HSP12, HSP26, and PIR3. We found that wild-type POP2 efficiently complemented the increased expression of HSP12 and HSP26 caused by pop2Δ mutation, but POP2-S39A did not (Fig 3B and 3C). Additionally, POP2-S39D also efficiently complemented the increased expression of HSP12 and HSP26 (Fig 3B and 3C). In contrast, introduction of Pop2S39A and Pop2S39D as well as wild-type Pop2 fully recovered the increased expression of PIR3 observed in pop2Δ mutant cells (Fig 3D). Thus, S39 phosphorylation is involved in the regulation of HSP12 and HSP26, but not in that of PIR3.
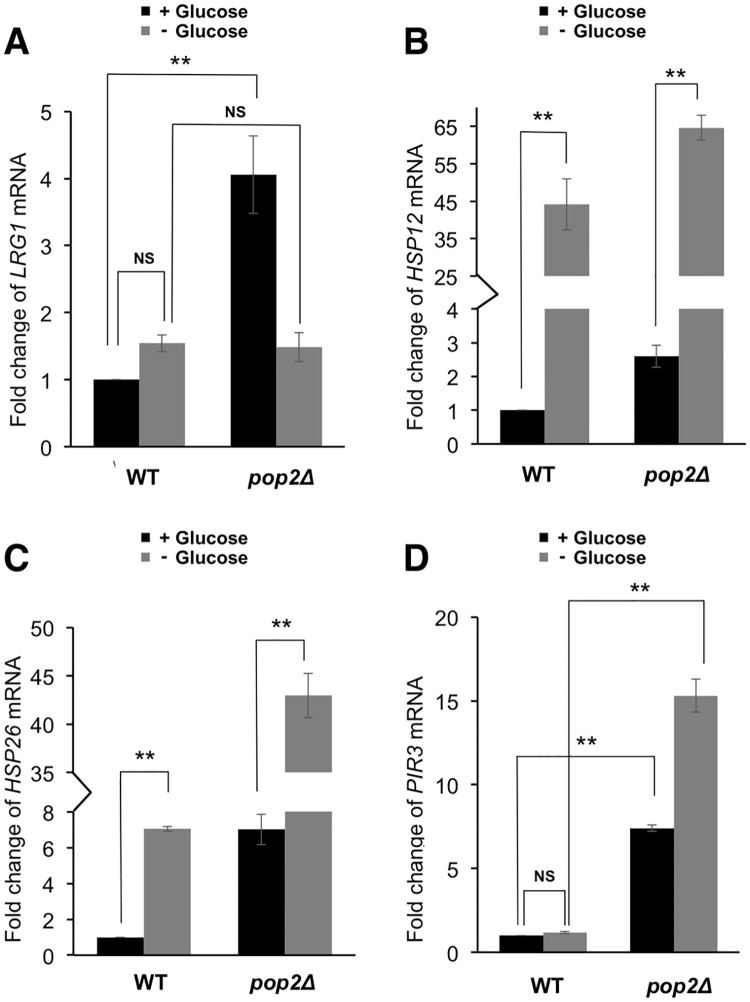
Since the phosphorylation of Pop2 at S39 is regulated by glucose availability as described above (Fig 2), we next examined whether expression of HSP12 and HSP26 are affected by glucose availability. The levels of HSP12 and HSP26 mRNAs were upregulated by glucose starvation (Fig 4B and 4C). On the other hand, the expression of LRG1 and PIR3 was similar in the presence and absence of glucose (Fig 4A and 4D). These data are consistent with the idea that the phosphorylation of Pop2 at S39 in glucose medium is involved in the glucose repression of HSP12 and HSP26. Since the mRNA levels of HSP12 and HSP26 in pop2Δ mutant cells were further upregulated by glucose starvation (Fig 4B and 4C), the POP2-independent mechanisms should be involved in the glucose repression of HSP12 and HSP26. The mRNA levels of HSP12 and HSP26 in POP2-S39A and POP2-S39D cells were also upregulated by glucose starvation (data not shown). The POP2-independent mechanisms include Hsf1 and Msn2/4 transcription factors [26–28], which are involved in multiple glucose sensing pathways in yeast [29]. Although the effect of the POP2-S39A mutation is much smaller than the effect of glucose depletion (3-fold in Fig 3B vs 45-fold in Fig 4B), Pop2 phosphorylation still contributes to glucose repression of HSP12 and HSP26 in some extent. Our results also showed that the impacts of Pop2 regulation on gene expression are different between LRG1 and PIR3. LRG1 mRNA might be degraded in a POP2-dependent manner in glucose medium but not in glucose starvation, while expression of PIR3 is dependent on POP2 in both conditions (Fig 4A and 4D).
Fig 4. The expression of LRG1, HSP12, HSP26 and PIR3 in wild-type and pop2Δ in glucose and glucose-deprived conditions.
(A, B, C, D) Expression of LRG1, HSP12, HSP26, PIR3 mRNAs in wild-type cells and pop2Δ mutant cells. mRNA levels were quantified by qRT-PCR. Wild-type, pop2Δ cells were grown at 30°C until exponential phase, washed once with medium lacking glucose and re-suspended in medium lacking glucose for 1 h. Cells were collected before and after glucose starvation for RNA isolation. The relative mRNA levels were calculated using delta delta Ct method normalized to ACT1 reference gene. The data show mean ± the standard deviation (n = 3). NS, not significant, **P < 0.01 as determined by Tukey’s test.
Pop2 might be phosphorylated by Pho85
The amino acid sequence surrounding S39 of Pop2 is matched to the phosphorylation consensus sequence of CDK substrates, [(K)-S/T-P-X-(K/R)], in which S/T is a phosphorylation site [30]. In the PhosphoPep database (http://www.phosphopep.org/), Pop2 phosphorylation at S39 is shown to be dependent on Pho85 [31]. Therefore, we examined Pop2 phosphorylation in the pho85Δ mutant. The signals of the shifted-bands numbered 1 and 3, corresponding to S39 phosphorylation (Fig 1), were substantially reduced and almost disappeared in pho85Δ (Fig 2B). These data support the idea that Pop2 is phosphorylated at S39 by Pho85. PHO85 is a homolog of CDC28 [32]. Therefore, we also examined Pop2 phosphorylation in temperature-sensitive cdc28 mutants, cdc28-4 and cdc28-13. However, Pop2 phosphorylation was not altered in these mutants even at non-permissive temperature (data not shown), suggesting that Cdc28 is not involved in S39 phosphorylation.
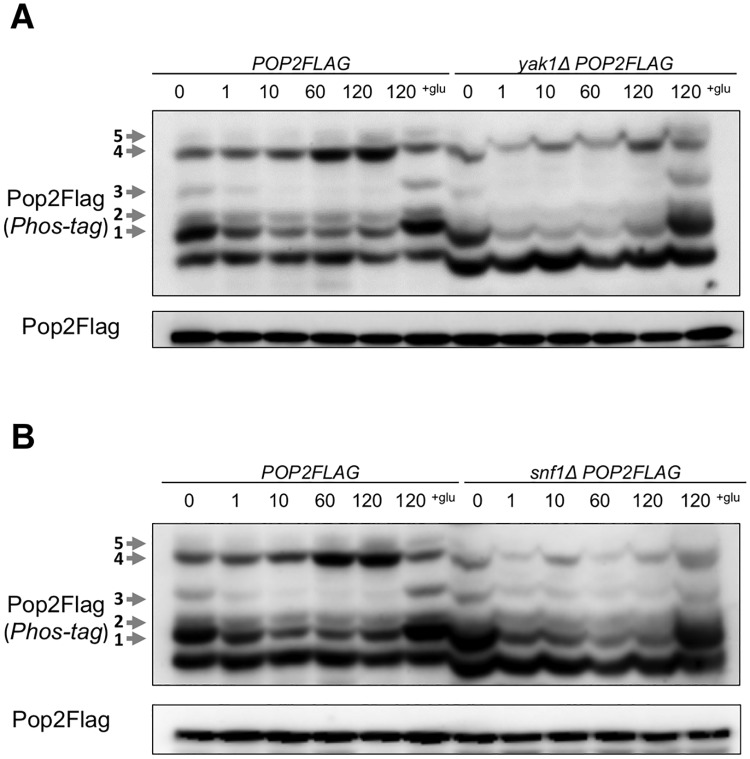
We also found that glucose deprivation-mediated changes of shifted-band 1 and shifted-band 3 were hardly observed in pho85Δ (Fig 2B), but still observed in yak1Δ and snf1Δ cells (Fig 5A and 5B). Thus, Yak1 and Snf1 may not be involved in generation of these two phosphorylation forms of Pop2. Taken together, our results suggest that phosphorylation of Pop2 at S39 is dependent on Pho85 kinase.
Fig 5. The phosphorylation of Pop2 in yak1Δ and snf1Δ.
(A) WT and snf1Δ cells harboring YCplac33-POP2FLAG (POP2FLAG, snf1ΔPOP2FLAG) were collected as in Fig 2A. Extracts prepared from each strain were run on phos-tag and conventional gels, then immunoblotted with anti-Flag antibody. Phosphorylated Pop2Flag is indicated with arrows and numbers according to the positions. Representative data are shown. (B) WT and yak1Δ cells harboring YCplac33-POP2FLAG (POP2FLAG, yak1ΔPOP2FLAG) were collected as in Fig 2A. Samples were analyzed as in (A). Representative data are shown.
Expression of HSP12 and HSP26 is upregulated by pho85 deletion in glucose condition
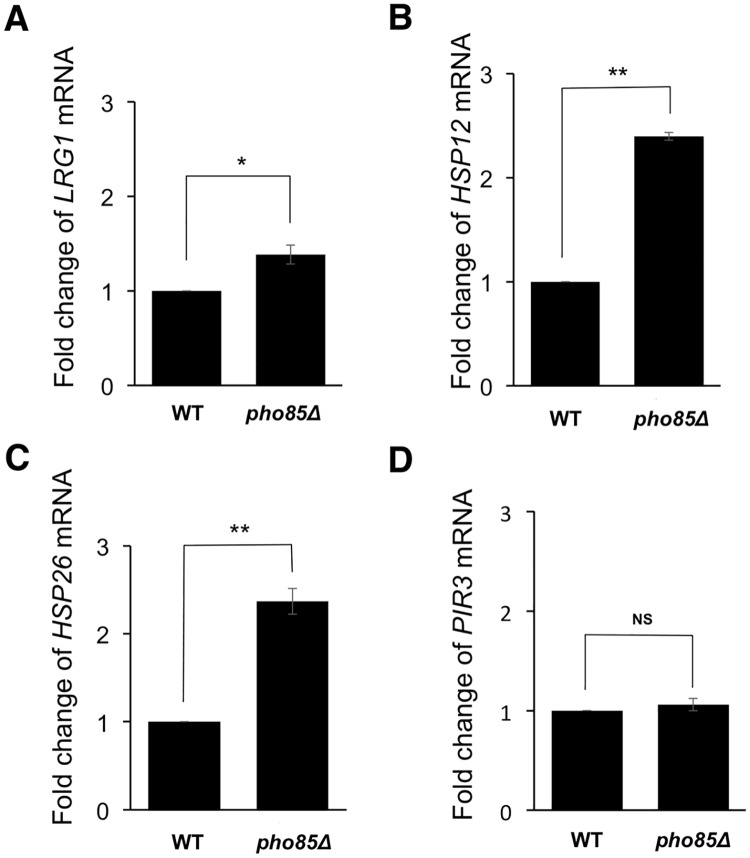
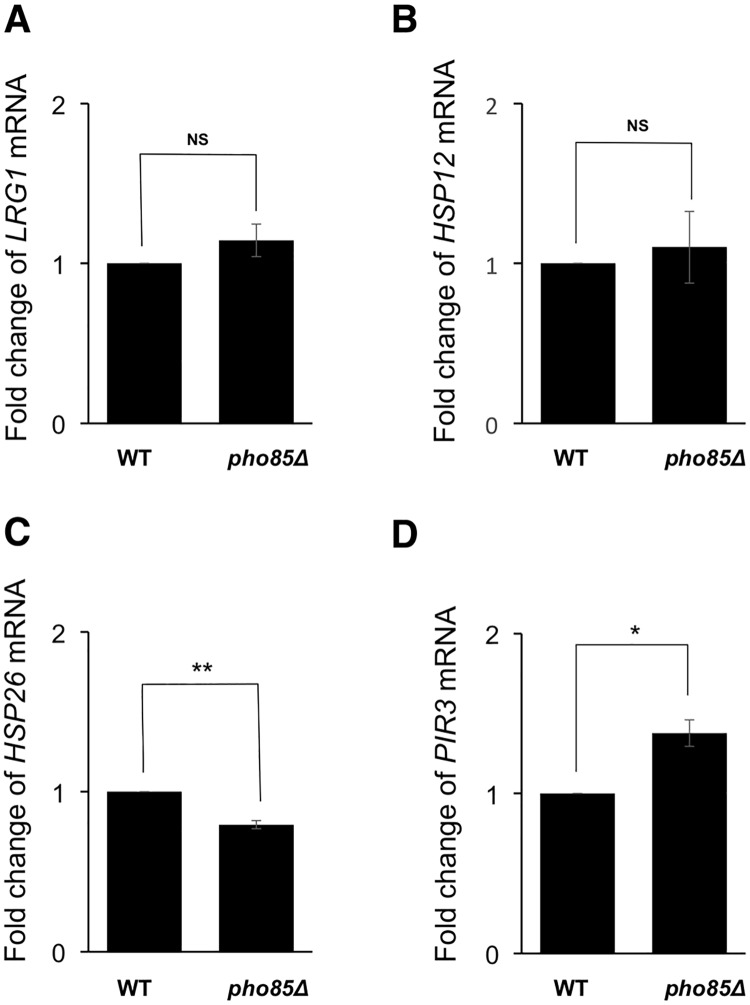
Pho85 kinase functions in the suppression of stress response genes under glucose-rich conditions [15]. Therefore, we examined the effect of Pho85 on LRG1, HSP12, HSP26, and PIR3 expression in the medium supplemented with glucose. As shown in Fig 6B and 6C, HSP12 and HSP26 expression in pho85Δ cells were more than 2-fold higher than wild-type cells. On the other hand, pho85 deletion slightly increased and did not change the LRG1 mRNA and PIR3 mRNA levels, respectively (Fig 6A and 6D). Moreover, pho85 deletion did not significantly increase HSP12 and HSP26 expression when the cells were moved to glucose starvation conditions (Fig 7). Our data suggest that Pho85 partly contributes to repress HSP12 and HSP26 expression, probably through the phosphorylation of Pop2 at S39 in glucose medium.
Fig 6. Expression of HSP12 and HSP26 is elevated by PHO85 deletion in glucose medium.
(A, B, C, D) Expression of LRG1, HSP12, HSP26, PIR3 mRNAs in wild-type cells and pho85Δ mutant cells. mRNA levels were quantified by qRT-PCR. WT, pho85Δ cells were grown at 30°C until exponential phase. The cells were collected for RNA isolation. The relative mRNA levels were calculated using delta delta Ct method normalized to ACT1 reference gene. The data show mean ± the standard deviation (n = 3). NS, not significant, *P < 0.05, **P < 0.01 as determined by student’s t-test.
Fig 7. Expression of LRG1, HSP12, HSP26 and PIR3 is not significantly affected by PHO85 deletion in glucose starvation.
(A, B, C, D) Expression of LRG1, HSP12, HSP26, PIR3 mRNAs in wild-type cells and pho85Δ mutant cells. mRNA levels were quantified by qRT-PCR. WT, pho85Δ cells were grown at 30°C until exponential phase, washed once with medium lacking glucose and re-suspended in medium lacking glucose for 1 h. The cells were collected after glucose starvation for RNA isolation. The relative mRNA levels were calculated using delta delta Ct method normalized to ACT1 reference gene. The data show mean ± the standard deviation (n = 3). NS, not significant, *P < 0.05, **P < 0.01 as determined by student’s t-test.
Discussion
Multiple migration forms of Pop2
In this study, we showed that Pop2 is phosphorylated at S39 in a Pho85 kinase-dependent manner in the normal growth condition. Our results suggest that three phosphorylated forms of Pop2, shifted-band 1, 3, and 5, are dependent on S39 (Fig 1), while only two phosphorylated forms, shifted-band 1 and 3, are dependent on PHO85 (Fig 2B, 2C and 2D). A previous study showed that Pop2 is phosphorylated at T97 by Yak1 in response to glucose limitation [6]. Consistently, the shifted-band 2 seen in our phos-tag SDS-PAGE was dependent on T97 and YAK1: the shifted-band 2 was disappeared by T97A mutation (Fig 1A) and in yak1Δ mutant (Fig 5A). Either S39A or T97A mutation of POP2 leads to the loss of shifted-band 3, suggesting that this band is derived from Pop2 phosphorylated at both S39 and T97. We also found that the shifted-band 4 and 5 were significantly increased upon glucose depletion in wild-type cells (Fig 2B), but not in snf1Δ and yak1Δ mutants (Fig 5A and 5B). Thus, these two bands are dependent on Snf1 and Yak1. Furthermore, S39A also causes the loss of shifted-band 5 (Fig 1A); therefore, the shifted-band 5 likely results from Pop2 phosphorylated not only at S39 but also at other unknown sites. Thus, Pop2 is phosphorylated and regulated by multiple kinases.
Glucose-dependent phosphorylation of Pop2-S39
When exponentially growing cells were transferred to the glucose-deprived medium, the shifted-band 1 and 3 of Pop2, which correspond with S39 phosphorylation, rapidly disappeared (Fig 2B, 2C and 2D). After glucose addition to the glucose-starved culture, those phosphorylated forms were recovered. The signals of shifted-band 1 and 3 were substantially decreased but not completely disappeared in the pho85Δ mutant. Additionally, the shifted-band 5, which is also dependent on S39 phosphorylation (Fig 1A), was increased upon glucose starvation in wild-type as well as pho85Δ (Fig 2B and 2E). This suggests that the phosphorylation of Pop2 at S39 is dependent not only on Pho85 but also on other kinases, which might be glucose-activated Snf1 and Yak1 (Fig 5A and 5B). S39 phosphorylation dependent on Pho85 is decreased upon glucose depletion and reappeared upon glucose addition (Fig 2B, 2C and 2D). Furthermore, pho85Δ mutation has almost no effect on the expression of HSP12 and HSP26 in glucose starvation (Fig 7), suggesting that Pho85 kinase activity is regulated by glucose availability. Previous studies showed that Pho85 has multiple cyclin partners, which determine the substrate specificity [16]. Investigating whether Pho85 and its cyclin partners directly phosphorylate Pop2 will be the focus of future studies.
S39-phosphorylated Pop2 contributes to glucose repression of HSP12 and HSP26
Our previous studies showed that Pop2, together with Ccr4, Dhh1, and Puf5, is involved in regulation of LRG1 expression [4, 33, 34]. This regulation is mediated by mRNA stability control. S39 phosphorylation of Pop2 is not involved in regulation of LRG1 expression, since POP2-S39A efficiently suppressed the increased level of LRG1 mRNA caused by pop2Δ mutation (Fig 3A). We searched other targets for Pop2 and then examined whether Pop2 phosphorylation at S39 is involved in their expression. We found that Pop2 phosphorylation at S39 specifically regulates Pop2 function to repress the expression of certain stress response genes, HSP12 and HSP26 (Fig 3B and 3C). In the pop2Δ mutant, these mRNA levels are increased in glucose-containing media, indicating that Pop2 is involved in glucose repression of HSP12 and HSP26. Wild-type POP2 efficiently complemented the increased expression of HSP12 and HSP26 caused by pop2Δ mutation, but POP2-S39A did not (Fig 3B and 3C). Interestingly, POP2-S39A completely complemented the increased expression of PIR3 caused by pop2Δ mutation (Fig 3D). Thus, the effect of S39A mutation is gene specific. How does Pop2 phosphorylated at S39 regulate the expression of HSP12 and HSP26? Since S39 phosphorylation of Pop2 is not involved in the regulation of LRG1 expression, it might be unrelated to regulation of Pop2 deadenylase activity. Pop2 was originally found as a regulator of PGK1 expression [3]. The Ccr4-Not complex is known to regulate gene expression not only through mRNA degradation but also through other mechanisms including transcriptional initiation and elongation [35]. One possibility is that S39 phosphorylation specifically influences the interaction of Pop2 with other factors involved in transcriptional initiation and elongation. Further studies are required to reveal this regulation.
Conclusions
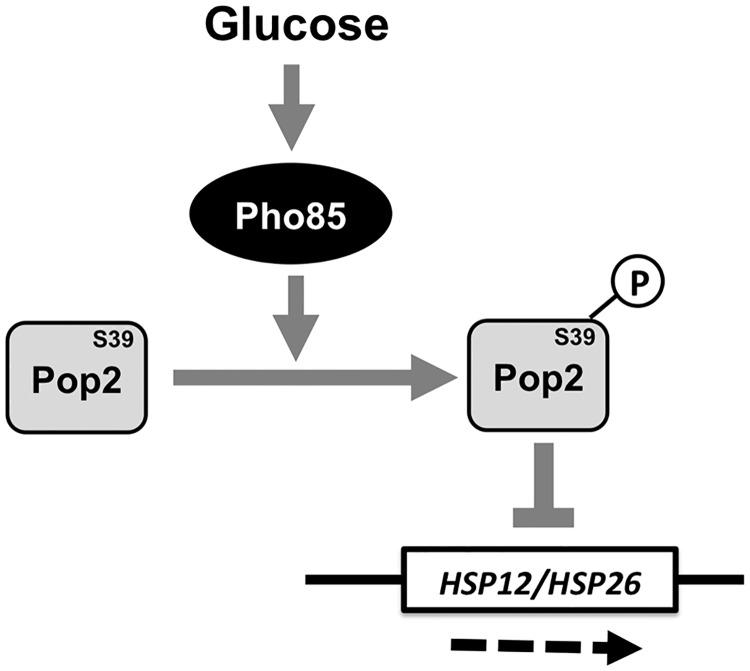
Taken together, we have shown that Pop2 is phosphorylated in a Pho85-dependent manner and this phosphorylation contributes to glucose repression of stress response genes, HSP12 and HSP26. Based on the present data, Fig 8 shows our proposed model. Our study provides valuable insights the role of Pop2 in glucose repression of stress responses in yeast. The molecular mechanism how Pop2 regulates HSP12 and HSP26 mRNAs through S39 phosphorylation will be the focus of future studies.
Fig 8. Proposed model of Pho85-dependet phosphorylation of Pop2 to repress the expression of stress response genes, HSP12 and HSP26.
In glucose medium, Pho85 is activated by glucose to phosphorylate Pop2 at S39. This phosphorylation regulates Pop2 function to repress the expression of stress response genes, HSP12 and HSP26.
Supporting information
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We thank the members of Molecular Cell Biology Laboratory for valuable discussions. This research was supported by JSPS KAKENHI Grant Numbers 15K06944 and 18K06053 (to K.I.).
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This research was supported by JSPS KAKENHI Grant Numbers 15K06944 and 18K06053 (to K.I.).
References
- 1.Parker R. RNA degradation in Saccharomyces cerevisae. Genetics. 2012;191(3):671–702. 10.1534/genetics.111.137265 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Collart MA. Global control of gene expression in yeast by the Ccr4-Not complex. Gene. 2003;313:1–16. 10.1016/S0378-1119(03)00672-3 . [DOI] [PubMed] [Google Scholar]
- 3.Sakai A, Chibazakura T, Shimizu Y, Hishinuma F. Molecular analysis of POP2 gene, a gene required for glucose-derepression of gene expression in Saccharomyces cerevisiae. Nucleic Acids Research. 1992;20(23):6227–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li X, Ohmori T, Irie K, Kimura Y, Suda Y, Mizuno T, et al. Different regulations of ROM2 and LRG1 expression by Ccr4, Pop2, and Dhh1 in the Saccharomyces cerevisiae cell wall integrity pathway. mSphere. 2016;1(5). 10.1128/mSphere.00250-16 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hata H, Mitsui H, Liu H, Bai Y, Denis CL, Shimizu Y, et al. Dhh1p, a putative RNA helicase, associates with the general transcription factors Pop2p and Ccr4p from Saccharomyces cerevisiae. Genetics. 1998;148(2):571–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moriya H, Shimizu-Yoshida Y, Omori A, Iwashita S, Katoh M, Sakai A. Yak1p, a DYRK family kinase, translocates to the nucleus and phosphorylates yeast Pop2p in response to a glucose signal. Genes & Development. 2001;15(10):1217–28. 10.1101/gad.884001 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ueda Y, To-E A, Oshima Y. Isolation and characterization of recessive, constitutive mutations for repressible acid phosphatase synthesis in Saccharomyces cerevisiae. Journal of Bacteriology. 1975;122(3):911–22. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaffman A, Herskowitz I, Tjian R, O’Shea EK. Phosphorylation of the transcription factor PHO4 by a cyclin-CDK complex, PHO80-PHO85. Science. 1994;263(5150):1153 . [DOI] [PubMed] [Google Scholar]
- 9.Measday V, Moore L, Retnakaran R, Lee J, Donoviel M, Neiman AM, Andrews B. A family of cyclin-like proteins that interact with the Pho85 cyclin-dependent kinase. Molecular and Cellular Biology. 1997;17(3):1212–23. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carroll AS O’Shea EK. Pho85 and signaling environmental conditions. Trends in Biochemical Sciences. 2002;27(2):87–93. 10.1016/S0968-0004(01)02040-0 [DOI] [PubMed] [Google Scholar]
- 11.Tennyson CN, Lee J, Andrews BJ. A role for the Pcl9-Pho85 cyclin–cdk complex at the M/G1 boundary in Saccharomyces cerevisiae. Molecular Microbiology. 2002;28(1):69–79. 10.1046/j.1365-2958.1998.00773.x [DOI] [PubMed] [Google Scholar]
- 12.Timblin BK, Tatchell K, Bergman LW. Deletion of the gene encoding the cyclin-dependent protein kinase Pho85 alters glycogen metabolism in Saccharomyces cerevisiae. Genetics. 1996;143(1):57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilliquet V, Berben G. Positive and negative regulators of the Saccharomyces cerevisiae ‘PHO system’ participate in several cell functions. FEMS Microbiology Letters. 1993;108(3):333–9. 10.1111/j.1574-6968.1993.tb06124.x [DOI] [PubMed] [Google Scholar]
- 14.Toh-e A, Tanaka K, Uesono Y, Wickner RB. PHO85, a negative regulator of the PHO system, is a homolog of the protein kinase gene, CDC28, of Saccharomyces cerevisiae. Molecular and General Genetics MGG. 1988;214(1):162–4. 10.1007/BF00340196 . [DOI] [PubMed] [Google Scholar]
- 15.Timblin BK, Bergman LW. Elevated expression of stress response genes resulting from deletion of the PHO85 gene. Molecular Microbiology. 2003;26(5):981–90. 10.1046/j.1365-2958.1997.6352004.x . [DOI] [PubMed] [Google Scholar]
- 16.Huang D, Friesen H, Andrews B. Pho85, a multifunctional cyclin-dependent protein kinase in budding yeast. Molecular Microbiology. 2007;66(2):303–14. 10.1111/j.1365-2958.2007.05914.x . [DOI] [PubMed] [Google Scholar]
- 17.Adams A, Gottschling DE, Kaiser CA (1997). Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 18.Longtine MS, McKenzie AIII, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14(10):953–61. [DOI] [PubMed] [Google Scholar]
- 19.Sakumoto N, Matsuoka I, Mukai Y, Ogawa N, Kaneko Y, Harashima S. A series of double disruptants for protein phosphatase genes in Saccharomyces cerevisiae and their phenotypic analysis. Yeast. 2002;19(7):587–99. 10.1002/yea.860 [DOI] [PubMed] [Google Scholar]
- 20.Kushnirov VV. Rapid and reliable protein extraction from yeast. Yeast. 2000;16(9):857–60. [DOI] [PubMed] [Google Scholar]
- 21.Kinoshita E, Kinoshita-Kikuta E, Takiyama K, Koike T. Phosphate-binding Tag, a new tool to visualize phosphorylated proteins. Molecular & Cellular Proteomics. 2006;5(4):749–57. 10.1074/mcp.T500024-MCP200 [DOI] [PubMed] [Google Scholar]
- 22.Albuquerque CP, Smolka MB, Payne SH, Bafna V, Eng J, Zhou H. A multidimensional chromatography technology for in-depth phosphoproteome analysis. Molecular & Cellular Proteomics. 2008;7(7):1389–96. 10.1074/mcp.M700468-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swaney DL, Beltrao P, Starita L, Guo A, Rush J, Fields S, Krogan NJ, Villén J. Global analysis of phosphorylation and ubiquitylation crosstalk in protein degradation. Nature methods. 2013;10(7): 10.1038/nmeth.2519 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holt LJ, Tuch BB, Villén J, Johnson AD, Gygi SP, Morgan DO. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science. 2009;325(5948):1682 10.1126/science.1172867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lenssen E, Oberholzer U, Labarre J, De Virgilio C, Collart MA. Saccharomyces cerevisiae Ccr4–Not complex contributes to the control of Msn2p-dependent transcription by the Ras/cAMP pathway. Molecular Microbiology. 2002;43(4):1023–37. 10.1046/j.1365-2958.2002.02799.x [DOI] [PubMed] [Google Scholar]
- 26.Yamamoto A, Mizukami Y, Sakurai H. Identification of a novel class of target genes and a novel type of binding sequence of heat shock transcription factor in Saccharomyces cerevisiae. Journal of Biological Chemistry. 2005;280(12):11911–9. 10.1074/jbc.M411256200 [DOI] [PubMed] [Google Scholar]
- 27.MacIsaac KD, Wang T, Gordon DB, Gifford DK, Stormo GD, Fraenkel E. An improved map of conserved regulatory sites for Saccharomyces cerevisiae. BMC bioinformatics. 2006;7:113-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Venters BJ, Wachi S, Mavrich TN, Andersen BE, Jena P, Sinnamon AJ, et al. A comprehensive genomic binding map of gene and chromatin regulatory proteins in Saccharomyces. Molecular Cell. 2011;41(4):480–92. 10.1016/j.molcel.2011.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Busti S, Coccetti P, Alberghina L, Vanoni M. Glucose signaling-mediated coordination of cell growth and cell cycle in Saccharomyces cerevisiae. Sensors. 2010;10(6). . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pines J. Cyclins and cyclin-dependent kinases: Theme and variations In: Vande Woude GF, Klein G, editors. Advances in cancer research. 66: Academic Press; 1995. p. 181–212. [DOI] [PubMed] [Google Scholar]
- 31.Bodenmiller B, Campbell D, Gerrits B, Lam H, Jovanovic M, Picotti P, Schlapbach R, Aebersold R. PhosphoPep—a database of protein phosphorylation sites in model organisms. Nature Biotechnology. 2008;26:1339 10.1038/nbt1208-1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Toh-e A, Tanaka, Uesono Y, Wickner RB. et al. PHO85, a negative regulator of the PHO system, is a homolog of the protein kinase gene, CDC28, of Saccharomyces cerevisiae. Molecular and General Genetics MGG. (1988) 214: 162 org/10.1007/BF00340196. [DOI] [PubMed] [Google Scholar]
- 33.Duy DL, Suda Y, Irie K. Cytoplasmic deadenylase Ccr4 is required for translational repression of LRG1 mRNA in the stationary phase. PLOS ONE. 2017;12(2):e0172476 10.1371/journal.pone.0172476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Viet NTM, Duy DL, Saito K, Irie K, Suda Y, Mizuno T, Irie K. Regulation of LRG1 expression by RNA binding protein Puf5 in the budding yeast cell wall integrity pathway. Genes to Cells. 0(ja). 10.1111/gtc.12646 [DOI] [PubMed] [Google Scholar]
- 35.Collart MA. The Ccr4-Not complex is a key regulator of eukaryotic gene expression. Wiley Interdisciplinary Reviews: RNA. 2016;7(4):438–54. 10.1002/wrna.1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.