Abstract
Mammalian cells carefully control their cholesterol levels by employing multiple feedback mechanisms to regulate synthesis of cholesterol and uptake of cholesterol from circulating lipoproteins. Most of a cell’s cholesterol (~80% of total) is in the plasma membrane (PM), but the protein machinery that regulates cellular cholesterol resides in the endoplasmic reticulum (ER) membrane, which contains a very small fraction (~1% of total) of a cell’s cholesterol. How does the ER communicate with PM to monitor cholesterol levels in that membrane? Here, we describe a tool, ALOD4, that helps us answer this question. ALOD4 traps cholesterol at the PM, leading to depletion of ER cholesterol without altering total cell cholesterol. The effects of ALOD4 are reversible. This tool has been used to show that the ER is able to continuously sample cholesterol from PM, providing ER with information about levels of PM cholesterol.
Keywords: Cholesterol trafficking, Plasma membrane, Endoplasmic reticulum, Anthrolysin O
1. Introduction
Cholesterol levels in mammalian cells are tightly regulated to lie within narrow limits. Regulation is achieved by controlling the two pathways by which cells acquire cholesterol. Cells can either synthesize cholesterol in the ERor uptake circulating cholesterol-rich low-density lipoprotein (LDL) via the LDL receptor [1]. When cholesterol levels in a cell rise above optimal levels, Scap, an ER membrane protein and cholesterol sensor, binds cholesterol and blocks proteolytic activation of a transcription factor, sterol regulatory element binding protein (SREBP). This leads to a decrease in expression of SREBP gene targets, which include cholesterol bio-synthetic enzymes for endogenous production of cholesterol and the LDL receptor which uptakes lipoprotein particles containing exogenous cholesterol [2]. As a result, cholesterol levels decline and return to optimal levels. Cholesterol levels are highest in the PM, which contains ~80% of total cellular cholesterol [3]. To meet the needs of PM, most of the cholesterol synthesized in ER or LDL-derived cholesterol from lysosomes must be ultimately transported to the PM. However, the cholesterol control machinery that regulates this cholesterol synthesis and uptake resides in the ER, which contains only ~1% of the cell’s total cholesterol [3, 4]. This raises the question: How does the ER monitor cholesterol levels in PM?
Recent studies that focused on the organization of cholesterol in PM have provided some insights into this problem of organelle communication. These studies showed that cholesterol in PMs, which comprises 40–45 mol% of total PM lipids when cells are grown in lipoprotein-rich serum, is organized into three different pools [5, 6]. Two of these pools of cholesterol (~27 mol% of total PM lipids) are sequestered by sphingomyelin and other membrane factors, respectively, and are inaccessible to soluble proteins that bind membrane cholesterol [6]. PM cholesterol in excess of these two pools constitutes a third pool that is accessible to cholesterol-binding proteins and can be transported to ER to block Scap-mediated proteolytic activation of SREBP, thereby inhibiting cholesterol synthesis and uptake. Thus, controlling the accessibility of PM cholesterol allows regulated communication between PM and ER, ensuring that cholesterol uptake and synthesis is not prematurely shut down before the cell’s cholesterol needs are met. Further understanding of the regulation of intracellular cholesterol trafficking requires new tools that selectively modulate transport pathways.
Here, we provide a detailed description of one such method to monitor and modulate cholesterol transport from PM to ER [7]. The method uses a recently described non-lytic cholesterol-binding protein, designated as Anthrolysin O domain 4 (ALOD4), as a reversible inhibitor of cholesterol transport from PM to ER [7–9]. ALOD4 binds to accessible cholesterol in PMs of cells at 37 °C without lysing cells or getting internalized. In addition to serving as an effective tool to monitor PM cholesterol accessibility, ALOD4 also modulates transport of cholesterol from PM to ER. This provides a useful tool to decouple cholesterol levels in ER from those in PM, and to study cholesterol transport between these membranes. Compared to traditional reagents such as β-cyclodextrin that lower ER cholesterol and trigger SREBP activation by decreasing total cellular cholesterol, ALOD4 traps PM cholesterol and triggers SREBP activation by lowering ER cholesterol without changing levels of cholesterol in the PM or the whole cell. The use of ALOD4 has shed light on how ER continuously monitors PM cholesterol levels and ensures cellular cholesterol homeostasis [7].
2. Materials
2.1. Plasmid Transformation
pALOD4—pRSET B expression vector encoding the domain 4 of Anthrolysin O (amino acids 404–512 with S404C and C472A mutations) along with an NH2-terminal hexahistidine tag followed by an enterokinase cleavage site (Addgene).
pALOD4(Mut)—pRSET B expression vector encoding the domain 4 of Anthrolysin O (amino acids 404–512 with S404C, C472A, G501A, T502A, T503A, L504A, Y505A, and P506A mutations) along with an NH2-terminal hexahistidine tag followed by an enterokinase cleavage site (Addgene).
BL21 (DE3) pLysS Escherichia coli competent cells (Invitrogen).
SOC medium (Invitrogen).
LB-Agar (RPI) plates containing 100 μg/mL ampicillin (RPI).
2.2. ALOD4 Expression
LB broth (RPI) containing 100 μg/mL ampicillin (LB-Amp).
1 M Isopropyl β-p-thiogalactopyranoside (IPTG) stock solution: Dissolve 2.38 g of IPTG in 10 mL of water.
2.3. ALOD4 Purification
Buffer A: 50 mM Tris-HCl, pH 7.5, 1 mM Tris(2-carboxyethyl)phosphine (TCEP).
Buffer B: Buffer A with 150 mM NaCl.
Buffer C: Buffer B with 500 mM imidazole.
Buffer D: Buffer A with 500 mM NaCl.
EDTA-free protease inhibitor tablets (Roche).
20 mg/mL Phenyl methanesulfonyl fluoride (PMSF) stock solution: Dissolve 1 g of PMSF in 50 mL of ethanol.
Lysozyme from chicken egg white (Sigma-Aldrich).
Lysis buffer: Buffer B containing 1 mg/mL lysozyme, 400 μg/mL PMSF, and one EDTA-free protease inhibitor tablet per 20 mL.
100-mL Dounce homogenizer (Kimble).
Branson Digital Sonifier (S-250, Fisher Scientific).
0.22 μm 250 mL Stericup filter unit (Millipore).
5-mL HisTrap HP Ni column (GE Healthcare).
1-mL HiTrap Q HP anion exchange column (GE healthcare).
Tricorn 10/300 Superdex 200 Increase column (GE healthcare).
Amicon Ultra-4 10-kDa cutoff centrifugal filters (Millipore).
15% SDS-PAGE gels.
5× gel loading buffer: 10% (w/v) SDS, 10 mM β-mercaptoethanol, 20% (v/v) glycerol, 0.2 M Tris–HCl, pH 6.8, 0.05% (w/v) bromophenol blue.
Precision Plus Protein Kaleidoscope Standards (BioRad).
Coomassie staining solution (Bio-Rad).
De-staining solution: Water/ethanol/acetic acid (5:4:1, v/v/v).
Spectrometer (e.g., NanoDrop, Thermo Fisher Scientific).
2.4. ALOD4 Labeling
10 mM stocks of Alexa Fluor 488 C5 maleimide (Invitrogen) and Alexa Fluor 647 C2 maleimide (Invitrogen) in DMSO.
Nickel-nitrilotriacetic acid (Ni-NTA) agarose beads (Qiagen).
1 M Dithiothreitol (DTT) stock solution: Dissolve 6.48 mg of DTT in 1 mL of water.
Glycerol for molecular biology (>99%).
2.5. PM Cholesterol Sequestration by ALOD4
CHO-K1 cells (American Type Culture Collection, CCL-61).
SV-589 cells (NIGMS Human Genetic Cell Repository).
48-well plates (Fisher Scientific).
Medium A: 1:1 mixture of Ham’s F-12 and Dulbecco’s modified Eagle’s medium (Sigma-Aldrich) supplemented with 100 units/mL penicillin and 100 μg/mL streptomycin sulfate (Corning) and 5% (v/v) fetal bovine serum (FCS, Sigma-Aldrich).
Medium B: Dulbecco’s modified Eagle’s medium (low glucose) (Sigma-Aldrich) supplemented with 100 units/mL penicillin and 100 μg/mL streptomycin sulfate and 5% (v/v) FCS.
Medium C: 1:1 mixture of Ham’s F-12 and Dulbecco’s modified Eagle’s medium supplemented with 5% (v/v) FCS.
Medium D: Dulbecco’s modified Eagle’s medium (low glucose) supplemented with 5% (v/v) FCS.
Dulbecco’s Phosphate buffered saline solution (PBS) (Sigma-Aldrich). The formulation of this buffer is 8 g of NaCl, 0.2 g of potassium phosphate (monobasic), 1.15 g of sodium phosphate (dibasic), and 0.2 g of KCl, all in 1 L water.
Buffer E: 10 mM Tris-HCl, pH 6.8,100 mM NaCl, 1% (w/v) SDS, 1 mM EDTA, 1 mM EGTA, 20 μg/mL PMSF, and EDTA-free protease inhibitor tablets (one tablet per 20 mL).
10% and 15% SDS-PAGE gels.
Bio-Rad Trans Blot Turbo System and nitrocellulose transfer packs.
Antibodies: IgG-20B12 against hamster SREBP1 [10], IgG-7D4 against hamster SREBP2 [11], IgG-4H4 against Scap [12], anti-His tagged antibody (Millipore), donkey anti-mouse secondary IgG antibody (Jackson ImmunoResearch), and goat anti-rabbit secondary IgG antibody (Jackson ImmunoResearch).
3. Methods
3.1. Plasmid Transformation
Add 100 ng of pALOD4 or pALOD4(Mut) plasmid to 50 μL of BL21 (DE3) pLysS cells and incubate the mixture on ice for 30 min.
Heat shock the cells by placing in a 42 °C water bath for 45 s.
Immediately transfer cells to ice for 2 min.
Recover cells by adding 450 μL of SOC medium to cells and incubate for 1 h at 37 °C.
Plate 100 μL of transformed and recovered cells on LB-Agar plates with ampicillin (100 μg/mL). Incubate plates for 16 h at 37 °C in a bacterial incubator.
3.2. ALOD4 Expression
The protocol is the same for ALOD4 and ALOD4(Mut).
Pick a single colony from above plates, transfer into a 250 mL flask containing 160 mL of LB-Amp, and incubate the flask at 37 °C in a shaker-incubator (225 rpm) until OD600 = 0.8–1 (see Note 1).
Transfer 12 mL of the above starter culture to a 2 L flask containing 1 L of LB-Amp. Incubate flasks at 37 °C in a shaker-incubator (225 rpm).
Once the OD600 = 0.4–0.6 (see Note 2), reduce the temperature to 18 °C and continue shaking for 1 h at 225 rpm.
Add IPTG to a final concentration of 1 mM to induce ALOD4 expression and continue shaking at 18 °C for 16 h.
Harvest the cells by centrifugation at 3220 × g for 10 min at 4 °C. Pellets can be stored at −80 °C after flash freezing in liquid nitrogen.
3.3. ALOD4 Purification
The protocol is the same for ALOD4 and ALOD4(Mut).
Resuspend cell pellet from 1 L of bacterial culture in 20 mL of lysis buffer. Homogenize the cell suspension using a Dounce homogenizer. We typically use cell pellets from 6 L of bacterial cultures which yields 1–2 mg of protein (see Note 3).
For lysozyme disruption, incubate the homogenized cell suspension at 4 °C for 3 h while stirring on a rotator.
Lyse the lysozyme-disrupted cells using a tip sonicator (see Note 4).
Centrifuge the lysed cell suspension at 220,000 × g for 1 h.
Filter the resultant supernatant using a 0.22 μm Millipore filter.
Connect a pre-packed 5-mL HisTrap-HP Ni column to an FPLC system (ÄKTA, GE Healthcare) and pre-equilibrate column with 10 column volumes (50 mL) of buffer B. Use a flow rate of 0.5 mL/min for all Ni chromatography operations.
Load the resultant filtered supernatant from step 5 onto the Ni column.
Wash column with 20 column volumes (100 mL) of a mixture of 90% buffer B and 10% buffer C (50 mM imidazole). Elute bound ALOD4 using a linear gradient starting from 90% buffer B/10% buffer C (50 mM imidazole) and ending at 40% buffer B/60% buffer C (300 mM imidazole) over 20 column volumes (100 mL) (see Note 5).
Subject an aliquot (~20 μL) of eluted fractions to 15% SDS-PAGE followed by Coomassie staining to assay for fractions containing purified ALOD4 (MW ~16 kDa). Use Precision Plus Protein Kaleidoscope Standards as molecular weights.
Pool fractions containing ALOD4 and concentrate using an Amicon Ultra-4 10 kDa cutoff centrifugal filter to ~10 mL (see Note 6).
Add ~140 mL of NaCl-free Buffer A to the protein solution for decreasing NaCl concentration to ~15 mM.
Connect a pre-packed 1-mL HiTrap Q HP anion exchange column to an FPLC and pre-equilibrate with 10 column volumes (10 mL) of buffer A. Use a flow rate of 0.5 mL/min for all ion exchange chromatography operations.
Load ALOD4 in low-NaCl buffer on the HiTrap Q column. Wash the column with 10 column volumes (10 mL) of buffer A.
Elute bound ALOD4 using buffer D into a single 2 mL fraction.
Connect a Tricorn 10/300 Superdex 200 gel filtration column to an FPLC and pre-equilibrate with 2 column volumes (60 mL) of buffer B. Use a flow rate of 0.5 mL/min for all gel filtration chromatography operations.
Load the elution from step 14 on to the Superdex 200 column followed by 50 mL of buffer B.
Pool protein-rich fractions and concentrate using an Amicon Ultra-4 10 kDa cutoff centrifugal filter to a concentration of 1–2 mg/mL and store at 4 °C for use over the next 4 weeks. Measure protein concentration using a NanoDrop instrument (see Note 7). For long-term storage, supplement concentrated protein with 20% (v/v) glycerol, flash-freeze in liquid nitrogen, and store at −80 °C (use within 6 months). See Fig. 1 for an example of the homogeneity and monodisperse nature of purified ALOD4 (lane 1 and blue curve).
Fig. 1.
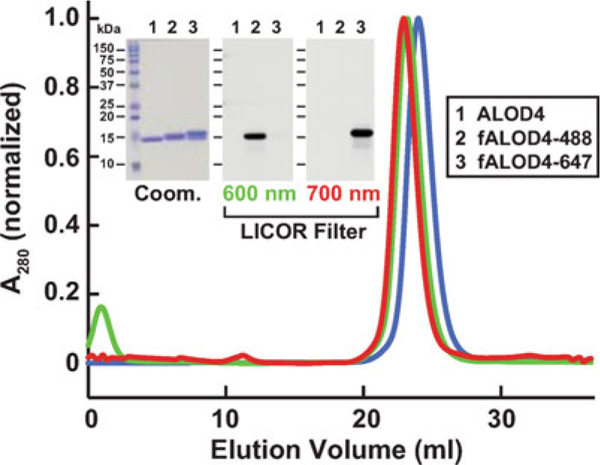
Biochemical characterization of ALOD4. Gel filtration chromatography of purified proteins. Recombinant ALOD4 was purified and labeled with Alexa Fluor 488 (fALOD4-488) or Alexa Fluor 647 (fALOD4-647) fluorescent dyes as described in Subheading 3. Buffer B (1 mL) containing 0.8 mg of ALOD4, fALOD4-488, or fALOD4-647 was loaded onto a Tricorn 10/300 Superdex 200 column and chromatographed at a flow rate of 0.5 mL/min. Absorbance at 280 nm (A280) was monitored continuously to identify ALOD4(blue), fALOD4-488(green), or fALOD4-647(red) proteins. Maximum A280 values for each protein (ALOD4: 390 mAU, fALOD4-488: 231 mAU, and fALOD4-647: 279 mAU) are normalized to 1. (Inset) 3 μg of each protein was subjected to 15% SDS/PAGE and stained with Coomassie (left) or imaged with the 600 nm filter (middle) or the 700 nm filter (right) on a LICOR instrument. Coom, Coomassie. Figure adapted from [7]
3.4. ALOD4 Labeling
In many experiments, it is informative to quantify the amount of ALOD4 bound to plasma membranes of cells. Quantification can be easily carried out using ALOD4 labeled with fluorescent dyes. These reagents can also be used for cellular labeling studies using fluorescence microscopy. The labeling protocol is the same for ALOD4 and ALOD4(Mut).
Combine 20 nanomoles (320 μg) of purified ALOD4 with 200 nanomoles of Alexa Fluor maleimide in a total volume of 300 μL of buffer B in 1.5 mL Eppendorf tubes. Protect the tubes from light by wrapping with aluminum foil.
Place tubes on a rotator at 4 °C for 16 h.
Quench labeling reactions by addition of DTT to a final concentration of 10 mM and then load the reaction onto a column containing 500 μL of Ni-NTA beads equilibrated in buffer B.
Wash column with 10 column volumes of buffer B containing 50 mM imidazole.
Elute bound labeled ALOD4 using buffer B containing 300 mM imidazole into a single 2 mL fraction.
Connect a Tricorn 10/300 Superdex 200 gel filtration column to an FPLC and pre-equilibrate with two column volumes (60 mL) of buffer B. Use a flow rate of 0.5 mL/min for all gel filtration chromatography operations.
Load fraction containing labeled ALOD4 from step 6 onto the Superdex 200 column followed by 50 mL of buffer B. Pool protein-rich fractions and concentrate using Amicon Ultra-4 10 kDa cutoff centrifugal filter to a concentration of 1–2 mg/mL. Measure concentration and degree of labeling of ALOD4 using a spectrometer (see Note 8).
Subject an aliquot to 15% SDS-PAGE followed by Coomassie staining and fluorescence scanning (using an Odyssey FC Imager and LI-COR analysis) to verify protein purification and labeling.
For long-term storage, supplement concentrated and labeled ALOD4 with 20% (v/v) glycerol, flash-freeze in liquid nitrogen, and store at −80 °C. See Fig. 1 for examples of the homogeneity and mono disperse nature of ALOD4 labeled with Alexa Fluor 488 or Alexa Fluor 647 (lanes 2, 3 and green, red curves). Either fluorescent reagent can be used for quantification and labeling studies.
3.5. PM Cholesterol Sequestration by ALOD4
3.5.1. PM Cholesterol Sequestration in CHO-K1 Cells
All experiments with CHO-K1 cells shouldl be performed at 37 °C in 8.8% CO2.
Set up CHO-K1 cells in medium A at a density of 6 × 104 cells/well in 48-well plates (day 0, see Note 9).
One day after, remove medium from each well and wash cells with 500 μL of PBS.
Add 200 μL of medium C containing varying amounts of purified ALOD4 or ALOD4(Mut) to each well. Either unlabeled or fluorescently labeled proteins can be used in this step.
Incubate 48-well plates for 1 h at 37 °C.
Collect medium from each well.
Wash each well twice with 500 μL of PBS.
Add 200 μL of Buffer E to each well and place the 48-well plate on a shaker at room temperature for 20 min for lysis.
Mix equal aliquots of medium collected in step 5 and lysed cells from step 7 with 5× loading buffer in Eppendorf tubes and incubate tubes at 95 °C for 10 min.
Subject above mixtures to either 10% or 15% SDS-PAGE (see Note 10).
Transfer electrophoresed proteins to nitrocellulose membranes using the Bio-Rad Trans Blot Turbo system.
Subject the nitrocellulose membranes to immunoblot analysis with an anti-His antibody (1:1000 dilution) for ALOD4, IgG-20B12 (2 μg/mL) for SREBP1, IgG-7D4 (10 μg/mL) for SREBP2, and IgG-4H4 (0.2 μg/mL) for Scap.
Visualize bound antibodies by chemiluminescence using a 1/5000 dilution of donkey anti-mouse IgG or a 1/2000 dilution of goat anti-rabbit IgG conjugated to horseradish peroxidase.
Expose membranes to Phoenix Blue X-Ray Film for 1–300 s or scan using an Odyssey FC Imager (Dual-Mode Imaging System; 2 min integration time) and analyze using Image Studio ver. 5.0. See Fig. 2 for an example of SREBP2 activation in CHO-K1 cells by ALOD4, but not by ALOD4(Mut). Increase in the cleaved nuclear form of SREBP-2 is a measure of activation of SREBP-2.
Fig. 2.
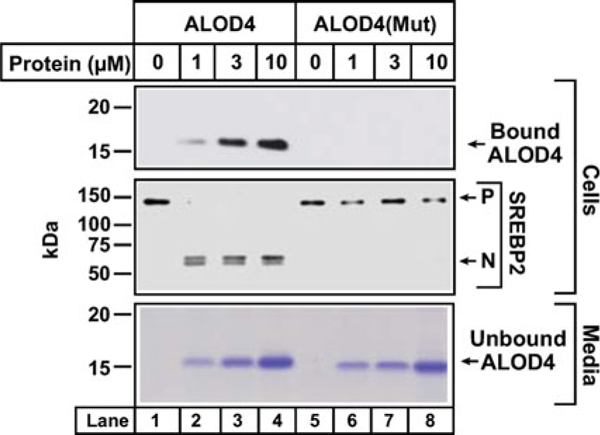
ALOD4 triggers activation of SREBP transcription factors. Immunoblot analysis of CHO-K1 cells after incubation with ALOD4 or ALOD4(Mut) proteins. On day 0, CHO-K1 cells were set up in medium A at a density of 3 × 104 cells/well of 48-well plates. On day 2, medium was removed, cells were washed with 500 μL of PBS and then incubated with 200 μL of medium C with indicated concentrations of ALOD4 or ALOD4(Mut). After incubation for 1 h at 37 °C, the medium was collected, and cells were harvested as described in Subheading 3. Equal aliquots of cells and media (10% of total) were subjected to immunoblot analysis or Coomassie staining as described in Subheading 3. P precursor form of SREBP2, N cleaved nuclear form of SREBP2. Figure adapted from [7]
3.5.2. PM Cholesterol Sequestration in SV-589 cells
All experiments with SV-589 cells should be performed at 37 °C in 5% CO2.
Day 0: Set up SV-589 cells in medium B at a density of 4 × 104 cells/well in 48-well plates.
Day 2: Remove medium from each well and wash the cells with 500 μL of PBS.
Add 200 μL of medium D containing varying amounts of purified ALOD4 or ALOD4(Mut) to each well.
Perform steps 3–8 as described in Subheading 3.5.1.
Subject the nitrocellulose membranes to immunoblot analysis with anti-His antibody (1/1000 dilution) for ALOD4, IgG-20B12 (2 μg/mL) for SREBP1, IgG-7D4 (10 μg/mL) for SREBP2, and IgG-4H4 (0.2 μg/mL) for Scap.
Perform steps 12 and 13 as described in Subheading 3.5.1. Typical results are similar to those observed for CHO-K1 cells in Fig. 2.
4. Notes
Care should be taken to avoid using a starter culture at OD600 > 0.8. Higher OD600 of starter cultures hinders growth rate of expression cultures and subsequent protein expression. Starter culture usually reaches OD600 = 0.8 in 12 h.
OD600 = 0.8 is optimal for protein expression. Care should be taken to not induce expression cultures at OD600 > 0.8 as this reduces protein yield. Culture usually reaches OD600 = 0.5 in 4 h, at which point temperature can be reduced to 18 °C. During an ~1 h cool-down period, the culture reaches OD600 = 0 8.
6 L of bacterial culture yields ~2 mg of protein. Number of liters of bacterial cultures used can be increased or reduced depending on the desired yield of protein.
We use a Branson Digital Sonifier (S-250, Fisher Scientific) to subject lysates to a 3 s on/3 s off sonication cycle at amplitude of 40% for 3 min. Following a 6 min cool-down period, repeat the cycle two more times.
Elution of ALOD4 with 300 mM imidazole (instead of a linear gradient) after washing with 50 mM imidazole results in protein yields with significant amounts of a nonspecific contaminant protein that migrates at ~50 kDa.
Concentrating the protein to lower volumes using Amicon Ultra-4 10 kDa cutoff centrifugal filter leads to protein aggregation. Employing the anion exchange column in the purification allows for further concentration without aggregation.
Extinction coefficients (ε280) for proteins can be obtained using “ProtParam tool” (expasy.org). Calculated ε280 values for ALOD4 and ALOD4(Mut) are 34,940 M−1 cm−1 and 33,630 M−1 cm−1, respectively.
Extinction coefficients of Alexa Fluor dyes can be found on Invitrogen website: Alexa Fluor 488 C5 maleimide (ε495 = 71,000 M−1 cm−1), Alexa Fluor 647 C2 maleimide (ε651 = 265,000 M−1 cm−1).
48-well plates are a convenient format for setup of cells so that multiple variables can be tested at the same time.
Use 10% SDS-PAGE for analyzing SREBP and Scap and 15% SDS-PAGE for analyzing ALOD4.
References
- 1.Brown MS, Goldstein JL (1997) The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 89:331–340 [DOI] [PubMed] [Google Scholar]
- 2.Brown MS, Goldstein JL (2009) Cholesterol feedback: from Schoenheimer’s bottle to Scap’s MELADL. J Lipid Res 50(Suppl): S15–S27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lange Y, Steck TL (2016) Active membrane cholesterol as a physiological effector. Chem Phys Lipids 199:74–93 [DOI] [PubMed] [Google Scholar]
- 4.Lange Y, Steck TL (1997) Quantitation of the pool of cholesterol associated with acyl-CoA: cholesterol acyltransferase in human fibroblasts. J Biol Chem 272:13103–13108 [DOI] [PubMed] [Google Scholar]
- 5.Das A, Goldstein JL, Anderson DD et al. (2013) Use of mutant 125I-perfringolysin O to probe transport and organization of cholesterol in membranes of animal cells. Proc Natl Acad Sci U S A 110:10580–10585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Das A, Brown MS, Anderson DD et al. (2014) Three pools of plasma membrane cholesterol and their relation to cholesterol homeostasis. elife 3:e02882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Infante RE, Radhakrishnan A (2017) Continuous transport of a small fraction of plasma membrane cholesterol to endoplasmic reticulum regulates total cellular cholesterol. elife 6:e25466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gay A, Rye D, Radhakrishnan A (2015) Switch-like responses of two cholesterol sensors do not require protein oligomerization in membranes. Biophys J 108:1459–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chakrabarti RS, Ingham SA, Kozlitina J et al. (2017) Variability of cholesterol accessibility in human red blood cells measured using a bacterial cholesterol-binding toxin. elife 6:e23355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rong S, Cortes VA, Rashid S et al. (2017) Expression of SREBP-1c requires SREBP-2-mediated generation of a sterol ligand for LXRin livers of mice. elife 6:e25015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang J, Brown MS, Ho YK et al. (1995) Three different rearrangements in a single intron truncate sterol regulatory element binding protein-2 and produce sterol-resistant phenotype in three cell lines. Role of introns in protein evolution. J Biol Chem 270:12152–12161 [DOI] [PubMed] [Google Scholar]
- 12.Ikeda Y, DeMartino GN, Brown MS et al. (2009) Regulated endoplasmic reticulum-assisted degradation of a polytopic protein: p97 recruits proteasomes to Insig-1 before extraction from membranes. J Biol Chem 284:34889–34900 [DOI] [PMC free article] [PubMed] [Google Scholar]


