SUMMARY
Establishing the balance between positive and negative innate immune mechanisms is crucial for maintaining homeostasis. Here we uncover the regulatory crosstalk between two, previously unlinked, innate immune receptor families: RIG-I, an anti-viral cytosolic receptor activates type I interferon, and NLR (nucleotide-binding domain, leucine repeat domain containing protein). We show that NLRP12 dampens RIG-I-mediated immune signaling against RNA viruses by controlling RIG-I’s association with its adaptor MAVS. The nucleotide-binding domain of NLRP12 interacts with the ubiquitin ligase TRIM25 to prevent TRIM25-mediated, Lys63-linked, ubiquitination and activation of RIG-I. NLRP12 also enhances RNF125-mediated, Lys48-linked, degradative ubiquitination of RIG-I. Vesicular stomatitis virus (VSV) infection downregulates NLRP12 expression to allow RIG-I activation. Myeloid cell specific Nlrp12-deficient mice display a heightened interferon and TNF response and are more resistant to VSV infection. These results indicate that NLRP12 functions as a checkpoint for anti-viral RIG-I activation.
In Brief
Chen et al. show that the nucleotide-binding domain, leucine repeat domain containing protein NLRP12 associates with the ubiquitin ligase TRIM25 to reduce K63-linked ubiquitination of the anti-viral innate immune receptor RIG-I. This prevents RIG-I association with MAVS and thus serves as a checkpoint for interferon and cytokines induction in response to RNA viruses.
Graphical Abstract

INTRODUCTION
NLR (nucleotide-binding oligomerization domain and leucine rich repeat-containing protein, also known as NOD-like receptors) proteins have garnered attention as a large family of intracellular pattern recognition receptors/sensors. These proteins trigger inflammation in response to pathogen infection and sterile activators including self (host) and environment-derived molecules (Davis et al., 2011). This function is illustrated by a number of NLR proteins that display inflammasome function. The inflammasome NLRs such as NLRP1, NLRP3, NLRC4 and NLRP6 assemble with ASC (apoptotic speck-containing protein with CARD), and procaspase-1 to form an inflammasome complex, leading to caspase-1 activation to cause the maturation and subsequent release of mature IL-1β and IL-18 (Broz and Dixit, 2016). However NLRs also have functions that are independent of the inflammasome. The most prominent examples are CIITA and NLRC5, which respectively act as master transcriptional activators that regulate class II and I major histocompatibility complex (MHC) gene expression (Kufer and Sansonetti, 2011; Ting et al., 2010). Most relevant to this work, several NLR proteins have been found to act as negative regulators of crucial innate immune pathways, including the canonical and non-canonical nuclear factor κB (NF-κB), mitogen-activated protein kinase (MAPK), cytokine production and type I interferon (IFN-I) pathways (Allen et al., 2012; Cui et al., 2012; Moore et al., 2008; Wang et al., 2015).
NLRP12 (NLR family pyrin domain containing 12) is a pyrin containing NLR protein that is primarily expressed by dendritic cells (DC) and neutrophils, with low to undetectable expression level in macrophages (Arthur et al., 2010; Vladimer et al., 2012; Zaki et al., 2011). In an overexpression system, NLRP12 is co-localized with the inflammasome adaptor, ASC, leading to caspase-1 activation (Wang et al., 2002). In Nlrp12−/− cells, the protein is implicated in inflammasome function in response to Yersinia (Vladimer et al., 2012) and malaria (Ataide et al., 2014). However, deletion or reduction of Nlrp12 is correlated with increased IL-1β plus other cytokines during obesity (Truax et al., 2018). This implies that its inflammasome-inducing function may be restricted to specific stimuli, while the protein also may negatively regulate inflammasome- and non-inflammasome-associated cytokine production (Papale et al., 2016; Silveira et al., 2017; Zaki et al., 2014). Its negative regulatory functions are exerted through the suppression of non-canonical NF-κB activation by associating with NF-κB inducing kinase (NIK) to promote NIK degradation in a proteasome-dependent pathway (Lich et al., 2007). In a colitis-associated colorectal cancer model and osteoclast differentiation, Nlrp12−/− mice show exaggerated non-canonical and canonical NF-κB activation in addition to ERK phosphorylation (Allen et al., 2012; Krauss et al., 2015; Zaki et al., 2011). Nlrp12−/− mice also exhibit enhanced disease in a mouse model of multiple sclerosis (Gharagozloo et al., 2015; Lukens et al., 2015) and enhanced inflammatory response to Salmonella and Brucella (Silveira et al., 2017; Zaki et al., 2014). Finally, it is required for neutrophil and dendritic cell migration to an inflammatory site (Arthur et al., 2010; Cai et al., 2016; Ulland et al., 2016) primarily through the regulation of CXCL1 (Hornick et al., 2018) although one study found the opposite finding in culture (Zamoshnikova et al., 2016). Recently, NLRP12 is reported to regulate gut microbiota, favoring commensal bacteria that suppress colitis and metabolic diseases (Chen et al., 2017; Truax et al., 2018). While the roles of NLRP12 in sterile inflammation and bacterial infection have been investigated, the biological role of NLRP12 in host immune response against virus infection is less studied. A recent study by Hornick et al. demonstrated a detrimental role for NLRP12 in influenza infections through excessive neutrophil recruitment (Hornick et al., 2018). The present report shows that in a different route of infection with a different virus, NLRP12 serves as a checkpoint against anti-viral immunity, specifically by blocking IFN-I production through its impact on ubiquitin ligases. Checkpoint proteins are found in multiple biologic processes, with the earliest examples described during cell cycle regulation (Hartwell and Weinert, 1989). Our report shows that NLRP12-mediated function is achieved by interference with the E3 ligase, TRIM25 (Gack et al., 2007), which is required for the Lys63-linked polyubiquitin-activation of retinoic-acid-inducible gene-I (RIG-I), a cytosolic pattern recognition receptor for viral RNA (Yoneyama et al., 2004). Oligomerized RIG-I interacts with and activates the mitochondria antiviral signaling adaptor, MAVS (VISA, IPS-1 or Cardif) (Tan et al., 2018), which then associates with adaptor proteins to recruit TRAF3 and TRAF family member-associated NF-κB activator (TANK) and trigger the activation of TANK binding kinase-1 (TBK1) and the IκB kinase (IKK-epsilon), leading to activated IRFs (Mao et al., 2010; Saha et al., 2006; Seth et al., 2005) and IFN (Kawai et al., 2005). During the resolution phase of viral infection, RIG-I signaling is degraded after conjugation of Lys48 ubiquitin by the ring-finger protein 125 (RNF125), (Arimoto et al., 2007). This report shows that Nlrp12 deficiency in myeloid cells leads to increased immune signaling to RNA viruses and 5’ triphosphate double stranded RNA by affecting TRIM25 and RNF125 to alter RIG-I activation and degradation.
RESULTS
Nlrp12 deletion in bone marrow-derived DC enhances IFN-I induction by an RNA virus and double-stranded RNA.
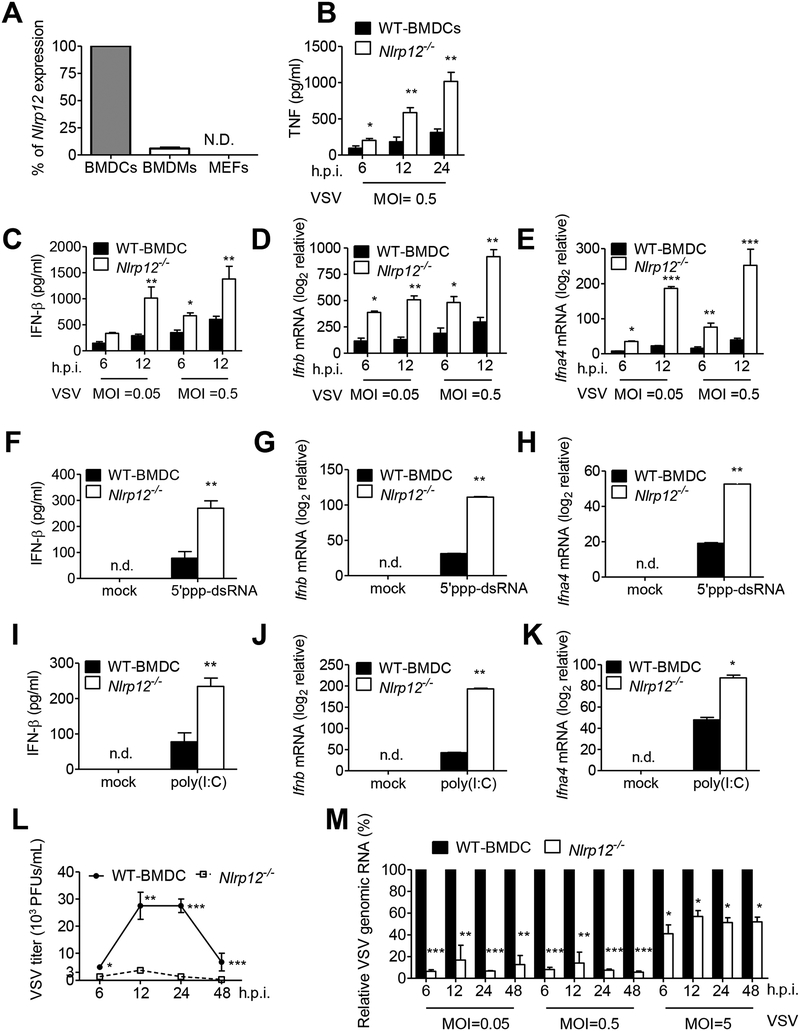
We investigated the function of NLRP12 during a viral infection. Human NLRP12 gene is abundantly expressed by CD14+-derived macrophages, DC and neutrophils, while mouse Nlrp12 is more highly expressed by DC (Arthur et al., 2010). Nlrp12 is lowly expressed in mouse bone marrow-derived macrophages (BMDMs) and negligible in mouse embryonic fibroblasts (MEFs), when compared to its expression in DC (Figure 1A), thus we focused on its role in DC. We infected BMDC with vesicular stomatitis virus (VSV), a negative single strand RNA virus [(−) ssRNA], and measured cytokine production by ELISA. Significantly higher amounts of TNF (Figure 1B), IFN-β (Figure 1C), Ifnb and Ifna4 transcripts (Figure 1D and 1E) were observed in Nlrp12−/− DC than in WT DC after VSV infection of different multiplicities of infection (MOI) and times after infection.
Figure 1. NLRP12 suppresses VSV and short dsRNA-induced production of IFN-I.
(A) RNAs were harvested from bone marrow derived dendritic cells (BMDCs), bone marrow derived macrophages (BMDCs) and mouse embryonic fibroblasts (MEFs) and assayed for Nlrp12 mRNA expression. Nlrp12 expression level in BMDCs was denoted as 100%, and the relative Nlrp12 expression in BMDMs and MEFs was compared to that expressed by BMDCs. BMDC from wild type (WT) and Nlrp12−/− mice were infected with VSV (MOI 0.05 and 0.5). Supernatants and cell lysates were measured for (B) TNF, (C) IFN-β protein, (D) Ifnb and (E) Ifna4 transcripts by ELISA and real time qPCR, respectively. WT and Nlrp12−/− BMDC were transfected with (F-K) 5’ppp-dsRNA or (G-I) low molecular weight (LMW) poly(I:C). BMDC were infected with VSV (MOI 0.5), with (L) virus titer and (M) relative amount of VSV genomic RNA determined by plaque assay and real time qPCR, respectively. WT level at each MOI and timepoint was set to 100% and levels in Nlrp12−/− cells compared to these values. Representative data were collected and expressed as mean ± s.e.m. from at least three independent experiments. Student’s t test was performed, with *P<0.05 or **P<0.01 for WT versus Nlrp12−/− mice.
Non-self ligands or PAMPs of viruses, including uncapped 5’ triphosphate containing short double-stranded RNA (5’ppp-dsRNA) and short double-stranded RNA polymers polyinosinic-polycytidylic acid [poly(I:C)], are recognized by RIG-I (Hornung et al., 2006; Pichlmair et al., 2006; Yoneyama et al., 2004). Significantly higher amounts of IFN-β protein and Ifnb and Ifna4 transcripts were detected in Nlrp12−/− DC compared to WT DC after transfection of 5’ppp-dsRNA or low molecular weight (LMW) poly(I:C) (Figures 1F-1K). Poly(I:C) can be recognized by both RIG-I and TLR3, however 5’ppp-dsRNA is only recognized by RIG-I (Lu et al., 2010). These findings suggest that NLRP12 negatively regulates IFN-β stimulated by a RIG-I ligand. During viral infection, viral titers (Figure 1L) and relative viral genomic copy numbers (Figure 1M) were significantly reduced in Nlrp12−/− DC at different times and MOIs compared to WT DC. The inhibitory effect of NLRP12 was most prominently observed at lower MOIs (0.05 and 0.5) compared to a higher MOI (5.0), suggesting that there may be threshold where NLRP12 can no longer attenuate an anti-viral response. In addition to the Nlrp12−/− strain that has been previously reported (Allen et al., 2012; Zaki et al., 2011), a second Nlrp12−/− strain referred to as Nlrp12−/−#2 was generated using CRISPR/cas in Taiwan in the C57BL/6J background (Figure S1A–S1D and STAR Methods) and produced similar outcome (Figure S1E, F).
NLRP12 inhibits the RIG-I dependent pathway.
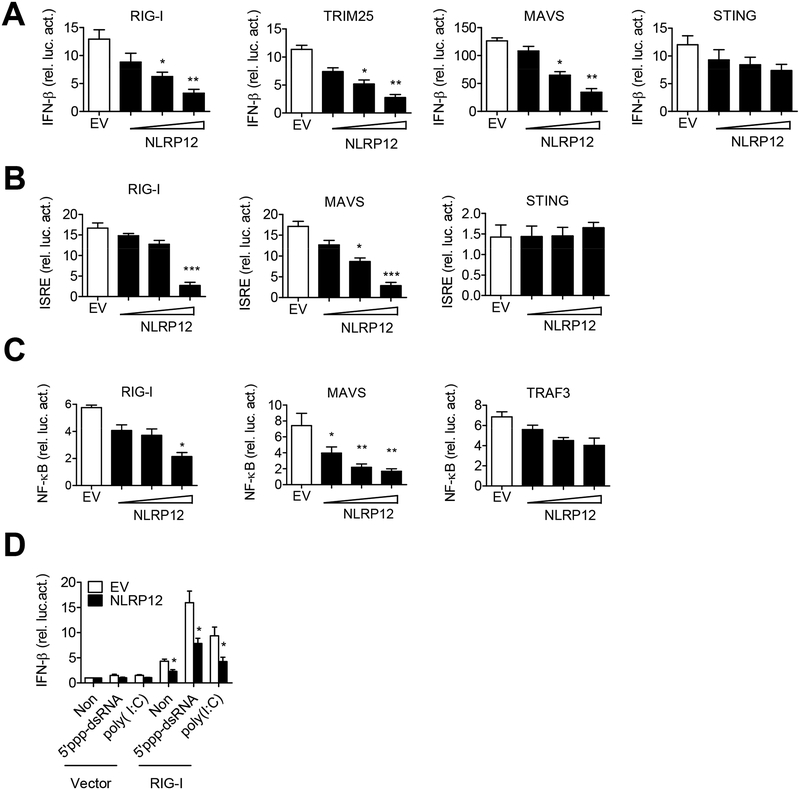
Since 5’ppp-dsRNA activates RIG-I, we investigated if NLRP12 affects RIG-I function. Overexpression of NLRP12 in HEK293 cells attenuated the activation of IFN-β promoter luciferase reporter activity by RIG-I, TRIM25 and MAVS in a dose-dependent manner, but not by STING (stimulator of interferon genes), a receptor of cyclic dinucleotides produced after DNA binding to cyclic GAMP synthase (cGAS) (Figure 2A). NLRP12 overexpression also inhibited the activation of the interferon-stimulated responsive element (ISRE) promoter by RIG-I and MAVS but not by STING (Figure 2B). NLRP12 significantly suppressed NF-κB promoter activated by RIG-I and MAVS, but not by TRAF3 (Figure 2C). Finally, NLRP12 reduced RIG-I-dependent IFN-β promoter-driven luciferase activity induced by 5’ppp-dsRNA or poly(I:C) (Figure 2D). These results suggest that a major effect of NLRP12 is the inhibition of RIG-I-mediated signalings.
Figure 2. NLRP12 suppresses RIG-I-mediated IFN-β, ISRE and NF-κB reporter activities.
HEK293 cells were transfected with 100 ng of (A) IFN-β, (B) ISRE or (C) NF-κB luciferase reporter with the internal control Renilla luciferase reporter pLR-TK plasmid and indicated plasmids (RIG-I, TRIM25, MAVS, STING or TRAF3) in the presence of empty vector (EV, pCDNA3) or NLRP12-encoding plasmid (pCDNA3/HA-NLRP12, 30, 100, 300 ng/sample). Luciferase assays were performed 24 hr post-transfection. (D) HEK293 cells were transfected with IFN-β luciferase reporter (30 ng), pLR-TK (3ng), and pFlag-CMV2-RIG-I plasmid (300 ng) or pFlag-CMV2 (Vector) with EV or NLRP12 expression plasmid (300 ng). 5’ppp-dsRNA and LMW poly(I:C) were then transfected into these cells at 24 hr, and luciferase assays were performed 12 hr later. Data were collected and expressed as mean ± s.e.m. from at least three independent experiments. Student’s t test was performed, *P<0.05; **P<0.01; ***P<0.001 for EV versus NLRP12 transfectants.
Nlrp12−/− DC exhibit elevated immune signaling after VSV infection and 5’ppp-dsRNA stimulation.
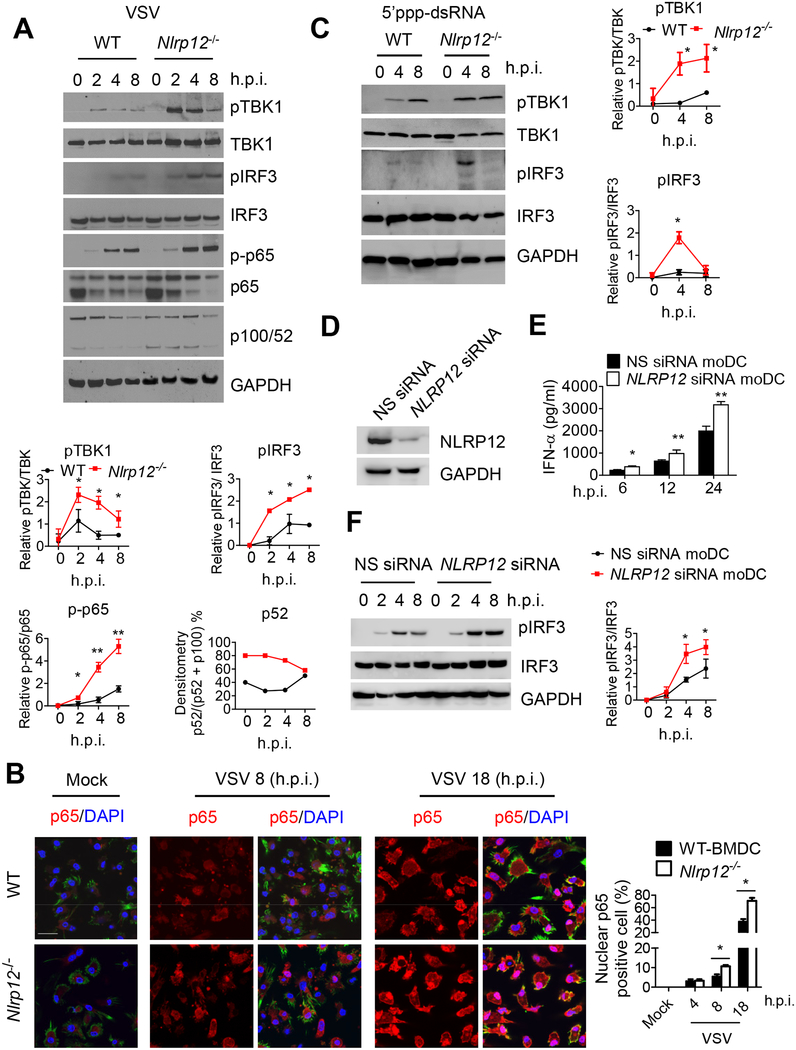
To assess changes in RIG-I-mediated signaling cascades upon viral infection, we analyzed changes in the phosphorylation of effectors that lie downstream of RIG-I. Phosphorylated TBK1 (pTBK1) and IRF3 (pIRF3) were induced 2–8 hr after VSV infection in WT DC, while these signals were significantly higher in Nlrp12−/− DC (Figure 3A). A compilation of results from three experiments is shown (Figure 3A, bottom). NF-κB lies downstream of the RIG-I signaling cascade and is a key transcriptional factor for IFN and proinflammatory cytokine genes. Phosphorylated NF-κB p65 (p-p65) was induced 2–8 hr after VSV infection in WT cells, and was enhanced significantly in Nlrp12−/− DC (Figure 3A). VSV did not induce the processing of p100 into p52 in WT cells, but Nlrp12−/− DC exhibited constitutive non-canonical NF-κB activation without VSV stimulation (Figure 3A). We also used microscopy to analyze p65 nuclear translocation. At 4 hr post-infection, 3% of p65 nuclear translocation was detected in WT and Nlrp12−/− DC, while this number was increased to 5–7% and 38% respectively by 8 and 18 hr post VSV infection in WT cells and to 11% and 65% in Nlrp12−/− DC at these two timepoints (Figure 3B). As expected, the microscopic changes of nuclear translocation lagged behind p-p65 in Figure 3A.
Figure 3. Nlrp12 deficiency promotes innate immune signaling.
(A) WT and Nlrp12−/− BMDC were infected with VSV (MOI 0.5), cell lysates were harvested and assayed for pTBK1, pIRF3, NF-κB p-p65 and non-canonical NF-κB processing of p100 to p52. For the densitometric analysis throughout this figure, bands were normalized with individual GAPDH and % phosphorylated protein over total amount of that protein was determined from at least three experiments, expressed as mean ± s.e.m and analyzed by Student’s t test. *P<0.05 for WT versus Nlrp12−/− cells. (B) WT and Nlrp12−/− BMDC were infected with VSV (MOI 0.5), stained with antibody to NF-κB p65 (red), DAPI (blue, nucleus) and Alexa Fluor-488 Phalloidin (green, cytoplasm), and visualized by confocal microscopy, scale bar 20 m. Arrowhead: representative p65 nuclear translocation (pink). Right, quantification of nuclear p65 from five fields (30 cells per field) collected from three experiments and Student’s t test shows *P<0.05 for WT versus Nlrp12−/− cells. (C) WT and Nlrp12−/− BMDC were transfected with 5’ppp-dsRNA, cell lysates were analyzed for pTBK1 and pIRF3. Graphs to the right, *P<0.05 for WT versus Nlrp12−/− cells. (D) Human DC (mo DC) was transfected with NLRP12 siRNA or scrambled siRNA (nonspecific, NS) for 48 h, and validated for reduced NLRP12 expression. (E) Cells were infected with 1 MOI of VSV, and assayed for IFN-α by ELISA. (F) VSV-infected cells were harvested for immunoblotting. A representative blot from three is shown. Densitometry of pIRF3 (right) with *P<0.05; **P<0.01.
We next investigated the impact of NLRP12 on immune signaling caused by the RIG-I ligand, 5’ppp-dsRNA. pTBK1 was slightly induced at 4 hr following 5’ppp-dsRNA transfection and peaked at 8 hr post transfection in WT DCs, while pTBK1 was significantly higher in Nlrp12−/− DC at 4 hr and sustained through 8 hr post-transfection (Figure 3C). pIRF3 was modestly induced in WT cells 4–8 hr post-transfection with 5’ppp-dsRNA, while the signal was significantly augmented in Nlrp12−/− DC 4 hr post transfection (Figure 3C, right is composite data). Studies described in Figures 3A–3C were also performed in Nlrp12−/−#2 with similar outcomes (Figure S1G). To validate these data in humans, we transfected human monocyte-derived DC (moDC) with pooled NLRP12 siRNA and the efficacy of knockdown was confirmed by immunoblot (Figure 3D). NLRP12 siRNA resulted in increased IFN-α secretion (Figure 3E) and enhanced pIRF3 (Figure 3F, right is composite data) after VSV infection, indicating that human NLRP12 also reduced an IFN response.
NLRP12 interacts with TRIM25 to interfere with RIG-I-mediated signaling cascade.
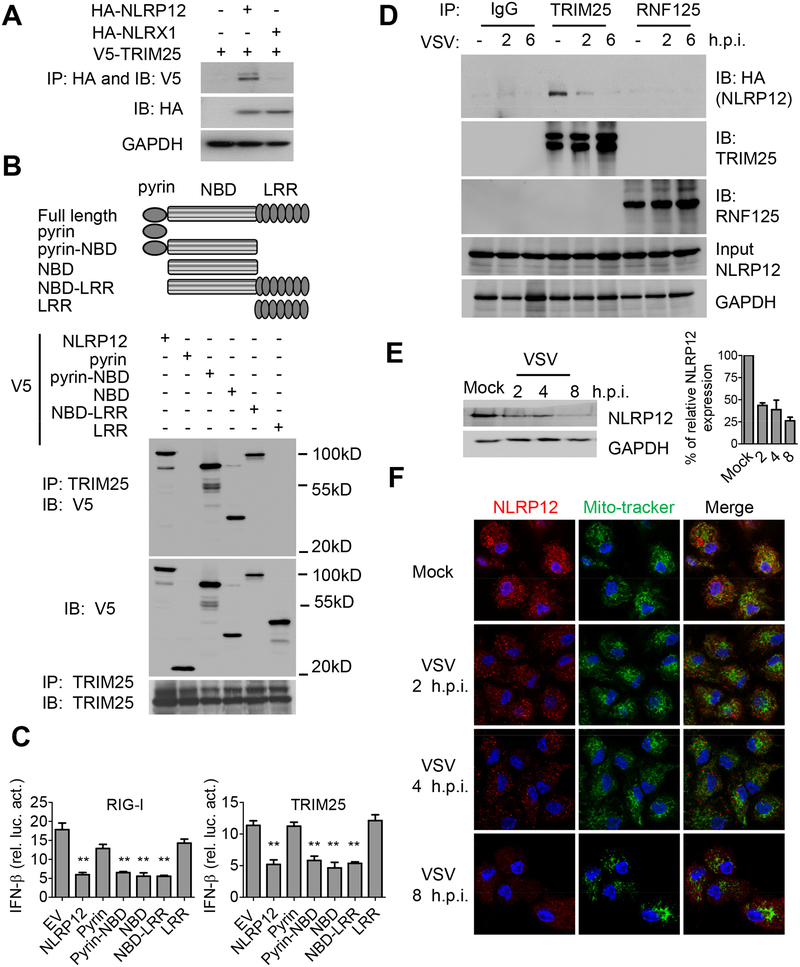
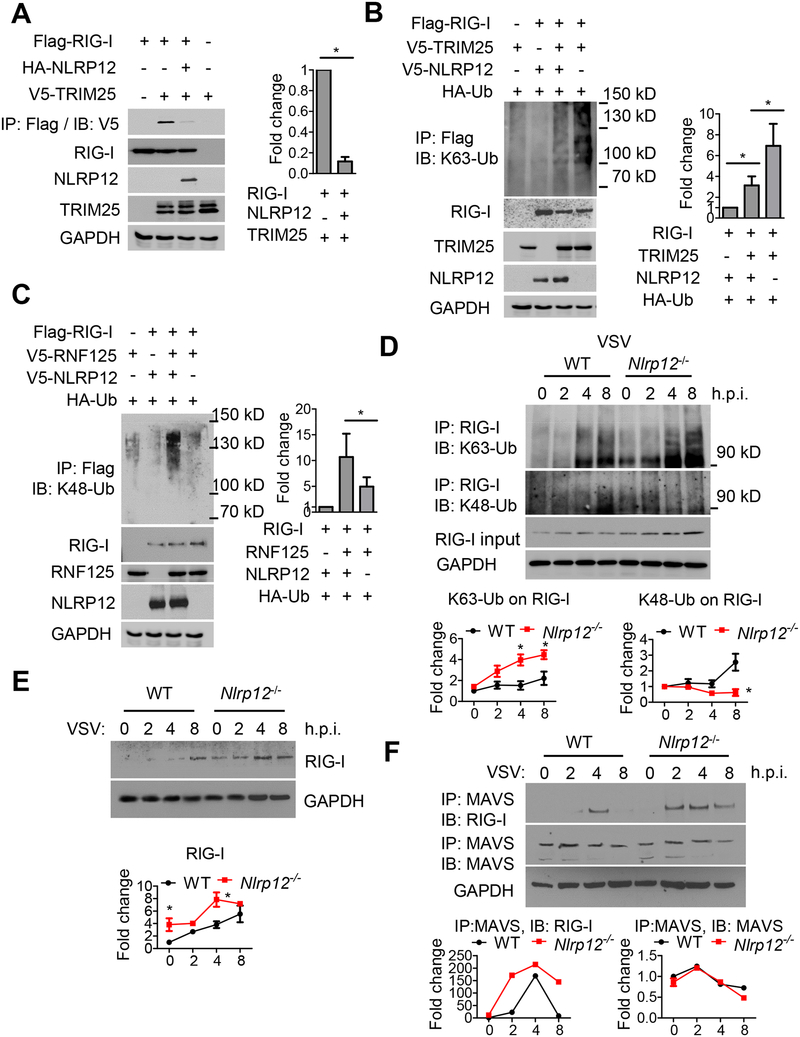
To explore the mechanism by which NLRP12 reduced the RIG-I-mediated signaling cascade, we tested if NLRP12 physically interacted with key adaptor proteins in the RIG-I signaling pathway including RIG-I, TRIM25, TBK1, TRAF3, TRAF6 and IKKα, by co-immunoprecipitation with NLRP12. Endogenous TRIM25 but not others interacted with overexpressed NLRP12 protein but this association was reduced by VSV (Figure S2A, B). In addition, overexpressed TRIM25 interacted with overexpressed NLRP12 but not with the negative control NLR, NLRX1 (Moore et al., 2008) (Figure 4A). Domain mapping analysis indicated that protein derived from constructs expressing full-length NLRP12, pyrin-NBD, NBD and NBD-LRR associated with endogenous TRIM25, whereas the pyrin or LRR domain of NLRP12 did not (Figure 4B). Thus NBD alone or combined with either pyrin or LRR can interact with TRIM25 while pyrin or LRR alone cannot.
Figure 4. NLRP12 NBD domain interacts with TRIM25 to reduce RIG-I-TRIM25 association.
(A) HEK293 cells were transfected with plasmids as indicated, and immunoprecipitation (IP) followed by immunoblotting (IB) with the indicated antibodies 24 hr after transfection. (B) HEK293 cells were transfected with domain-deletion constructs of NLRP12 and co-immunoprecipitation with endogenous TRIM25 24 hr later. (C) HEK293 cells were transfected with 100 ng of IFN-β luciferase reporter with pLR-TK and indicated plasmids (RIG-I or TRIM25) in the presence of empty vector (EV, pCDNA3-V5) or domain constructs of NLRP12. Luciferase assays were performed 24 hr post transfection. (D) HEK293 cells were infected with VSV (MOI 1) following transfection of the HA-NLRP12 plasmid. Endogenous TRIM25 and RNF125 were pulled down with anti-TRIM25 and anti-RNF125 antibodies respectively and immunoblotted with anti-HA for associated NLRP12. (E) Human DC were infected with VSV (MOI 1), and subjected to immunoblotting for endogenous NLRP12 protein. Densitometry indicates that the protein level went from 100% (mock) to 43% (2 hr timepoint), 38% (4 hr) and 26% (8 hr). (F) Confocal microscopic images of VSV-infected human DC, endogenous NLRP12 (red), mito-tracker (green) and DAPI (blue). Scale bar, 10 μm.
We next tested the functional effects of these deletion constructs. Truncated NLRP12 mutants with pyrin-NBD, NBD and NBD-LRR domains suppressed the IFN-β promoter luciferase activity induced by RIG-I and TRIM25 as efficiently as full-length NLRP12 (Figure 4C), while pyrin and LRR alone had no effect. This indicates that the NBD domain is needed for the suppression of RIG-I-mediated IFN-β expression. NLRP12 interacted with TRIM25 at the basal level but this interaction was gradually diminished upon VSV infection, suggesting that viral infection relieved TRIM25 from this inhibitory NLR (Figure 4D). Another E3 ligase, RING-finger protein (RNF125), causes the degradation of ubiquitin-conjugated RIG-I in a proteasome-dependent manner but it did not associate with NLRP12 (Figure 4D).
Interestingly, the level of endogenous NLRP12 in human DC was gradually reduced after a VSV infection. NLRP12 mRNA (Figure S2C) and protein (Figure 4E) were both reduced from 2–8 hours post-infection. Similarly, murine Nlrp12 mRNA was reduced by VSV infection (Figure S2D). We then assessed human NLRP12 protein expression in DC due to the availability of a good commercial antibody for human but not mouse NLRP12 protein. In an immunofluorescence assay, NLRP12 protein was abundantly expressed in human DC, but the expression level was gradually diminished over time (2–8 h.p.i.) following VSV infection (Figure 4F). NLRP12 was not located at the mitochondria at resting state nor was its localization altered by VSV infection (Figure 4F). These results indicate that NLRP12 attenuates the RIG-I pathway by targeting TRIM25, while viral infection reduces NLRP12 expression, thus providing one mechanism to remove its blockade of RIG-I function.
NLRP12 reduces Lysine 63-linked, but enhances Lysine 48-linked ubiquitination of RIG-I.
Lysine 48 (Lys48)-linked polyubiquitin chains target proteins for destruction in a proteosome-dependent process, while Lysine 63 (Lys63)-linked polyubiquitin chains are considered activation modifications not associated with proteasomal degradation. TRIM25 interaction with RIG-I leads to Lys63-linked, activation ubiquitination of RIG-I (Gack et al., 2007) which increases RIG-I-MAVS interaction, RIG-I tetramerization, stabilization and subsequent signaling transduction (Peisley et al., 2014).
TRIM25 associated with RIG-I as reported, but NLRP12 reduced this association (Figure 5A) and TRIM25-mediated Lys63-linked ubiquitination of RIG-I (Figure 5B). In contrast, RNF125-mediated Lys48-linked ubiquitination of RIG-I was increased by NLRP12 (Figure 5C). This suggests that NLRP12 reduces TRIM25-mediated Lys63-linked activation ubiquitination while enhancing RNF125-mediated Lys48-linked degradative ubiquitination of RIG-I.
Figure 5. NLRP12 reduces Lys63-linked but enhances Lys48-linked ubiquitination of RIG-I.
(A-C) HEK293 cells were transfected with plasmids as indicated, and co-immunoprecipitation was performed 24 hr post transfection. Right panels and all graphical data in this figure are composite densitometry from three experiments expressed as mean ± s.e.m. (A) Right graph, densitometry of TRIM25-RIG-I interactions. (B) and (C) right graphs, densitometry of RIG-I ubiquitination. RIG-I ubiquitination signal was normalized to total RIG-I levels. The relative fold change is shown in the histogram. Student’s t test was performed. * is P<0.05. (D) WT and Nlrp12−/− BMDC were infected with VSV (MOI 0.5), and cell lysates were subjected to immunoprecipitation with antibody against RIG-I. Levels of Lys63-linked (upper) and Lys48-linked ubiquitin (lower) on RIG-I were measured by immunoblotting with antibodies against K63-Ub or K48-Ub. RIG-I ubiquitination signals were normalized to GAPDH, and the basal RIG-I ubiquitination signal for WT group at 0 hr was set as “1”. Relative fold change is shown in graphic form. (E) VSV-infected WT and Nlrp12−/− BMDC were harvested and immunoblotted for RIG-I levels. Densitometry of RIG-I expression was measured as described in (D). (F) VSV-infected WT and Nlrp12−/− BMDC lysates were immunoprecipitated with antibody against MAVS. RIG-I-MAVS interactions and MAVS levels were measured by immunoblot. The densitometry of RIG-I-MAVS interactions and MAVS expression was measured as described in (D) and expressed in graphic form below each immunoblot.
As these experiments involved an overexpression system, we next investigated these same pathways in Nlrp12−/− cells. WT DC exhibited increased Lys63-linked ubiquitin on RIG-I, with maximal ubiquitination detected by 4–8 hr post-VSV infection, while Nlrp12−/− cells showed even greater Lys63-linked ubiquitination of RIG-I at these time points (Figure 5D). Conversely, Lys48-linked degradative ubiquitin on RIG-I was enhanced at 8 hr post-VSV infection in WT DC, while Nlrp12−/− cells did not exhibit this enhancement (Figure 5D). Consistent with these findings, RIG-I gradually increased over time after VSV infection in WT cells. In Nlrp12−/− cells, a higher basal level (0 hr) was observed, and a further increase of RIG-I protein was detected post-infection (4–8 hr, Figure 5E). Studies described in Figures 5D–5E were also performed in Nlrp12−/−#2 with similar outcomes (Figure S2E, S2F). Enhanced Lys63-linked ubiquitination of RIG-I and increased RIG-I protein in Nlrp12−/− cells are predicted to promote RIG-I interaction with MAVS. Indeed, earlier and longer-lasting RIG-I-MAVS interaction was observed in Nlrp12−/− cells (Figure 5F). In contrast to the reduction of RIG-I by the presence of NLRP12 (Figure 5E), NLRP12 did not affect MAVS protein levels (Figure 5F).
Nlrp12 deficiency augments host response to VSV infection.
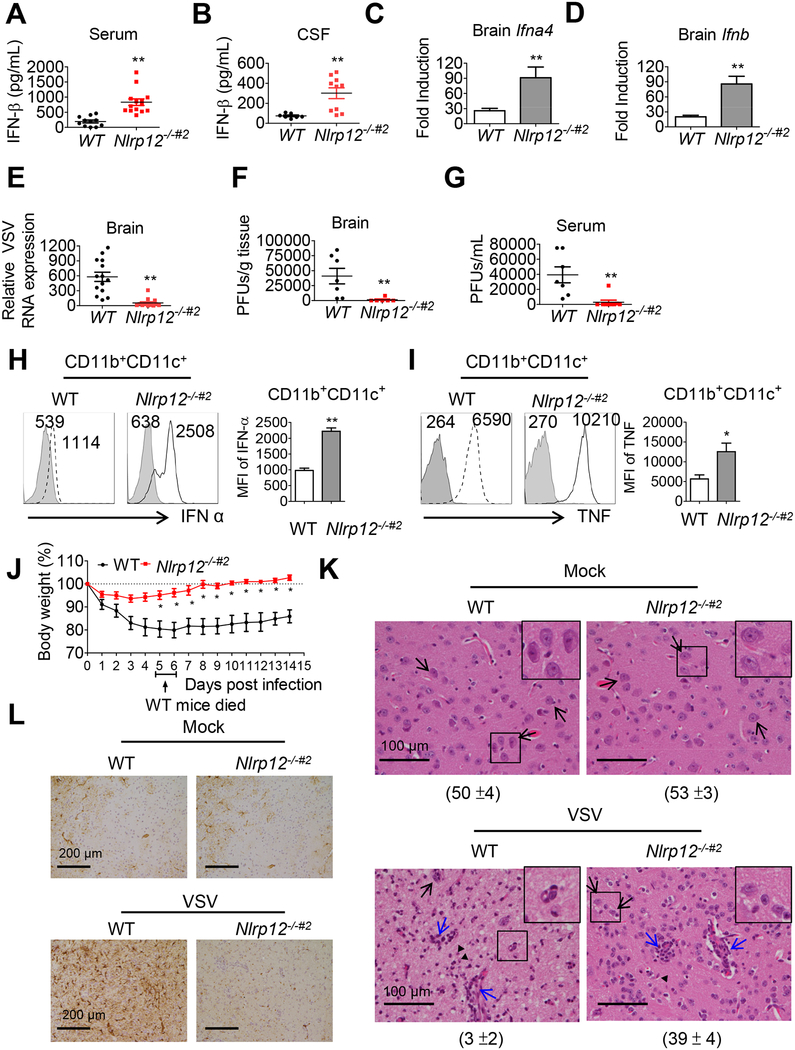
We next examined the in vivo impact of NLRP12. A preliminary experiment was performed in the Nlrp12−/− strain to show that this strain had increased IFN and lower viral load, however due to the difficulty of obtaining proper material transfer documents, Nlrp12−/−#2 mice were later generated in Taiwan for additional in vivo studies. Intranasal (i.n.) infection with VSV triggers type I IFN in the central nervous system and periphery of mice, and this response is critical for viral control (Detje et al., 2015). Serum and cerebral spinal fluid (CSF) levels of IFN-β (Figure 6A and 6B) as well as Ifnb and ifna4 transcripts (Figure 6C and 6D) were significantly higher in Nlrp12−/−#2 mice at 24 hr post-VSV infection than in WT controls. Relative brain VSV genomic RNA expression (Figure 6E), viral titers (Figure 6F) and viremia (Figure 6G) were significantly reduced in Nlrp12−/−#2 mice. To assess the impact of NLRP12 in brain DC, we isolated brain mononuclear cells (MNC) from VSV-infected WT and Nlrp12−/−#2 mice 12 hr post-infection. DC (CD11b+CD11c+) constituted approximately 2.4 % of total brain MNC in WT and Nlrp12−/−#2 mice (Figure S3A). Nlrp12−/−#2 DC from VSV infected brain tissues expressed significantly higher IFN-α (mean fluorescence intensity (MFI): 2508) than the WT DC (MFI: 1114) (Figure 6H), and the same was observed with TNF (MFI 10210 vs. 6590) (Figure 6I). Nlrp12−/−#2 brain DC (Figure S3B) and macrophages (Figure S3C) as well as splenic DC (Figure S3D) and macrophages (Figure S3E) also had higher RIG-I expression than WT counterparts. These data demonstrate that Nlrp12 deletion results in greater type I IFN production and RIG-I protein.
Figure 6. Nlrp12−/−#2 mice are more resistant to VSV infection.
WT and Nlrp12−/−#2 mice were inoculated with VSV (1×106 PFU) intranasally. (A) Serum and (B) cerebral spinal fluid (CSF) were assays for IFN-β by ELISA 24 hr post-infection. (C) Brain Ifna4 and (D) Ifnb transcripts were measured by real-time qPCR. (E) Brain-localized VSV genomic RNA was determined by qPCR. Viral titers in (F) brain and (G) serum were determined by plaque assays. (H, I) Brains were harvested 12 hr post-infection, and IFN-α (H) and TNF (I) in CD11b+CD11+ DC measured by FACS. Histogram is from a representative sample (left) and data compilation from at least five mice (right) are expressed as mean ± s.e.m for each group. Student’s t test shows *P<0.05. Shaded histogram is the isotype control. Numbers represent mean fluorescence intensity (MFI). (J) Body weight was monitored (n=15 per group). WT mice (20%) died between days 5–6 (arrow). Student’s t test shows *P<0.05 or **P<0.01. See Movies S1 and S2 for activity of VSV-infected WT and Nlrp12−/−#2 mice. (K) Mouse brain section was stained with H&E and (L) anti-GFAP by day 9 post-VSV infection. Five fields of H&E stained section were randomly selected and the numbers of neuron counted and represented as mean ± S.D (see number under each panel). Black arrow: representative neuron with intact nuclear membrane and smooth and round contour. Blue arrows, inflammatory foci. (L) shows brown horseradish peroxidase immunostaining: reactive astrocyte.
In concert with these observations, some of the WT mice presented with dyskinesia and significant lethargy following VSV infection (Movie S1), whereas Nlrp12−/−#2 mice were energetic (Movie S2). WT mice showed noticeable weight loss starting from day 2 post-infection, with all subjects exhibiting significantly more weight loss than Nlrp12−/−#2 mice from days 5 to 14 post-infection (Figure 6J) and one-fifth of the infected WT mice died between days 5 and 6 (Figure 6J, arrow). In contrast, Nlrp12−/−#2 mice lost only 3 % of their body weight during the first 3 days following infection and showed full recovery (100% or more) by day 6. Pathology examination showed rare healthy neurons and marked neuronal loss in the ventral striatum and hypothalamus of VSV-infected WT mice compared to uninfected mice (Figure 6K). By contrast, VSV infected Nlrp12−/−#2 mice showed significantly milder neuronal loss. Reactive astrocytes (Figure 6L, brown immunoperoxidase staining) indicative of enhanced expression of a reactive astrocytic biomarker, glial fibrillary acidic protein (GFAP), was found in WT but reduced in Nlrp12−/−#2 mice (Figure 6L). Increased astrogliosis typically accompanies CNS insult in VSV-infected WT mice (Sofroniew and Vinters, 2010). Taken together, Nlrp12−/−#2 mice, showed enhanced host response against VSV, had less neuron loss and astrogliosis than WT controls following VSV infection.
We next investigated if Nlrp12 deficiency altered immune infiltration in brain tissue during VSV infection. Lymphocytes infiltration was observed in both WT and Nlrp12−/−#2 brain tissue during VSV infection (Figure 6K, blue arrows). Brain mononuclear cells were isolated and analyzed. At day 5 post infection, about 0.1% of cells express myeloid DC markers (CD11b+CD11c+) (Figure S4A). Among these, monocytes (CD11b+Ly6Chi) represented approximately 20–21% of the CD11b+ (Figure S4B), neutrophils (CD11b+Ly6G+) were negligible (Figure S4B). CD3+ T cells were the most abundant immune cells (Figure S4C). The proportions of myeloid cells, monocytes, CD3+ T cells, and CD4+/CD8+ T cells in brain tissue were similar in WT and Nlrp12−/−#2 mice.
Because Nlrp12 was reported to be expressed by mouse CD4+ and CD8+ T cells (Lukens et al., 2015), we investigated the T-cell intrinsic role of NLRP12 during VSV infection at an early and late timepoints. At day 3 (Figure S5) and day 9 (Figure S6) post-infection, the proportion of CD3+, CD4+ or CD3+, CD8+ T cells from WT and Nlrp12−/−#2 was similar in the brain (Figure S5A and S6A) and the submandibular draining lymph node (SLN) (Figure S5B and S6B). By day 3 and day 9 post-infection, WT and Nlrp12−/−#2 CD4+ and CD8+ T cells expressed similar level of IFN-γ protein in the brain tissue (Figure S5C–D and S6C–D) and SLN (Figure S5E–F and S6E–F). These results suggest that NLRP12 did not affect T-cells derived IFN-γ production during VSV infection.
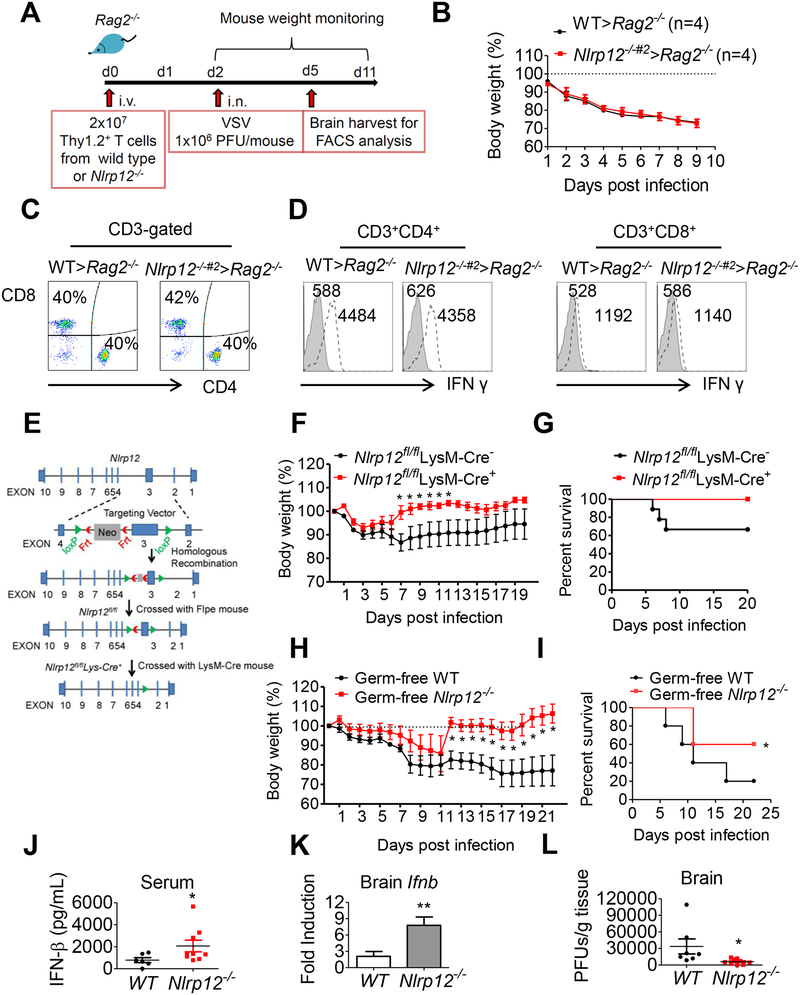
To determine if NLRP12 has a T cell-intrinsic role during viral infection, we reconstituted the Rag2−/− mice with Thy1.2+ T cells isolated from WT or Nlrp12−/−#2 mice. Reconstituted mice were then infected with VSV and weight loss and immune responses were monitored (Figure 7A). WT > Rag2−/− mice had WT Nlrp12-expressing T cells while Nlrp12−/−#2 > Rag2−/− mice had Nlrp12−/−#2 T cells. These two chimeric Rag2−/− strains showed similar weight loss (Figure 7B), infiltrated CD4+ and CD8+ T cells (Figure 7C) and IFN-γ levels in CD4+ or CD8+ cells in brain tissues by day 3 (Figure 7D) and day 9 (Figure S7) post VSV infection. These data indicate that Nlrp12 in T cells did not affect VSV infection outcome.
Figure 7. Nlrp12-deficiency in myeloid but not T cells protects host from VSV infection in a microbiota-independent manner.
(A) Reconstitution of Rag2−/− mice with T cells followed by VSV inoculation. (B) Body weight was monitored (n=4 per group). At day 3 post-infection, (C) Brain MNC were assayed for CD4+ and CD8+ T cells and (D) expression of IFN-γ by brain CD4+ and CD8+ T cells as determined by FACS analysis. (E) Scheme of Nlrp12flox/flox production using a targeting construct, which contained loxP sites flanking exon 3 of Nlrp12, next to a FRT flanked neomycin-resistance cassette in intron 2. (F) Control Nlrp12flox/flox LysM-Cre− and Nlrp12 KO (Nlrp12flox/flox LysM-Cre+) mice were inoculated with VSV (1×106 PFU) intranasally, and body weight loss was monitored (n=9 per group). Multiple t test shows *P<0.05. (G) Survival rate is displayed as Kaplan–Meier survival curves with log rank test (n = 9 per group). (H) Germ-free WT and Nlrp12−/− mice were inoculated with VSV intranasally. Body weight was monitored (n=5 per group). (I) Same as (G), except germ-free mice were used (n = 5 per group), *P < 0.05. (J) Serum IFN-β was measured by ELISA at 24 hr post-infection. (K) Ifnb transcript in brain tissue was measured by real time qPCR analysis. (L) Brain-localized viral titers were determined by plaque assays. For H, J, K and L, Mann-Whitney test was performed, *P<0.05.
To test the role of NLRP12 in myeloid cells, we generated a tissue-specific, myeloid conditional Nlrp12 knockout mice (Nlrp12flox/flox LysM-Cre+) (Figure 7E) and littermate controls (Nlrp12flox/flox LysM-Cre−) and infected these two groups with VSV. Mouse weight and survival were monitored for 22 days. The control mice had one weight drop at day 2 and another weight drop at day 6–7 post-infection, and the weight loss persisted for the duration of the study. By contrast, Nlrp12flox/flox LysM-Cre+ mice did not display this second weight drop and recovered from day 7 onward (Figure 7F). In addition, one-third of the control mice died from the infection while all of the Nlrp12flox/flox LysM-Cre+ mice survived and recovered (Figure 7G). This indicates that myeloid-specific Nlrp12 deficiency augments the host response and accelerates recovery during VSV infection.
We recently demonstrated that NLRP12 attenuates colon inflammation by maintaining the colonic microbial diversity and promoting anti-inflammatory commensals such as Lachnospiraceae (Chen et al., 2017). To assess if the microbiota played a role in the resistant phenotypes observed in the Nlrp12-deficient mice, germ-free WT and Nlrp12−/− littermate mice were infected with VSV. Germ-free WT mice were more sensitive to VSV infection than germ-free Nlrp12−/− mice by exhibiting more weight loss (Figure 7H), more death (Figure 7I), and significantly lower levels of serum IFN-β and ifnb transcript (Figure 7J and 7K), but significantly higher VSV genomic RNA expression (Figure 7L) 24 hr post-VSV infection. In composite, these results indicate that myeloid NLRP12 attenuates host response against VSV infection but this occurs regardless of the microbiota.
DISCUSSION
While several NLR proteins exhibit negative regulatory function, the precise molecular mechanism by which they mediate this inhibitory function in vivo or in primary cells is less clear. In this report, we show that Nlrp12-deficiency in human and mouse DC leads to elevated amounts of cytokines such as IFN-β and TNF in response to VSV infection, the RIG-I agonists 5’ppp-dsRNA and short poly(I:C). Mechanistically, NLRP12 interacts with TRIM25 to disrupt RIG-I and TRIM25 association and TRIM25-mediated Lys63-linked ubiquitination of RIG-I. The latter is required for RIG-I-mediated IFN-β production, achieved through its interaction with, and activation of, MAVS. In vivo infection shows that Nlrp12−/− mice are more resistant to VSV infection, have lower viral loads and recover faster than WT mice. This is attributed to higher amounts of IFN-α and IFN-β in the peripheral blood and CNS of infected Nlrp12−/− mice than in WT controls and the effect is mediated by myeloid NLRP12. These in vivo outcomes provide evidence that NLRP12 is a negative regulator of the anti-viral innate immunity activated by RIG-I.
Several NLRs have been reported to reduce the production of IFN-I in response to diverse stimuli; however in most cases, the precise pathway has not been identified, or the studies were limited to in vitro analysis. For example, NLRC3 associates with STING and TBK1 to impede the STING-TBK1 interaction, thereby reducing the downstream DNA virus mediated-type I IFN production (Zhang et al., 2014). NLRC5 interacts with RIG-I and MDA5 to inhibit RLR-mediated IFN-I responses (Cui et al., 2010), although reversed findings have been found by others (Benko et al., 2017). NLRP4 regulates double-stranded RNA- or DNA–mediated production of IFN-I by recruiting the E3 ubiquitin ligase DTX4, thus targeting TBK1 for degradation in vitro (Cui et al., 2012). NLRX1 is a negative regulator of RIG-I-mediated IFN-I signaling pathway by binding to MAVS and blocking its interaction with RIG-I (Moore et al., 2008). Its interference with MAVS has also been observed in myocardial ischemia (Li et al., 2016) while NLRX1 itself can be a target of Fas-associated factor 1 (FAF1), which prevents its interaction with MAVS (Kim et al., 2017). However its negative effect on IFN-I is not always observed and its role during a viral infection may vary, depending on the time course, infectious dose and/or viral proteins involved (Jaworska et al., 2014). Finally, NLRP14 expressed by germ cell has been reported to interact physically with STING, RIG-I, MAVS and TBK1, leading to the ubiquitination and degradation of TBK1 (Abe et al., 2017). This suggests the intriguing possibility that NLRP14 inhibits nucleic acid sensing to prevent germ cells from inappropriate responses to exogenous nucleic acid.
This report shows that NLRP12 interacts with the E3 ubiquitin ligase TRIM25 to reduce TRIM25-mediated Lys63-linked ubiquitination of RIG-I, but it also enhances Lys48-linked ubiquitination of RIG-I mediated by RNF125, leading to its degradation. We demonstrate that an NLR protein modifies the post-translational modification of RIG-I by TRIM25, resulting in reduced IFN-I production in both cells and intact animals.
While the expression of most NLR mRNA and/or proteins is enhanced by microbial infection, human NLRP12 protein (Figure 4D and 4E) and transcript and murine Nlrp12 transcript (Figure S2C, D) are down-regulated by VSV infection. The reduction of NLRP12/Nlrp12 expression may be one strategy by which the inhibitory effects of NLRP12 are relaxed when cells encounter a viral invasion thus unleashing TRIM25 and RIG-I. The down-regulation of NLR expression by microbial stimuli is also observed for another inhibitory NLR, NLRC3 (Schneider et al., 2012). Thus down-regulation of Nlrp12 and Nlrc3 expression provides a common mechanism by which the functions of these inhibitory NLRs are controlled. However, reduced NLRP12 protein and mRNA expression cannot be the only mechanism that reduces NLRP12-RIG-I interaction, as VSV reduced NLRP12-TRIM25 association even when NLRP12 is under the control of an exogenous promoter (Fig. 4D). Our study also shows that the impact of NLRP12 is most obvious at a lower MOI, while its influence wanes at higher MOI. This potentially may have relevance during a cytokine storm elicited by a severe/vigorous viral infection, where inflammatory cytokines are produced at high and potentially harmful levels and cannot be attenuated. Thus a cytokine storm may partly be attributed to the inability of inhibitors such as NLRP12 to mitigate this inflammatory response at high MOI.
Others reported that NLRP12 is an intrinsic regulator of T cells that negatively regulates T cell activation to augment autoreactivity (Gharagozloo et al., 2018). However we did not observe an impact of NLRP12 in T cell-mediated antiviral response. Instead, deletion of Nlrp12 expression in myeloid cells augmented host resistance against VSV infection. These results indicate that Nlrp12 in myeloid cells but not T cells is important for host response in the viral infection model tested here. In another paper, the presence of Nlrp12 was found to enhance neutrophil migration upon influenza viral infection. Thus NLRP12 may exhibit different outcomes depending on the virus, route of infection, the dosage of virus used to cause pathology and the genetic background (Hornick et al., 2018). For example, in our study, we noted that the attenuating effect of NLRP12 diminishes with greater viral load (Fig. 1L). In both the colitis model reported previously (Allen et al., 2012; Zaki et al., 2011) and the viral infection model studied here, NLRP12 serves to attenuate inflammatory response. The colitis model showed increased inflammatory cytokines such as TNF and IL-6, while the current work focuses on increased type I interferons (IFN-I) in Nlrp12−/− mice. However, we also find that TNF is elevated (Figure 1B and Figure 6I) in Nlrp12−/− mice infected with the virus. Thus findings in the colitis model and the viral infection study reported here are consistent. However a major difference between the two models is the involvement of the microbiome. It has been reported that NLRP12 attenuates colitis by promoting protective commensal bacterial growth (Chen et al., 2017). Our experiment did not show a role for the microbiota during VSV infection. In the colitis study, we observed elevated basal immune signaling in Nlrp12−/− mice that is independent of the gut microbiota. Elevated immune signals observed at the basal stage in Nlrp12−/− mice likely contribute to greater immune activation observed in viral infection. Finally, accumulating evidence demonstrates that sustained and abnormal activation of RLRs can cause autoimmune diseases resulting from elevated interferon (Kato et al., 2017). In addition to inflammatory disorders such as colitis, NLRP12 may also offer clues for patients suffering from interferonopathies-based autoimmunity.
In summary, a balance between positive and negative regulatory loops is crucial for maintaining homeostasis. This work supports the model that under resting conditions NLRP12 in the myeloid lineage interacts with TRIM25 to prevent RIG-I activation by inhibiting TRIM25-RIG-I interaction and Lys63-linked ubiquitination of RIG-I. During a viral infection, NLRP12-mediated inhibition is relieved by its reduced expression and possibly other mechanisms which unleash RIG-I-mediated signaling. In the absence of NLRP12, phosphorylated NF-κB p65 and IRF3 are increased, and host anti-viral response is freed and/or accelerated. Other works have shown that NLRP12 is essential for the maintenance of tumor-suppression, gut commensals and the prevention of autoimmunity. Thus, the presence of NLRP12 is important for maintaining homeostasis and serves as a checkpoint of innate immune response.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jenny P-Y Ting (jenny_ting@med.unc.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals and housing:
All studies with Nlrp12–/– were conducted in accordance with the UNC Chapel Hill IACUC approved animal protocol (#12–106) and Guide for the Care and Use of Laboratory Animals, while studies involving Nlrp12–/–#2 were in compliance with the Guide for the Care and Use of Laboratory Animals of the Taiwanese Council of Agriculture in a protocol approved by the Institutional Animal Care and Use Committee (IACUC) of National Yang-Ming University (IACUC #1060201). All mice were housed under specific-pathogen-free (SPF) conditions. Nlrp12–/– mice were originally provided by Millenium Pharmacetical and previously studied by us; these were backcrossed for at least nine generation onto the C57BI/6/J background (Allen et al., 2012). WT and Nlrp12−/− mice in this study were generated from common Nlrp12+/− parents, and experiments used littermate controls, or their immediate descendants. Germ-free mice were derived by sterile embryo transfer by the National Gnotobiotic Rodent Resource Center at UNC- Chapel Hill (Chen et al., 2017). A second Nlrp12-deficient (Nlrp12−/−#2) strain was generated at the Transgenic Mouse Model Core under the National Core Facility Program for Biotechnology in Taiwan using the CRISPR/Cas9 system and housed at the SPF animal core of National Yang-Ming University. The generation of this second gene-deficient strain was necessitated by our inability to obtain a Material Transfer agreement from the original source to ship these mice to Taiwan. A single guide RNA (sgRNA) containing a targeting sequence (crRNA sequence) at exon 1 of the Nlrp12 allele and tracer RNA sequence were designed and generated, and the donor vector containing homologous sequence and a new insertion of three stop codons at exon 1 were injected into C57BL/6J blastocysts with Cas9 mRNA and sgRNA. Genotyping was performed by PCR analysis with Nlrp12 3×stop primer, followed by digesting with Ase I restriction enzyme to determine the homozygosity and heterozygosity of stop codons knock-in Nlrp12 allele (Figure S1). The sequence of sgRNA was input into the website http://benchling.com/sign to check for potential off-target sites. The off-target test was carried out in two founders, and no off-target effect was seen at all potential risk sites. For the myeloid-specific Nlrp12-deficient mice, Nlrp12flox/flox mice were generated at the UNC Animal Models Core using a targeting construct, which contained the loxP sites flanking exon 3 of Nlrp12, next to a FRT flanked neomycin resistance cassette inserted in the intron 2. Targeted ES cells were injected into C57BL/6-albino blastocysts, and the resulting male chimera offsprings were mated with female Rosa26-Flpe mice on a C57BL/6J-albino background to create heterozygous Nlrp12flox/+ mice and to remove the neomycin cassette. The Nlrp12flox/+ offsprings were then bred with C57BL/6J mice for at least another seven generations before they were inbred to obtain the Nlrp12flox/flox control mice. Nlrp12flox/flox mice were then crossed with LysM-Cre mice to generate the F1 generation. Nlrp12flox/flox LysM-Cre+ and Nlrp12flox/flox LysM-Cre− (control) littermates from the F2 generation were used for future breeders or experiments. Myeloid-specific Nlrp12 KO (Nlrp12flox/flox LysM-Cre+) mice and control littermates (Nlrp12flox/flox LysM-Cre−) were genotyped and cohoused before the VSV infection experiment. Rag2−/− mice were originally from the Jackson Laboratory and were housed under SPF conditions before animal experiment at National Yang Ming University.
Human subjects:
Human whole blood was obtained from healthy donors at the Taipei Blood Center of the Taiwan Blood Services Foundation, under a protocol (AS-IRB02–103202) approved by the IRB of the Clinical Center of the Department of Health, Taiwan. Written informed consent was obtained from all donors.
Virus:
Vesicular stomatitis virus-Indiana strain (VSV), originally obtained from ATCC (ATCC VR-1419), was grown in the Vero cell. Vero cells were infected with VSV (multiplicity of infection, MOI of 0.01), and virus was harvested from culture supernatants 48 hr post-infection. Virus titers were determined by plaque formation in Vero cells with an overlay medium containing 1% methyl cellulose in complete DMEM medium (Life Technologies). VSV-infected cells were fixed with 0.2% of crystal violet in a 20% ethanol buffer to visualize plaques by 4 days post-infection. VSV-Indiana strain was used for in vitro and in vivo experiments throughout this study.
METHOD DETAILS:
Cell preparation:
Peripheral blood mononuclear cells (PBMCs) were isolated from whole blood of healthy human donors by standard density-gradient centrifugation with Ficoll-Paque (Amersham Biosciences). CD14+ cells were purified from PBMCs by high-gradient magnetic sorting, using the VarioMACS technique with anti-CD14 microbeads (Miltenyi Biotec GmbH). Cells were then cultured in complete RPMI 1640 medium (Life Technologies) supplemented with 10 % (v/v) fetal calf serum (FCS), 20 ng/ml human GM-CSF (R&D Systems) and 10 ng/ml human IL-4 (R&D Systems) for 6 days (immature DC) (Chen et al., 2008). Human whole blood was obtained from healthy donors at the Taipei Blood Center of the Taiwan Blood Services Foundation, under a protocol (AS-IRB02–103202) approved by the IRB of the Clinical Center of the Department of Health, Taiwan. Written informed consent was obtained from all donors. For mouse bone marrow-derived DC (BMDC), bone marrow cells were isolated from femurs and tibias and cultured in RPMI 1640 complete medium supplemented with 10% (v/v) FCS, L-Glutamine, pen/strep and 40 ng/ml recombinant mouse GM-CSF (R&D Systems) for 9 days (Chen et al., 2008).
Virus infection and ligand stimulation of human moDC and mouse BMDM:
Human monocyte-derived dendritic cells (moDC) or mouse bone marrow derived dendritic cells (BMDC) were seeded at cell culture plates with a confluency of 80% in complete RPMI medium overnight. The cells were then washed with PBS and infected with VSV at the indicated MOI at 37°C in serum-free RPMI for 1 hr. The cells were then washed with PBS and cultured in the complete RPMI. For ligand stimulation, 5’ppp-dsRNA and LMW poly(I:C) were suspended in LyoVec™ transfection reagent (InvivoGen) for 15 min at room temperature according to the manufacturer’s instruction. Mouse BMDC were seeded in 24-well plates at a density of 3×105 per well, and 25μl of 5’ppp-dsRNA/LyoVec™ (0.01 μg/μl) or LMW poly(I:C)/LyoVec™ transfection reagent (0.03 μg/μl) was then transfected into BMDC, respectively. Supernatants and cells were harvested at 12 hr. IFN-β protein and Ifnb/Ifna4 transcripts were measured by ELISA and real time qPCR, respectively.
RNA isolation and mRNA analysis:
For mRNA detection, total RNA was extracted from GWAT lysate using TRIzol™(Thermo Scientific). cDNA was generated from the total RNA by using ReverAid first strand cDNA Synthesis Kit (Cat. K1622, Thermo Scientific, Inc.). Quantitative RT-PCR was performed on a StepOnePlus™ 7 Real-Time PCR System with SYBR™ Green PCR Master Mix (Cat. BIO-98005, Thermo Scientific, Inc.).
Transfection and luciferase reporter analysis:
HEK293 cells were seeded in 24-well plates at a density of 3 ×105 per well and transfected using lipofectamine™ 2000 (Cat. 11668019, Thermo Scientific, Inc.) according to the manufacturer’s instruction. Ten ng of pRT-TK Renilla luciferase reporter plasmid and 100 ng of firefly luciferase reporter plasmids were co-transfected with indicated plasmids. Luciferase activity was measured 24 hr after transfection using the Dual-Glo Luciferase Assay System (Cat. E2920, Promega, Inc.).
Human NLRP12 knockdown experiment:
Human moDC seeded in 12-well plates at a density of 5 ×105 per well were transfected with either 40 nM of pre-validated NLRP12 pool siRNA (sc-45388, Santa Cruz, CA) or a control siRNA using HiPerFect Transfection reagent (Cat. 301704, Qiagen) for 48 h. Cells were then stimulated with VSV (MOI 1). Supernatants and cell lysates were at indicated time points for cytokine determination and immune blots analysis.
Co-immunoprecipitation and ubiquitination assays:
Cells were lysed in 1 % Triton X-100 isotonic buffer. Co-immunoprecipitations were performed using anti-V5-agarose gel, anti-HA-agarose beads and anti-Flag-M2 gel (all from Sigma) or 1μg antibody specific for TRIM5, RNF125, RIG-I, TRAF3, TRAF6 or IKKα with UltraLink Protein A/G beads (Pierce). For in vitro ubiquitination assays, mouse BMDC or HEK293 cells were lysed in 1% SDS isotonic buffer. Lysates were boiled for 15 min to remove any non-covalent interactions, followed by centrifugation for 10 min at 15,000 rpm. Supernatants were transferred to a new tube, and diluted with 1% Triton X-100 buffer for immunoprecipitation with anti-Flag M2 or 1μg of anti-RIG-I antibody and UltraLink Protein A/G beads. Immunocomplex was harvested and washed with 0.1 % Triton X-100 isotonic buffer and resuspended with protein loading buffer.
Viral genome quantification and determination of virus titer:
Total RNA was isolated from homogenized cells or brain tissues using QIAamp RNA Mini Kit (Cat. 52904, QIAGEN, Inc) according to manufacturer’s protocol. The RNA was reverse transcribed into cDNA and viral genome was quantified by SYBR green qPCR using VSV-specific primers. The relative VSV replication was obtained by setting the measured VSV expression in wild type cells to 100%. The relative percentage of VSV expression in Nlrp12−/− cells was calculated after comparing with wild type cells (100%). The infective VSV particles from cells, serum or brains were determined by plaque assay in Vero cells, and denoted as plaque forming units per sample volume (pfu/ml) or per sample gram tissue (pfu/g).
Confocal microscopy, histopathology and immunohistochemistry:
Wild type and Nlrp12−/− BMDC (5×105) were seeded onto coverslips in a 12-well culture dish and were grown overnight before inoculation with VSV at an MOI 0.5 for 1 hr. Virus was removed and cells were replenished with RPMI medium containing FCS, and then were harvested at indicated time points. Cells were fixed with 4% paraformaldehyde (PFA), followed by permeabilization with 0.1% Tritox-100. Cells were stained with anti-p65 (Cat. 8242, clone: D14E12; Cell Signaling, Inc.) and TRITC-conjugated anti-rabbit antibody (Cat: 111-225-003, Jackson ImmunoReasearch, Inc.) and then were counterstained for nucleic acids and F-actin with DAPI and Alexa Fluor 488 Phalloidin (Cat: A12379, Thermo Scientific, Inc.), respectively. For the detection of NLRP12 in human DC, cells were seeded and infected with VSV at an MOI of 1 for 1 hr. Virus was then removed, and cells were replenished with RPMI medium containing FCS and then harvested at indicated time points. The infected cells were further incubated with mito-tracker® Deep Red FM at the final concentration of 500 nM for 30 min at 37°C. After incubation, cells were fixed in ice-cold 100% methanol for 15 min at −20°C and rinse three times with PBS for 5 min. The immunostaining was performed with anti-NLRP12 antibody (Cat. PA5–21027, ThermoFisher Inc.) and TRITC-conjugated anti-rabbit antibody. Cells were analyzed using confocal microscopy (Leica: TCS-SP5-MP-SMD) and images were analyzed with Leica LAS AF software. For histopathology and immunohistochemistry, brains were harvested and fixed in 10% neutral buffered formalin fixation solution. Fixed tissue was embedded in paraffin, sectioned and stained with hematoxylin and eosin (H&E). For immunohistochemistry, formalin-fixed paraffin-embedded tissues were cut into 5 μm sections and slides were stained with anti-GFAP (Cat. 3670, clone: GA5; Cell Signaling, Inc.) and developed with chromogen diaminobenzidine (Cat. K3468, DAKO Inc.) and counterstained with Gills Haematoxylin.
Experimental VSV infection mouse model and T cell adoptive transfer:
Four-week-old WT and Nlrp12−/−#2 littermate mice originating from the same breeders were cohoused for an additional 4–6 weeks to normalize the microbiome difference. Age- and sex-matched WT and Nlrp12−/−#2 littermate mice (8- to10-week-old) from the above conditions were used for in vivo VSV infection (1×106 PFU). For the myeloid-specific Nlrp12-deficient mice, mice were cohoused for 4 weeks before the VSV infection experiment, and 8- to10-week-old Nlrp12flox/flox LysM-Cre+ mice and control littermates (Nlrp12flox/flox LysM-Cre−) which were age- and sex-matched were used for the VSV infection experiment. For the germ free (GF) mice, age-, weight-, and sex-matched GF WT and Nlrp12−/− mice were used for the VSV infection study. All of mice were anesthetized by injecting ketamine (150 mg/Kg) and rampone (30 mg/Kg) intraperitoneally, followed by administrating of 15 μl volume of VSV (1×106 PFU) in the left nostril. Mice were monitored and body weight was recorded daily, and mice were sacrificed when displaying clinical morbidity or frank hind limb paralysis. The body weight of dead mice was included using the weight at the time of death in successive time points. In some experiments, blood and brain were harvested by 24 hr post-infection to analyze the amount of IFN-β protein, transcript level of ifn-β gene and viral titer.
For the T cell adoptive transfer experiment, splenocytes and peripheral LNs were harvested from WT and Nlrp12−/−#2 mice, and bulk T cells were positively enriched using anti-Thy1.2 (CD90.2) magnetic beads (Miltenyi Biotec 130-049-101). Rag2-deficient male mice (10 to 12 week-old) were reconstituted with 2×107 T cells from WT or Nlrp12−/−#2 mice (8 week-old male mice). Two days later, mice were anesthetized by injecting ketamine and rampone intraperitoneally, followed by administrating of 15 μl volume of VSV (1×106 PFU) in the left nostril. Mice were monitored and body weight was recorded daily. Mouse brains were harvested for IFNγ expression in T cells at day 3 and day 9 post-infection.
Isolation of mononuclear cells from the CNS and flow cytometry analysis:
Mononuclear cells (MNCs) were isolated from brains of VSV-infected mice as described (Chen et al., 2012). Briefly, PBS-perfused brains were minced into pieces, and the cell suspension was digested with HBSS buffer containing 0.05 % (v/v) collagenase IV (Cat. C5138, Sigma Inc.) and 0.025 U/mL DNase I (Cat. D5025, Sigma Inc.) at room temperature for 30 min. Brain homogenates were passed through a cell strainer (40 μm) and resuspended in 40% (w/v) Percoll-HBSS solution and overlaid onto 70% (w/v) Percoll-HBSS. Thirty % (w/v) Percoll-HBSS and HBSS were added prior to centrifugation at 200 g at 20° C for 40 min. After centrifugation, brain mononuclear cells (MNC) were recovered from the 70 – 40% interphase and washed with PBS to remove the residual Percoll. Isolated brain mononuclear cells were re-suspended in FACS buffer and stained as follows. To determine the expression of TNF and IFN-α in conventional myeloid DC (CD11b+CD11c+), MNC (5 × 105/reaction) were incubated with a cocktail of surface marker antibodies including anti-CD11b-Alexa Fluor 647 (Cat. C101220, Biolegend Inc.) and anti-CD11c-BV421 (Cat. 5627852, BD Bioscience Inc.). The stained cells were then fixed and permeabilized with the CytoFix/perm solution (Cat. 554714, BD Bioscience Inc.), followed by washing with BD wash buffer and staining with anti-IFN-α-FITC (Cat. 22100–3, PBL Inc.) and anti-TNF-APC/Cy7 (Cat. 506344, Biolegend Inc.) for 30 min on ice in the dark. After washing with FACS buffer, cells were re-suspended in 1xPBS and analysed with the Beckman Coulter Cytoflex. To determine RIG-I expression in DC and macrophages at 12 hr post infections, MNC (5 × 105/reaction) were incubated with a cocktail of surface marker antibodies including anti-CD11b-PE (Cat. 101208, BD Bioscience Inc.), anti-CD11c-BV421(Cat. 5627852, BD Bioscience Inc.) and anti-F4/80-Alexa Fluor 647(Cat., Bio-Rad Inc.). Stained cells were then fixed and permeabilized with CytoFix/perm solution, followed by washing with BD wash buffer and staining with anti- RIG-I Alexa Fluor 488 (Cat. sc- 376845, Santa Cruz Inc.) for 30 min on ice in the dark. After washing with FACS buffer, cells were resuspended in 1xPBS and analysed with the Beckman Coulter Cytoflex. To determine myeloid infiltrating cells at day 5 post infection, MNC were incubated with a cocktail of surface markers including CD3-APC/Cy7, CD11b-Alexa Fluor 647, Ly6G-FITC, Ly6C-PE and CD11c-BV421. To determine IFNγ expression in T cells at day 3 and day 9 post- infection, MNC (5 × 105/reaction) were incubated with a cocktail of surface marker antibodies including anti-CD3-PE (Cat. 553064, BD Bioscience Inc.), anti-CD4-Alexa Fluor 647 (Cat. 553051, BD Bioscience Inc.) and anti-CD8-Alexa Fluor 488 (Cat. 100726, Biolegend Inc.). The stained cells were then fixed and permeabilized with CytoFix/perm solution, followed by washing with BD wash buffer and staining with anti-IFNγ-Alexa Fluor 700 (Cat. 505823, Biolegend Inc.). Stained cells were washed with FACS buffer and resuspended in PBS for analysis with the Beckman Coulter CytoFlex. All of the data were processed using FlowJo software (version 10, Treestar).
QUANTIFICATION AND STATISTICAL ANALYSIS:
All data were presented as mean ± s.e.m. and analyzed using GraphPad Prism software (Version 5.0). An unpaired, two-tailed student’s t test (for parametric data) or Mann-Whitney test (for nonparametric data) was used to determine the significance between two sets of data. Error bar represents mean ± SEM with *p<0.05 and **p<0.01.
Supplementary Material
Movie S1. Mobility of WT mice after VSV infection, related to Figure 6.
Movie S2. Mobility of Nlrp12−/−#2 mice after VSV infection, related to Figure 6. WT (Movies S1) and Nlrp12−/−#2 (Movies S2) mice were intranasally inoculated with VSV (1x106 pfus). A 30s video recorded the mobility of WT (Movies S1) and Nlrp12−/−#2 (Movies S2) mice 4 days post-VSV intranasal infection. These videos are related to Figure 6.
Highlights.
NLRP12 reduces interferon and cytokine responses to RNA viruses and 5’ppp-dsRNA
NLRP12 associates with TRIM25 to disrupt Lys63-ubiquitination and activation of RIG-I
Vesicular stomatitis virus (VSV) infection downregulates NLRP12
Myeloid-specific Nlrp12-deficient mice have increased resistance to VSV
ACKNOWLEDGEMENTS
This work was supported by U19-AI109965, DK094779, CA156330 and AI029564 (J.P.-Y.T.), NSC 103–2320-B-038–004-MY2, MOST 104–2320-B-010–0440-MY3, “Cancer Progression Research Center, National Yang-Ming University” from the Ministry of Education (MOE) in Taiwan, “Development and Construction Plan” of the School of Medicine, National Yang-Ming University, 107F-M01 (S.-T.C.), NIH T32 AI07273 (J.P.-Y.T. and J.W.T.).
We thank the following for core and technical support: National Yang-Ming University Imaging Core Facility, Flow Cytometry Core Facility, Animal Studies Core Facility and Taiwan Animal Consortium, Taiwan Mouse Clinic, UNC Animal Models Core, Animal Histopathology Core Facility and Animal Studies Core Facility. We thank Dr. S.-L. H. for providing critical reagents.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- Abe T, Lee A, Sitharam R, Kesner J, Rabadan R, and Shapira SD (2017). Germ-Cell-Specific Inflammasome Component NLRP14 Negatively Regulates Cytosolic Nucleic Acid Sensing to Promote Fertilization. Immunity 46, 621–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen IC, Wilson JE, Schneider M, Lich JD, Roberts RA, Arthur JC, Woodford RM, Davis BK, Uronis JM, Herfarth HH, et al. (2012). NLRP12 suppresses colon inflammation and tumorigenesis through the negative regulation of noncanonical NF-kappaB signaling. Immunity 36, 742–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimoto K, Takahashi H, Hishiki T, Konishi H, Fujita T, and Shimotohno K (2007). Negative regulation of the RIG-I signaling by the ubiquitin ligase RNF125. Proc Natl Acad Sci U S A 104, 7500–7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur JC, Lich JD, Ye Z, Allen IC, Gris D, Wilson JE, Schneider M, Roney KE, O’Connor BP, Moore CB, et al. (2010). Cutting edge: NLRP12 controls dendritic and myeloid cell migration to affect contact hypersensitivity. J Immunol 185, 4515–4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ataide MA, Andrade WA, Zamboni DS, Wang D, Souza Mdo C, Franklin BS, Elian S, Martins FS, Pereira D, Reed G, et al. (2014). Malaria-induced NLRP12/NLRP3-dependent caspase-1 activation mediates inflammation and hypersensitivity to bacterial superinfection. PLoS Pathog 10, e1003885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benko S, Kovacs EG, Hezel F, and Kufer TA (2017). NLRC5 Functions beyond MHC I Regulation-What Do We Know So Far? Front Immunol 8, 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broz P, and Dixit VM (2016). Inflammasomes: mechanism of assembly, regulation and signalling. Nat Rev Immunol 16, 407–420. [DOI] [PubMed] [Google Scholar]
- Cai S, Batra S, Del Piero F, and Jeyaseelan S (2016). NLRP12 modulates host defense through IL-17A-CXCL1 axis. Mucosal Immunol 9, 503–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Wilson JE, Koenigsknecht MJ, Chou WC, Montgomery SA, Truax AD, Brickey WJ, Packey CD, Maharshak N, Matsushima GK, et al. (2017). NLRP12 attenuates colon inflammation by maintaining colonic microbial diversity and promoting protective commensal bacterial growth. Nat Immunol 18, 541–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ST, Lin YL, Huang MT, Wu MF, Cheng SC, Lei HY, Lee CK, Chiou TW, Wong CH, and Hsieh SL (2008). CLEC5A is critical for dengue-virus-induced lethal disease. Nature 453, 672–676. [DOI] [PubMed] [Google Scholar]
- Chen ST, Liu RS, Wu MF, Lin YL, Chen SY, Tan DT, Chou TY, Tsai IS, Li L, and Hsieh SL (2012). CLEC5A regulates Japanese encephalitis virus-induced neuroinflammation and lethality. PLoS Pathog 8, e1002655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Li Y, Zhu L, Liu D, Songyang Z, Wang HY, and Wang RF (2012). NLRP4 negatively regulates type I interferon signaling by targeting the kinase TBK1 for degradation via the ubiquitin ligase DTX4. Nat Immunol 13, 387–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Zhu L, Xia X, Wang HY, Legras X, Hong J, Ji J, Shen P, Zheng S, Chen ZJ, et al. (2010). NLRC5 negatively regulates the NF-kappaB and type I interferon signaling pathways. Cell 141, 483–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BK, Wen H, and Ting JP (2011). The inflammasome NLRs in immunity, inflammation, and associated diseases. Annu Rev Immunol 29, 707–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detje CN, Lienenklaus S, Chhatbar C, Spanier J, Prajeeth CK, Soldner C, Tovey MG, Schluter D, Weiss S, Stangel M, et al. (2015). Upon intranasal vesicular stomatitis virus infection, astrocytes in the olfactory bulb are important interferon Beta producers that protect from lethal encephalitis. J Virol 89, 2731–2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, et al. (2007). TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 446, 916–920. [DOI] [PubMed] [Google Scholar]
- Gharagozloo M, Mahmoud S, Simard C, Mahvelati TM, Amrani A, and Gris D (2018). The Dual Immunoregulatory function of Nlrp12 in T Cell-Mediated Immune Response: Lessons from Experimental Autoimmune Encephalomyelitis. Cells 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharagozloo M, Mahvelati TM, Imbeault E, Gris P, Zerif E, Bobbala D, Ilangumaran S, Amrani A, and Gris D (2015). The nod-like receptor, Nlrp12, plays an anti-inflammatory role in experimental autoimmune encephalomyelitis. J Neuroinflammation 12, 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell LH, and Weinert TA (1989). Checkpoints: controls that ensure the order of cell cycle events. Science 246, 629–634. [DOI] [PubMed] [Google Scholar]
- Hornick EE, Banoth B, Miller AM, Zacharias ZR, Jain N, Wilson ME, Gibson-Corley KN, Legge KL, Bishop GA, Sutterwala FS, et al. (2018). Nlrp12 Mediates Adverse Neutrophil Recruitment during Influenza Virus Infection. J Immunol 200, 1188–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, et al. (2006). 5’-Triphosphate RNA is the ligand for RIG-I. Science 314, 994–997. [DOI] [PubMed] [Google Scholar]
- Jaworska J, Coulombe F, Downey J, Tzelepis F, Shalaby K, Tattoli I, Berube J, Rousseau S, Martin JG, Girardin SE, et al. (2014). NLRX1 prevents mitochondrial induced apoptosis and enhances macrophage antiviral immunity by interacting with influenza virus PB1-F2 protein. Proc Natl Acad Sci U S A 111, E2110–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Oh SW, and Fujita T (2017). RIG-I-Like Receptors and Type I Interferonopathies. J Interferon Cytokine Res 37, 207–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, and Akira S (2005). IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol 6, 981–988. [DOI] [PubMed] [Google Scholar]
- Kim JH, Park ME, Nikapitiya C, Kim TH, Uddin MB, Lee HC, Kim E, Ma JY, Jung JU, Kim CJ, et al. (2017). FAS-associated factor-1 positively regulates type I interferon response to RNA virus infection by targeting NLRX1. PLoS Pathog 13, e1006398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss JL, Zeng R, Hickman-Brecks CL, Wilson JE, Ting JP, and Novack DV (2015). NLRP12 provides a critical checkpoint for osteoclast differentiation. Proc Natl Acad Sci U S A 112, 10455–10460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kufer TA, and Sansonetti PJ (2011). NLR functions beyond pathogen recognition. Nat Immunol 12, 121–128. [DOI] [PubMed] [Google Scholar]
- Li H, Zhang S, Li F, and Qin L (2016). NLRX1 attenuates apoptosis and inflammatory responses in myocardial ischemia by inhibiting MAVS-dependent NLRP3 inflammasome activation. Mol Immunol 76, 90–97. [DOI] [PubMed] [Google Scholar]
- Lich JD, Williams KL, Moore CB, Arthur JC, Davis BK, Taxman DJ, and Ting JP (2007). Monarch-1 suppresses non-canonical NF-kappaB activation and p52-dependent chemokine expression in monocytes. J Immunol 178, 1256–1260. [DOI] [PubMed] [Google Scholar]
- Lu C, Xu H, Ranjith-Kumar CT, Brooks MT, Hou TY, Hu F, Herr AB, Strong RK, Kao CC, and Li P (2010). The structural basis of 5’ triphosphate double-stranded RNA recognition by RIG-I C-terminal domain. Structure 18, 1032–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukens JR, Gurung P, Shaw PJ, Barr MJ, Zaki MH, Brown SA, Vogel P, Chi H, and Kanneganti TD (2015). The NLRP12 Sensor Negatively Regulates Autoinflammatory Disease by Modulating Interleukin-4 Production in T Cells. Immunity 42, 654–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao AP, Li S, Zhong B, Li Y, Yan J, Li Q, Teng C, and Shu HB (2010). Virus-triggered ubiquitination of TRAF3/6 by cIAP1/2 is essential for induction of interferon-beta (IFN-beta) and cellular antiviral response. J Biol Chem 285, 9470–9476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore CB, Bergstralh DT, Duncan JA, Lei Y, Morrison TE, Zimmermann AG, Accavitti-Loper MA, Madden VJ, Sun L, Ye Z, et al. (2008). NLRX1 is a regulator of mitochondrial antiviral immunity. Nature 451, 573–577. [DOI] [PubMed] [Google Scholar]
- Papale A, Kummer E, Galbiati V, Marinovich M, Galli CL, and Corsini E (2016). Understanding chemical allergen potency: role of NLRP12 and Blimp-1 in the induction of IL-18 in human keratinocytes. Arch Toxicol. [DOI] [PubMed] [Google Scholar]
- Peisley A, Wu B, Xu H, Chen ZJ, and Hur S (2014). Structural basis for ubiquitin-mediated antiviral signal activation by RIG-I. Nature 509, 110–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, and Reis e Sousa C (2006). RIG-I-mediated antiviral responses to single-stranded RNA bearing 5’-phosphates. Science 314, 997–1001. [DOI] [PubMed] [Google Scholar]
- Saha SK, Pietras EM, He JQ, Kang JR, Liu SY, Oganesyan G, Shahangian A, Zarnegar B, Shiba TL, Wang Y, et al. (2006). Regulation of antiviral responses by a direct and specific interaction between TRAF3 and Cardif. EMBO J 25, 3257–3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider M, Zimmermann AG, Roberts RA, Zhang L, Swanson KV, Wen H, Davis BK, Allen IC, Holl EK, Ye Z, et al. (2012). The innate immune sensor NLRC3 attenuates Toll-like receptor signaling via modification of the signaling adaptor TRAF6 and transcription factor NF-kappaB. Nat Immunol 13, 823–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth RB, Sun L, Ea CK, and Chen ZJ (2005). Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 122, 669–682. [DOI] [PubMed] [Google Scholar]
- Silveira TN, Gomes MT, Oliveira LS, Campos PC, Machado GG, and Oliveira SC (2017). NLRP12 negatively regulates proinflammatory cytokine production and host defense against Brucella abortus. Eur J Immunol 47, 51–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofroniew MV, and Vinters HV (2010). Astrocytes: biology and pathology. Acta Neuropathol 119, 7–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X, Sun L, Chen J, and Chen ZJ (2018). Detection of Microbial Infections Through Innate Immune Sensing of Nucleic Acids. Annu Rev Microbiol 72, 447–478. [DOI] [PubMed] [Google Scholar]
- Ting JP, Duncan JA, and Lei Y (2010). How the noninflammasome NLRs function in the innate immune system. Science 327, 286–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truax AD, Chen L, Tam JW, Cheng N, Guo H, Koblansky AA, Chou WC, Wilson JE, Brickey WJ, Petrucelli A, et al. (2018). The Inhibitory Innate Immune Sensor NLRP12 Maintains a Threshold against Obesity by Regulating Gut Microbiota Homeostasis. Cell Host Microbe 24, 364–378 e366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulland TK, Jain N, Hornick EE, Elliott EI, Clay GM, Sadler JJ, Mills KA, Janowski AM, Volk AP, Wang K, et al. (2016). Nlrp12 mutation causes C57BL/6J strain-specific defect in neutrophil recruitment. Nat Commun 7, 13180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vladimer GI, Weng D, Paquette SW, Vanaja SK, Rathinam VA, Aune MH, Conlon JE, Burbage JJ, Proulx MK, Liu Q, et al. (2012). The NLRP12 inflammasome recognizes Yersinia pestis. Immunity 37, 96–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Manji GA, Grenier JM, Al-Garawi A, Merriam S, Lora JM, Geddes BJ, Briskin M, DiStefano PS, and Bertin J (2002). PYPAF7, a novel PYRIN-containing Apaf1-like protein that regulates activation of NF-kappa B and caspase-1-dependent cytokine processing. J Biol Chem 277, 29874–29880. [DOI] [PubMed] [Google Scholar]
- Wang P, Zhu S, Yang L, Cui S, Pan W, Jackson R, Zheng Y, Rongvaux A, Sun Q, Yang G, et al. (2015). Nlrp6 regulates intestinal antiviral innate immunity. Science 350, 826–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, and Fujita T (2004). The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol 5, 730–737. [DOI] [PubMed] [Google Scholar]
- Zaki MH, Man SM, Vogel P, Lamkanfi M, and Kanneganti TD (2014). Salmonella exploits NLRP12-dependent innate immune signaling to suppress host defenses during infection. Proc Natl Acad Sci U S A 111, 385–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki MH, Vogel P, Malireddi RK, Body-Malapel M, Anand PK, Bertin J, Green DR, Lamkanfi M, and Kanneganti TD (2011). The NOD-like receptor NLRP12 attenuates colon inflammation and tumorigenesis. Cancer Cell 20, 649–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamoshnikova A, Gross CJ, Schuster S, Chen KW, Wilson A, Tacchini-Cottier F, and Schroder K (2016). NLRP12 is a neutrophil-specific, negative regulator of in vitro cell migration but does not modulate LPS- or infection-induced NF-kappaB or ERK signalling. Immunobiology 221, 341–346. [DOI] [PubMed] [Google Scholar]
- Zhang L, Mo J, Swanson KV, Wen H, Petrucelli A, Gregory SM, Zhang Z, Schneider M, Jiang Y, Fitzgerald KA, et al. (2014). NLRC3, a member of the NLR family of proteins, is a negative regulator of innate immune signaling induced by the DNA sensor STING. Immunity 40, 329–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1. Mobility of WT mice after VSV infection, related to Figure 6.
Movie S2. Mobility of Nlrp12−/−#2 mice after VSV infection, related to Figure 6. WT (Movies S1) and Nlrp12−/−#2 (Movies S2) mice were intranasally inoculated with VSV (1x106 pfus). A 30s video recorded the mobility of WT (Movies S1) and Nlrp12−/−#2 (Movies S2) mice 4 days post-VSV intranasal infection. These videos are related to Figure 6.