Abstract
Human Caprin-1 and Caprin-2 are prototypic members of the caprin (cytoplasmic activation/proliferation-associated protein) protein family. Vertebrate caprin proteins contain two highly conserved homologous regions (HR1 and HR2) and C-terminal RGG motifs. Drosophila caprin (dCaprin) shares HR1 and RGG motifs but lacks HR2. Caprin-1 and Caprin-2 have important and non-redundant functions. The detailed molecular mechanism s of their actions remain largely unknown. Previously, we determined the crystal structure of a ~120-residue fragment of Caprin-1 within the HR1 region. The structure has a noval all α-helical fold that self-associates to form a homodimer. In this study, the crystal structure of a corresponding fragment from Caprin-2 is reported. The Caprin-2 fragment has similar protein fold and dimeric structure as those of the Caprin-1 fragment. Structural comparison reveals that the molecular interactions mediating homodimerization of Caprin-1 and Caprin-2 are largely conserved in the two systems. Structural modelling study of the corresponding dCaprin fragment indicates that dCaprin may also adopt similar dimeric structure. Presence of a dimerization domain within HR1 may represent an evolutionarily conserved feature of the caprin protein family. Interestingly, while Caprin-1 and Caprin-2 adopt similar overall dimeric structures, the two structures have quite different molecular surface properties. In the Caprin-1 dimeric structure, some of the surface areas are known or suspected to function as binding sites for Carpin-1 interacting proteins. The different surface properties of the caprin dimeric structures may dictate their intermolecular interaction with specific protein partners.
Keywords: Caprin-1, Caprin-2, G3BP1, FMRP, RNA stress granule
Introduction
Caprin (cytoplasmic activation/proliferation-associated protein) proteins are characterized by the presence of two homologous regions (HR1 and HR2) that are highly conserved in vertebrates (Grill et al., 2004; Shiina, Shinkura, & Tokunaga, 2005). In human, there are two caprin proteins: Caprin-1 and Caprin-2 (Figure 1). The HR1 regions of Caprin-1 and Caprin-2 are 51% identical & 73% similar, while the HR2 regions are 36% identical & 54% similar. In both proteins, a less conserved E-rich region is present between HR1 and HR2. Caprin-2 harbours an identifiable protein domain at its C-terminus (CRD: C1q-related domain) which is homologous to the globular head domain of the complement protein C1q (Gaboriaud et al., 2003; Garlatti et al., 2010; Miao et al., 2014; Shapiro and Scherer, 1998; Venkatraman Girija et al., 2013). A Drosophila protein (dCaprin) contains a HR1 region homologous to those in Caprin-1 and Caprin-2. Caprin-1 and Caprin-2, as well as dCaprin, contain C-terminal RGG boxes and RG-rich sequences characteristic of some RNA-binding proteins.
Figure 1. Caprin proteins and structure of the dimerization domain of Caprin-2.
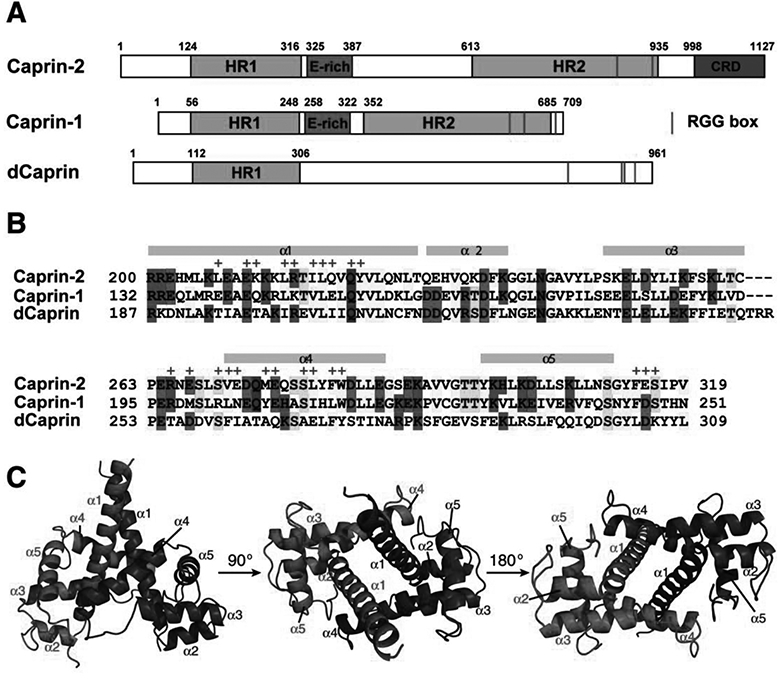
A) Schematic representation of human Caprin-2 and Caprin-1, as well as Drosoplila Caprin (dCaprin). HR1 and HR2: homologous region 1 and 2. CRD: C1q-related domain. The thin red lines indicate the locations of the RGG boxes. B) Sequence of the human Caprin-2 dimerization domain aligned with homologous sequences of Caprin-1 and dCaprin. The secondary structures are indicated above the Caprin-2 sequence. Residues involved in Caprin-2 dimerization are indicate with “+” sign above the sequence. C) Structure of the Caprin-2 homodimer rendered in cartoon mode, viewed from three different angles. The two protomers are coloured red and blue respectively.
Most studies on caprin proteins have been directed to Caprin-1. Caprin-1 assumes many different functions. Caprin-1 plays an essential role in normal cellular proliferation (Grill, et al., 2004; Wang, David, & Schrader, 2005). As a positive regulator of cell proliferation, Caprin-1 is highly expressed in proliferating T or B lymphocytes and hemopoietic progenitors in the thymus and spleen (Grill, et al., 2004). Caprin-1 is required for the G1/S transition in the cell cycle (Wang, et al., 2005). Caprin-1 is also involved in the regulation of synaptic plasticity. As a highly expressed protein in the brain, Caprin-1 is localized in the neuronal RNA granules in dendrites of hippocampal and neocortical pyramidal neurons (Shiina, et al., 2005). Caprin-1 containing granules bind to mRNAs of key proteins for synaptic plasticity and repress their local translation in dendrites of hippocampal and neocortical pyramidal neurons. This translational repression is released when Caprin-1 is dissociated from the neuronal RNA granules as a result of synaptic stimulation (Shiina, et al., 2005). Caprin-1 also regulates interferon (IFN)-mediated antiviral innate immune response. Caprin-1, in complex with Ras-GAP SH3 domain-binding protein 1 and 2 (G3BP1 and G3BP2), is required for the translation of mRNAs of antiviral IFN-stimulated genes (ISGs) (Bidet, Dadlani, & Garcia-Blanco, 2014). Caprin-1 also participates in cellular stress response as a component of cytoplasmic RNA stress granules (SGs). It was shown that Caprin-1 (together with G3BP1) localized into SGs in response to stress (Solomon et al., 2007). Formation of Caprin-1 containing SGs is linked to the pathogenesis of several human diseases, such as tumorigenesis and metastasis in osteosarcoma (OS), viral infection by Japanese encephalitis virus (JEV) (Katoh et al., 2013), hearing loss due to mutations in the Pou4f3 gene (Collin et al., 2008), and Huntington disease (HD) (Ratovitski et al., 2012).
Only a few studies on Caprin-2 have been published (Ding et al., 2008; Konopacka, Greenwood, Loh, Paton, & Murphy, 2015; Shiina and Tokunaga, 2010). All of these studies indicate that Caprin-2 have non-redundant functions from Caprin-1. Caprin-2 level was increased at the stage of differentiation (Aerbajinai, Lee, Wojda, Barr, & Miller, 2004), suggesting its involvement in the developmental transition from rapid proliferation toward terminal differentiation. Caprin-2 and Caprin-1 are both highly expressed in brain and localized to neuronal RNA granules (Shiina and Tokunaga, 2010). However, the Caprin-1 and Caprin-2 containing neuronal granules are different (e.g., G3BP1 and ribosome components are found in the Caprin-1 containing but not the Caprin-2 containing granules). Suppression of either Caprin-1 or Caprin-2by siRNA knockdown reduces dendrite length and spine density in neurons (Shiina and Tokunaga, 2010). Rescue experiments showed that Caprin-1 knockdown was rescued by Caprin-1 expression but not by Caprin-2 expression, and Caprin-2 knockdown was rescued by Caprin-2 expression but not by Caprin-1 expression. In another study, it was shown that Caprin-2 interacted with the low-density lipoprotein receptor-related proteins 5&6 (LRP5/6, co-receptors for Frizzled) and promoted the Wnt signaling pathway (Ding, et al., 2008). Caprin-2 was also found to play a pivotal role in regulating rin-2 enhances the abundance and poly-A tail length of the mRNA of arginine vasopressin (AVP), a hormone that regulates water and salt levels in the whole body (Konopacka, et al., 2015).
In spite of the emerging biological and physiopathological significance of Caprin-1 and Caprin-2, detailed molecular mechanisms of their actions remain largely unknown. Previously, we identified a homidimerization domain of Caprin-1 (residues 132–251) (Wu, Zhu, Huang, & Du, 2015, 2016). In the crystal structure we determined, the Caprin-1 homidimerization domain adopts a novel all α-helical protein fold that self-associates to form a homodimer. Homodimerization of Caprin-1 creates a large concave surface that is dominantly negatively charged and may function as a protein-protein binding groove. Given the fact that the sequence of the Caprin-1 dimerization domain locates within the HR1 region (Figure 1), it is interesting to see whether corresponding sequences in Caprin-2 and dCaprin also adopt similar dimeric structures. In this paper, we present the crystal structure of a Caprin-2 fragment (residues 199–329), which exists as a dimer that closely resembles the Caprin-1 dimer. Interestingly, although the overall dimeric structures of Caprin-1 and Caprin-2 are similar to each other, the two structures have quite different surface properties in terms of spatial distributionof hydrophobicity, hydrophilicity and charges. We also build a structural model of a dCaprin dimer. Insights from the structural and modelling studies are discussed.
Materials and methods
Protein sample preparation and crystallization
A cDNA clone for the full-length human Caprin-2 (GenBank:BC117672, IMAGE clone 40068103) was purchased from American Type Culture Collection (ATCC). The DNA encoding a fragment of Caprin-2 (residues 199–329) was amplified from the plasmid by polymerase chain reaction (PCR) using Phusion DNA polymerase. The PCR product was purified by agarose gel electrophoresis and subsequently processed by T4 DNA polymerase in the presence of 2.5 mM dATP. The processed DNA was inserted into an in-house developed cloning vector using ligation independent cloning (LIC) method. The vector contains DNA sequences encoding the Halo-tag (Los et al., 2008), His-tag, and an octapeptide recognition sequence for human rhinovirus (HRV) 3C protease preceding a specific LIC cloning sequence. Therefore the Caprin-2 fragment was expressed as a fusion protein containing N-terminal Halo and His tags, followed by a HRV 3C protease recognition motif, and the target protein.
The recombinant plasmid was transformed into NiCo21(DE3) E. coli cells (New England Biolabs). Cell culture was grown in LB media until it reached an OD600 of 0.6–0.8. Isopropyl β-D-1-thiogalactopyranoside (IPTG) was added to the culture (at 0.4 mM final concentration) to induce protein expression. After induction, the culture was allowed to grow for 4 hours at 37°C before harvest. The overexpressed proteins were purified by cobalt affinity resin. After elution from the resin with a buffer containing 200 mM imidazole, 25 mM Tris (pH=7.5), 200 mM NaCl, the fusion protein was processed by HRV 3C protease in a dialysis tubing (10 ml of protein solution against 5 liters of dialysis buffer containing 25 mM Tris, pH=7.5, 200 mM NaCl) at 4°C overnight. The cleaved tags were separated from the target protein by a reverse IMAC (immobilized-metal affinity chromatography) process with cobalt affinity resin. The purified proteins were concentrated to a concentration of ~10 mg/ml in a buffer containing 25 mM Tris (pH 7.5), 200 mM NaCl. For Se-Met labelled protein, the bacterial cells were grown in M9 minimal culture. When the cultures reached an OD600 of 0.5, six amino acids (leucine, isoleucine, lysine, phenylalanine, threonine, and valine) were added to the culture at a final concentration of 50–100 mg/L. After growth at 37°C for half hour, L-seleno-methionine was added to the cell cultures at a final concentration of 50 mg/L. The other steps for protein expression and purification were identical to those for the unlabelled protein described above.
Crystallization trials were carried out using seven sets of in-house prepared screening solutions each containing 96 different conditions. The solutions use various PEGs as the precipitants. Other variables include buffers, pH values, salts, and cryoprotectants. The crystallization trials were set up by using 96-well format plates. Multiple crystallization conditions were identified after overnight incubation of the plates at 22°C. After optimizing the crystallization conditions, diffracting crystals were obtained at 22°C by sitting-drop vapour diffusion against 50 microliters of well solution containing 20% PEG8000, 0.1 M Tris (pH8.0), 10% glycerol, using 96-well format crystallization plates.
Data collection, data processing, and structure determination
Data collection was carried out at beamline 21ID-F of LS-CAT at the Advanced Photon Source (Argonne National Laboratory). Data were processed, integrated, and scaled with the programs Mosflm and Scala in CCP4 (Battye, Kontogiannis, Johnson, Powell, & Leslie, 2011). The structure was solved by SAD using a data set collected at the peak wavelength of Se (0.97872 Å) on a single crystal containing Se-Met labelled proteins. Locating the selenium atoms and building the initial structure were performed using the PHENIX package (Adams et al., 2011). Interactive model building was carried out with Coot (Emsley, Lohkamp, Scott, & Cowtan, 2010). The structure was refined using PHENIX (Afonine et al., 2012). Structure determination statistics are shown in Table 1. The figures were prepared with the program PyMOL (The PyMOL Molecular Graphics System, Version 1.5.0.4 Schrödinger, LLC.). Atomic coordinates and diffraction data for the structures have been deposited in the Protein Data Bank with accession code 5J97.
Table 1:
Data collection and processing statistics
| SeMet | |
|---|---|
| Data Collection | |
| Beamline | ID-F, LS-CAT, APS |
| Wavelength (Å) | 0.9787 |
| Space group | P41212 |
| Cell dimensions | 70.91 |
| a,b,c (Å) | 70.91 |
| 133.64 | |
| α,β,γ (°) | 90.0 |
| 90.0 | |
| 90.0 | |
| Resolution (Å) | 48.6−2.55 |
| (2.69−2.55)a | |
| Total observations | 116859(16000) |
| Unique reflections | 11231 (1571) |
| Completeness (%)a | 96.6 (95.8) |
| Wilson B-factor (Å2) | 48.2 |
| Rmerge (%)b | 7.3 (72.9) |
| I/σ(I)a | 19.5 (3.3) |
| CC1/2 | 1.0 (0.88) |
| Multiplicity | 10.4 (10.2) |
| Anomalous completeness | 97.5 (96.6) |
| Anomalous multiplicity | 5.6 (5.4) |
| Refinement | |
| Resolution (Å) | 40.2−2.55 |
| No. reflections | 11261 |
| Rwork/Rfree (%)c | 21.1/24.4 |
| No. of atoms | |
| Protein | 2013 |
| Water | 77 |
| B factors | |
| Protein | 46.0 |
| Water | 41.7 |
| No. of protein residues | 247 |
| RMSD bonds (Å) | 0.005 |
| RMSD angles (o) | 0.99 |
| Ramachandran | |
| favored (%) | 98 |
| allowed (%) | 2 |
| outliers (%) | 0 |
| Clashscore | 5.9 |
| PDB code | 5J97 |
Values in parentheses are for the highest-resolution shell
Rmerge = Ʃhkl Ʃi |Ii(hkl) – <I(hkl)> | /Ʃhkl Ʃi Ii(hkl), where <I(hkl)> is the average intensity of reflection hkl.
Rwork = Ʃhkl ||Fobs|−|Fcalc|| /Ʃhkl |Fobs|, where Fobs and Fcalc are the observed and calculated structure factors respectively. Rfree is calculated as for Rwork but only use a randomly selected subset of data (6%) which were excluded from refinement.
Structural modelling of the dCaprin dimer
A dimeric structural model of the putative dimerization domain of dCaprin (residues 187–309) was generated by the program SWISS-MODEL (Biasini et al., 2014), using the structure of the Caprin-2 dimer (PDB code 5J97) as the template. Sequence alignments of the dimerization domains of Caprin-1, Caprin-2, and dCaprin were carried out by ClustalW.
Results
The HR1 region of Caprin-2 contains a dimerization domain structurally similar to that of Caprin-1
We had cloned several different constructs of human Caprin-2 that contain residues within the HR1 and flanking regions (with different N-terminal and C-terminal boundaries). Protein expression in E. coli was tested by using many different fusion tags and E. coli host strains. The protein construct containing residues 199–329 of Caprin-2 was successfully expressed in high yield as a soluble fusion protein with either Halo or MBP tag. This Caprin-2 fragment corresponds to the dimerization domain of Caprin-1in our previous study (Wu, et al., 2015, 2016) plus a C-terminal flanking sequence. After removal of the tag by HRV 3C protease digestion, the target protein (containing an artificial sequence of GPSSPS at the N-terminus as a cloning artefact) remained soluble and was stable. The purified target protein readily crystallized.
We decided to use SAD phasing method to solve the structure so that the structure of Caprin-2 is fully independently determined. We prepared SeMet-labelled proteins and collected diffraction data at the selenium peak wavelength using a single crystal containing SeMet-labelled proteins. Statistics for data collection, processing, and structure determination are shown in Table 1.
Electron densities were observed for residues 199–319, as well as three artificial amino acids (Ser-Pro-Ser) at the N-terminus. No electron density was observed for residues 320–329. This data indicates that residues 199–319 of human Caprin-2 contain a stable domain. This domain adopts an all α-helical protein fold consisting of five α-helices and the structure self-associates to form a head-to-head dimer (Figure 1C). The protein fold and overall dimeric structure of Caprin-2 are very similar to the corresponding Caprin-1 dimeric structures we previously determined (PDB codes 4WBP and 4WBE) (Wu, et al., 2016). The r.m.s.d for the superimposition of 1394 common atoms between the Caprin-2 and Caprin-1 dimeric structures is1.7 Å (Figure 2).
Figure 2.
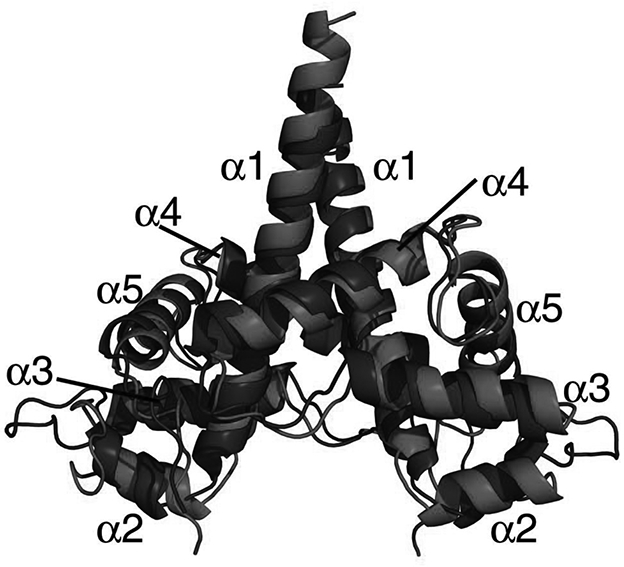
Superimposition of the dimeric structures of the Caprin-2 (in red, PDB code 5J97) and Caprin-1 (in blue, PDB code 4WBE).
The dimerization interface is defined by 48 residues (24 in each protomer) located in theα1 and α4 helices, the linker sequence between α3 and α4, and the C-terminus (Figures 1B and 1C). The interface buries a total of 2810 Å2 of solvent accessible surface area from the two protomers. In comparison, the dimerization interface in the Caprin-1 dimer buries a total of 3398 Å2 of solvent accessible surface area from the two protomers (Wu, et al., 2016). The larger dimerization interface in the Caprin-1 dimer is contributed by the fact that several more residues at the C-terminus of the protein participate in dimerization in the Caprin-1 dimer.
Molecular interactions mediating homodimerization are largely conserved in Caprin-1 and Caprin-2
The molecular interactions in the Caprin-2 dimerization interface are mainly contributed by residues from the two helices α1 and α4, as well as a few residues from two loop regions: one is N-terminal to the α4 helix and the other is C-terminal to the 5 helix (Figure 1B). The homodimerization surfaces show a high degree of shape complementarity (Figure 3A). Intermolecular hydrophobic contacts, electrostatic interactions between charged side-chains, and hydrogen bonds all contribute to formation of the dimeric structure (Figures 3 and 4). The side-chains of many hydrophobic residues (including Leu207, Leu214, Ile217, Leu218, Tyr222, Val271, Met275, Leu280, Phe282, Trp283 and Phe314), as well as the aliphatic parts of the side chains of many other residues (including Glu210, Lys211, Arg215, Gln219, Arg265, Glu267, Ser270, Glu272, Glu276, Ser279, and E315) mediate intermolecular hydrophobic interactions (Figures 3B and 4). In the dimerization interface, there are also several intermolecular hydrogen bonds that involve the side-chain or backbone atoms of Gln221, Arg265, Glu267, Val271, Glu315, and Ser316 (Figure 4).
Figure 3. Molecular interactions that mediate the homodimerization of Caprin-2.
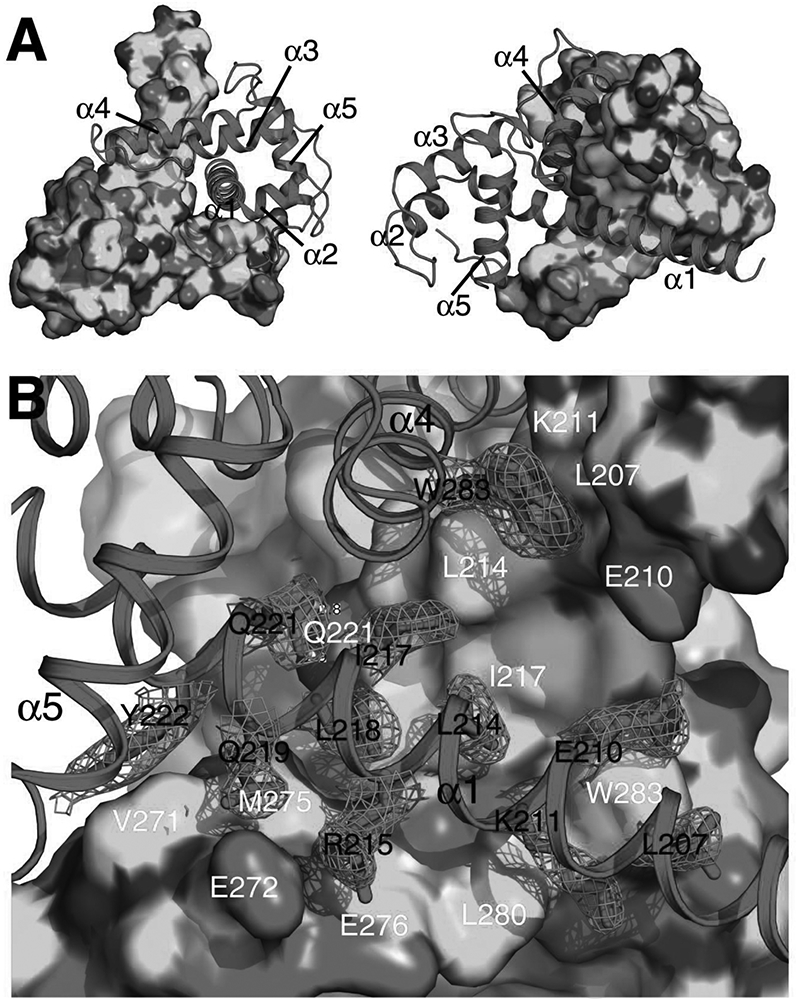
A) Shape complementarity at the homodimerization interface. The two protomers are rendered in surface mode (coloured by elements with C, N, O and S in green, blue, red and yellow respectively) and cartoon mode (coloured in magenta) respectively. B) Intermolecular hydrophobic contacts, salt bridges, and hydrogen bonds at the homodimerization interface. The side-chains for some of the key residues belonging to the protomer rendered in cartoon mode are shown in sticks. 2Fo-Fc electron density map (mesh) contoured at 1σ is shown for these side-chains. The residues involved in the intermolecular interactions are indicated differently for the two protomers: one letter abbreviation-residue number in black for residues belonging to the protomer rendered in cartoon mode and one letter abbreviation-residue number in white for residues belonging to the protomer rendered in surface mode. Hydrogen bonds are represented as yellow dashed lines.
Figure 4. Ligplot presentation of molecular interactions in the dimerization interface.
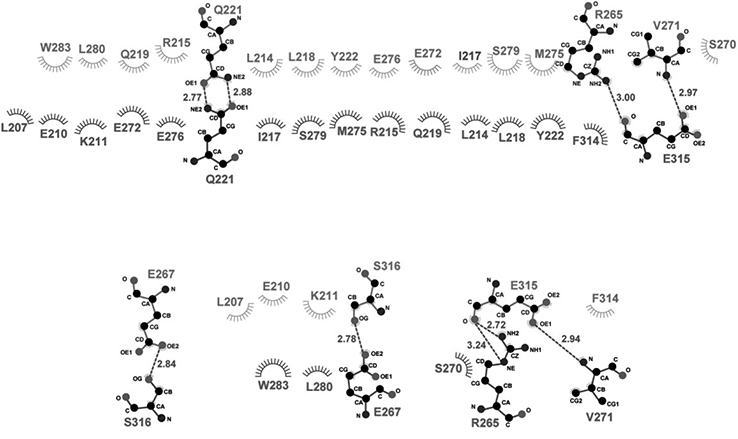
Hydrogen bonds are designated with green dashed lines (the distance between the two heavy atoms is indicated by a number in angstrom). Hydrophobic interactions are represented as starbursts.
Most of the residues involved in intermolecular hydrophobic interactions are conserved in the Caprin-2 and Caprin-1 sequences (Figure 1B). At the center of the dimerization interface, the side-chains of the two Gln221 residues (each from a protomer) form two hydrogen bonds (Figures 3B and 4). This residue is conserved in Caprin-2, Caprin-1 and dCaprin sequences (Figure 1B). Overall, the molecular interactions mediating homodimerization are largely conserved in Caprin-2 and Caprin-1.
Molecular surfaces of the Caprin-2 and Caprin-1 dimeric structures have very different charged properties
It was noted previously that the molecular surface of the Caprin-1 dimeric structure was dominantly negatively charged, especially in the V-shaped concave area (Wu, et al., 2016) (Figure 5). This surface is dominated by 24 negatively charged residues (12 residues from each protomer). Interestingly, comparing the Caprin-2 and Caprin-1 sequences, out of the 12 corresponding residues in Caprin-2, 9 residues in Caprin-2 are neutral or positively charged residues (Caprin-2 Q225 vs Caprin-1 D157, Q229 vs D161, H231 vs E163, S248 vs E180, K249 vs E181, I255 vs D187, K256 vs E188, C262 vs D194, and N266 vs D198) (Figure 1B).
Figure 5. Electrostatic surface representation of Caprin homodimeric structures (for Caprin-2 and Caprin-1) and a structural model (for dCaprin).

Blue and red represent regions of positive and negative potentials respectively.
Overall, the sequence of the dimerization domain of Caprin-2 has equal number of positively charged and negatively residues (17 Asp+Glu vs 17 Arg+Lys, each accounting for 14% of the residues in the sequence shown in Figure 1B). In comparison, the sequence of the dimerization domain of Caprin-1 has significantly more negatively residues than positively charged residues (26 Asp+Glu vs 17 Arg+Lys, accounting for 21.3% and 14% of the residues in the sequence shown in Figure 1B respectively). While Caprin-2 adopts a homodimeric structure that is overally similar to the Caprin-1 homodimeric structure, the electrostatic surface properties of the two structures are very different (Figure 5). The difference in the concave surfaces of the two structures is most pronounced. The large and continuous negatively charged area in the concave surface of Caprin-1 is not present in the Caprin-2 structure.
Residues within the dimerization domain of Caprin-1 are known to participate in interaction of Caprin-1 with its protein partners (El Fatimy et al., 2012). Different charged properties of the dimeric structures of Caprin-1 and Caprin-2 may dictate how these two homologous proteins interact with their respective partners.
Modelling of a dimeric structure of dCaprin
The Drosophila protein dCaprin is classified as a member of the caprin protein family because it contains a HR1 region at its N-terminus, as well as RGG boxes and RG-rich sequences at its C-terminus. Unlike Caprin-1 and Caprin-2, dCaprin does not contain a HR2 region. Within the HR1 region, the dCaprin-1 sequence corresponding to the dimerization domains of Caprin-1 and Caprin-2 has a high homology to the other two proteins (Figure 1B). The dCaprin sequence shown in Figure 1B shares 28% identity and 52% similarity with the Caprin-2 sequence (slightly lower similarity to the Caprin-1 sequence). With this high degree of sequence homology, it is expected that a reasonable structural model of a dCaprin dimeric structure can be built using the Caprin-2 dimeric structure as a template. Such a model was built by SWISS-MODEL (Biasini, et al., 2014).
Most of the residues that mediate the homodimerization of Caprin-2 are conserved in the dCaprin sequence (Figure 1B). In view of this, it is reasonable to expect that dCaprin may also contain a dimerization domain within its HR1 region. The electrostatic surface properties of the modeled dCaprin dimeric structure are more similar to those in the Caprin-2 structure than to those in the Caprin-1 structure (Figure 5). As mentioned earlier, the V-shaped concave surface of the Caprin-1 dimeric structure are dominantly negatively charged due to the presence of 12 Asp and Glu residues from each protomer. Out of the 12 corresponding residues in dCaprin, 7 residues are neutral or positively charged residues (N212 vs D157, Q218 vs E163, N235 vs E180, T236 vs E181, K243 vs E188, Q249 vs D194, and A256 vs D198) (Figure 1B). Overall, sequence of the putative dimerization domain of dCaprin has slightly more number of negatively charged residues than positively charged residues (20 Asp+Glu vs 17 Arg+Lys, accounting for 16% vs 14% of the residues in the sequence shown in Figure 1B). Results from the modeling study, combined with sequence data, suggest that while all caprin proteins may adopt overally similar dimeric structures, variations of the surface residues may lead to different surface properties and hence different protein-protein interactions.
Discussion
In this study, we had tested the bacterial expression of several protein constructs containing residues within the HR1 and flanking regions of human Caprin-2. Interestingly, none of the constructs containing the entire HR1 region had detectable expression. Similar scenario was observed in our previous study on human Caprin-1 (Wu, et al., 2015, 2016). In both cases of Caprin-1 and Caprin-2, a protein construct containin ~60% of the HR1 region (the C-terminal portion) and the linker sequence between the HR1 and E-rich regions (residues 199–329 in the case of Caprin-2) was successfully expressed. The structures determined in the previous study (for Caprin-1) and this study (for Caprin-2) show that the C-terminal 60% sequences of Caprin-1 and Caprin-2 HR1 regions harbour a stable dimerization domain. The linker sequences between the HR1 and E-rich regions are most likely flexible, because no electron density is observed for residues in these regions in both the Caprin-1 and Caprin-2 structures. It is not known whether the 40% N-terminal portion of the HR1 sequence (about 70 residues) contains a structural domain. Our efforts to bacterially express this sequence (either as an individual construct or within the entire HR1) were not successful for both Caprin-1 and Caprin-2.
The overall dimeric structures of Caprin-1 and Caprin-2 formed by their dimerization domains are similar to each other. In view of the extensive intermolecular interactions mediating the formation of the dimers and the significant degree of conservation of the interactions between Caprin-1 and Caprin-2, the dimeric structures should represent the native biological conformations of the Caprin proteins, instead of being crystallographic packing artifacts. The structures may mediate the dimerization of the full-length Caprin-1 and Caprin-2 proteins. At the present time, it is not known whether Caprin-1 and Caprin-2 function as a momomer, dimer, or multimer. Our structural results on the Caprin-1 and Caprin-2 proteins suggest that they may function as a dimer. We previously proposed that Caprin-1 might function as a scaffolding protein that recruits other proteins (such as FMRP, G3BP1 and the JEV core protein), as well as RNAs, into specific ribonucleoprotein complexes (Wu, et al., 2016). With the new structural insights on the Caprin-2 protein, it is reasonable to propose that Caprin-2 may also function as a scaffolding protein to mediate the formation of specific ribonucleoprotein complexes with its own partners. The structural similarities between the Caprin-1 and Caprin-2 dimers suggest that the two proteins may share some common features in their functional mechanisms, consistent with the observation that both proteins are localized to neuronal RNA granules (Shiina and Tokunaga, 2010).
Of course, Caprin-1 and Caprin-2 have distinct functions (Ding, et al., 2008; Konopacka, et al., 2015; Shiina and Tokunaga, 2010). The functional differences of the two proteins may depend on their specific protein and RNA partners. So far, Caprin-2 interacting protein or RNA molecules have not been identified. For Caprin-1, several protein partners that directly interact with Caprin-1 have been identified, including G3BP1 (Solomon, et al., 2007), FMRP (El Fatimy, et al., 2012), and the JEV core protein (Katoh, et al., 2013). The Caprin-1 sequences reported to be required for G3BP1 and FMRP interactions are mapped to different regions of Caprin-1. Caprin-1 residues 352–380 were reported to be required for G3BP1 interaction (Solomon, et al., 2007). This sequence is separated from the dimerization domain by ~100 residues. Caprin-1 residues 231 245 were reported to be required for interaction with FMRP (El Fatimy, et al., 2012). This sequence is located within the 5 helix of the dimerization domain. The 5 helix is not involved in the dimerization of the Caprin-1 or Caprin-2 proteins (Figures 1 and 2). One face of the 5 helix is fully exposed in the dimeric structure, readily accessible for protein-protein interaction. Protein-protein interaction through the 5 helix should not interfere with the homodimerization interaction of the Caprin proteins. The Caprin-1 residues required for interaction with the JEV core protein have not been mapped. The JEV core protein is a highly basic protein (105 residues, with 25 Arg/Lys residues and only 4 Asp/Glu residues). The negatively charged putative protein binding groove formed by homodimerization of Caprin-1 (Figure 5), and the E-rich region C-terminal to the dimerization domain (Figure 1), may potentially interact with the JEV core protein. Available biochemical and structural data of Caprin-1 indicate that the dimerization domain not only mediates the dimerization of the protein, the dimeric structure itself also participates in direct interaction with Caprin-1 partner proteins. It is not known whether the Caprin-2 dimeric structure determined in this study contains binding sites for Caprin-2 partner proteins. Based on structural comparison of the Caprin-1 and Caprin-2 dimeric structures (Figure 5), the two structures have quite different molecular surfaces. Despite the similarity in overall dimeric folds, the two structures should be different in their abilities to interact with specific protein partners.
Acknowledgement
We thank staff of the LS-CAT at the Advance Photon Source (Argonne National Laboratory) for assistance with data collection. The work was supported by the start-up fund and a seed grant from Southern Illinois University Carbondale, as well as a grant from the National Institute of Health (1R15GM116062–01) to Z. Du.
Reference
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Echols N, Headd JJ, … Zwart PH (2011). The Phenix software for automated determination of macromolecular structures. Methods, 55(1), pp. 94–106. doi: 10.1016/j.ymeth.2011.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aerbajinai W, Lee YT, Wojda U, Barr VA, & Miller JL (2004). Cloning and characterization of a gene expressed during terminal differentiation that encodes a novel inhibitor of growth. J Biol Chem, 279(3), pp. 1916–1921. doi: 10.1074/jbc.M305634200 [DOI] [PubMed] [Google Scholar]
- Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, … Adams PD (2012). Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr, 68(Pt 4), pp. 352–367. doi: 10.1107/s0907444912001308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battye TG, Kontogiannis L, Johnson O, Powell HR, & Leslie AG (2011). iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr, 67(Pt 4), pp. 271–281. doi: 10.1107/s0907444910048675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, … Schwede T (2014). SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res, 42(Web Server issue), pp. W252–258. doi: 10.1093/nar/gku340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidet K, Dadlani D, & Garcia-Blanco MA (2014). G3BP1, G3BP2 and CAPRIN1 are required for translation of interferon stimulated mRNAs and are targeted by a dengue virus non-coding RNA. PLoS Pathog, 10(7), p e1004242. doi: 10.1371/journal.ppat.1004242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin RW, Chellappa R, Pauw RJ, Vriend G, Oostrik J, van Drunen W, … Kremer H (2008). Missense mutations in POU4F3 cause autosomal dominant hearing impairment DFNA15 and affect subcellular localization and DNA binding. Hum Mutat, 29(4), pp. 545–554. doi: 10.1002/humu.20693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Xi Y, Chen T, Wang JY, Tao DL, Wu ZL, … Li L (2008). Caprin-2 enhances canonical Wnt signaling through regulating LRP5/6 phosphorylation. J Cell Biol, 182(5), pp. 865–872. doi: 10.1083/jcb.200803147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Fatimy R, Tremblay S, Dury AY, Solomon S, De Koninck P, Schrader JW, & Khandjian EW (2012). Fragile X mental retardation protein interacts with the RNA-binding protein Caprin1 in neuronal RiboNucleoProtein complexes [corrected]. PLoS One, 7(6), p e39338. doi: 10.1371/journal.pone.0039338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, & Cowtan K (2010). Features and development of Coot. Acta Crystallogr D Biol Crystallogr, 66(Pt 4), pp. 486–501. doi: 10.1107/s0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaboriaud C, Juanhuix J, Gruez A, Lacroix M, Darnault C, Pignol D, … Arlaud GJ (2003). The crystal structure of the globular head of complement protein C1q provides a basis for its versatile recognition properties. J Biol Chem, 278(47), pp. 46974–46982. doi: 10.1074/jbc.M307764200 [DOI] [PubMed] [Google Scholar]
- Garlatti V, Chouquet A, Lunardi T, Vives R, Paidassi H, Lortat-Jacob H, … Gaboriaud C (2010). Cutting edge: C1q binds deoxyribose and heparan sulfate through neighboring sites of its recognition domain. J Immunol, 185(2), pp. 808–812. doi: 10.4049/jimmunol.1000184 [DOI] [PubMed] [Google Scholar]
- Grill B, Wilson GM, Zhang KX, Wang B, Doyonnas R, Quadroni M, & Schrader JW (2004). Activation/division of lymphocytes results in increased levels of cytoplasmic activation/proliferation-associated protein-1: prototype of a new family of proteins. J Immunol, 172(4), pp. 2389–2400. [DOI] [PubMed] [Google Scholar]
- Katoh H, Okamoto T, Fukuhara T, Kambara H, Morita E, Mori Y, … Matsuura Y (2013). Japanese encephalitis virus core protein inhibits stress granule formation through an interaction with Caprin-1 and facilitates viral propagation. J Virol, 87(1), pp. 489–502. doi: 10.1128/jvi.02186-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopacka A, Greenwood M, Loh SY, Paton J, & Murphy D (2015). RNA binding protein Caprin-2 is a pivotal regulator of the central osmotic defense response. Elife, 4doi: 10.7554/eLife.09656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Los GV, Encell LP, McDougall MG, Hartzell DD, Karassina N, Zimprich C, … Wood KV (2008). HaloTag: a novel protein labeling technology for cell imaging and protein analysis. ACS Chem Biol, 3(6), pp. 373–382. doi: 10.1021/cb800025k [DOI] [PubMed] [Google Scholar]
- Miao H, Jia Y, Xie S, Wang X, Zhao J, Chu Y, … Li L (2014). Structural Insights into the C1q Domain of Caprin-2 in Canonical Wnt Signaling. J Biol Chem doi: 10.1074/jbc.M114.591636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratovitski T, Chighladze E, Arbez N, Boronina T, Herbrich S, Cole RN, & Ross CA (2012). Huntingtin protein interactions altered by polyglutamine expansion as determined by quantitative proteomic analysis. Cell Cycle, 11(10), pp. 2006–2021. doi: 10.4161/cc.20423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro L, & Scherer PE (1998). The crystal structure of a complement-1q family protein suggests an evolutionary link to tumor necrosis factor. Curr Biol, 8(6), pp. 335–338. [DOI] [PubMed] [Google Scholar]
- Shiina N, Shinkura K, & Tokunaga M (2005). A novel RNA-binding protein in neuronal RNA granules: regulatory machinery for local translation. J Neurosci, 25(17), pp. 4420–4434. doi: 10.1523/jneurosci.0382-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiina N, & Tokunaga M (2010). RNA granule protein 140 (RNG140), a paralog of RNG105 localized to distinct RNA granules in neuronal dendrites in the adult vertebrate brain. J Biol Chem, 285(31), pp. 24260–24269. doi: 10.1074/jbc.M110.108944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon S, Xu Y, Wang B, David MD, Schubert P, Kennedy D, & Schrader JW (2007). Distinct structural features of caprin-1 mediate its interaction with G3BP-1 and its induction of phosphorylation of eukaryotic translation initiation factor 2alpha, entry to cytoplasmic stress granules, and selective interaction with a subset of mRNAs. Mol Cell Biol, 27(6), pp. 2324–2342. doi: 10.1128/mcb.02300-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatraman Girija U, Gingras AR, Marshall JE, Panchal R, Sheikh MA, Gal P, … Wallis R (2013). Structural basis of the C1q/C1s interaction and its central role in assembly of the C1 complex of complement activation. Proc Natl Acad Sci U S A, 110(34), pp. 13916–13920. doi: 10.1073/pnas.1311113110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, David MD, & Schrader JW (2005). Absence of caprin-1 results in defects in cellular proliferation. J Immunol, 175(7), pp. 4274–4282. [DOI] [PubMed] [Google Scholar]
- Wu Y, Zhu J, Huang X, & Du Z (2015). Bacterial expression and preliminary crystallographic studies of a 149-residue fragment of human Caprin-1. Acta Crystallogr F Struct Biol Commun, 71(Pt 3), pp. 324–329. doi: 10.1107/s2053230x15002642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Zhu J, Huang X, & Du Z (2016). Crystal structure of a dimerization domain of human Caprin-1: insights into the assembly of an evolutionarily conserved ribonucleoprotein complex consisting of Caprin-1, FMRP and G3BP1. Acta Crystallogr D Struct Biol, 72(Pt 6), pp. 718–727. doi: 10.1107/s2059798316004903 [DOI] [PMC free article] [PubMed] [Google Scholar]


