Abstract
KRAS mutations are abnormalities widely found in genomic DNA and circulating tumor DNA (ctDNA) of various types of cancers. Thus, highly sensitive detection of KRAS mutations in genomic DNA is of great significance in disease diagnosis and personalized medicine. Here, we developed a ligation-initiated loop-mediated isothermal amplification (LAMP) assaying method for ultrasensitive detection of KRAS mutation. In the presence of mutant KRAS DNA (mutDNA), the dumbbell-shaped structure (DSS) is formed by the specific ligation of two substrates (SLS1 and SLS2), which act as a template to initiate the following LAMP amplification. Making use of the outstanding specificity of ligation reaction and superior amplification of LAMP, 10 aM mutDNA can be accurately determined. In addition, as low as 0.1% mutDNA can be detected in the presence of a large excess of wild-type KRAS DNA (wtDNA), indicating the high sensitivity and specificity of the method. Furthermore, this strategy has been successfully applied for detection of a KRAS mutation from tissue samples of colorectal cancer patients. Thus, the developed ligation-initiated LAMP fluorescence assaying strategy presents a promising prospect for ultrasensitive detection of mutations.
Introduction
Single nucleotide mutation is the permanent variation in a single nucleotide among the genomic DNA, thus altering the level of gene expression and linking to disease development1–3. As well known, the KRAS gene is a proto-oncogene which mainly participates in RAS-MAPK signaling pathway to control cell growth, proliferation and differentiation4–6. Moreover, single nucleotide mutation often occurs in and near codons of the KRAS gene, causing normal cells to become cancerous state7,8. Whole genome sequencing indicates that KRAS mutations are activated in various malignancies, such as colorectal cancer9, non-small-cell lung cancer10,11 and pancreatic adenocarcinoma12. Importantly, as a potential personalized biomarker, KRAS mutations can be used to not only predict the primary tumor but monitor minimal residual tumor during therapy13,14. Therefore, accurate and ultrasensitive detection of KRAS mutations will provide a robust evidence for early diagnosis of mutant KRAS-related cancer.
Various technology platforms have been applied to discriminate mutation, among which, DNA sequencing is a classic mutation detection technique that allows high-throughput screening of the individual nucleotides within a DNA sequence15,16. However, the sophisticated instrumentation and complicated procedures are the limiting factors for its extensive application17. The polymerase chain reaction (PCR)-based methods, such as wild-type blocking PCR18 and allele-specific PCR19, have received increasing attention owing to the advantages of simple assaying protocols and instrument automation, also these methods still require the careful design of specific probes for discrimination. Therefore, there is still an urgent need for the development of highly sensitive, cost-effective and rapid mutation detection strategy.
Currently, ligation reaction mediated by DNA ligase holds the capacity for characterization of mutation, fusion gene and other genetic alterations20,21. DNA ligase can efficiently catalyze the junction of the 3′-end of one DNA fragment with the 5′-end of another DNA fragment upon hybridization with a complementary DNA target22. Due to the excellent discrimination capacity of ligase, the ligation reaction demonstrates better specificity than primer extension reaction for single nucleotide polymorphisms23–25. For example, adopting the end-joining fidelity of DNA ligase on single nucleotide polymorphisms, a novel iLock assay with an improved specificity was developed for the detection of a mutation in mRNA26. Meanwhile, the various ligation based amplification strategies have been developed, which are extensively used to detect methylation and mutation at the single nucleotide level27–29. Thus, the ligation reaction mediated by DNA ligase can serve as a specific method to characterize single-base mutation in DNA strands with the merits of excellent discrimination capacity and simple detection process.
Loop-mediated isothermal amplification (LAMP), an outstanding isothermal amplification technique, has been proved to be a highly specific, sensitive and rapid detection method30–32. In LAMP reaction, owing to its high strand displacement activity, Bst DNA polymerase is widely used to efficiently amplify DNA sequences33, making LAMP suitable for point-of-care diagnostics. Recently, there have been a number of published upgrade strategies for LAMP, integrating LAMP with other DNA amplification techniques, such as PCR34, reverse transcription35 and ligation assays36. For instance, Du and coworkers have developed a ligation-LAMP strategy for the detection of miRNA with a low detection limit of 0.2 fM. With the upgrade LAMP strategies, the detection performance of LAMP changes from simplification to diversification, validating its potential as a promising technique in the application of diagnostic.
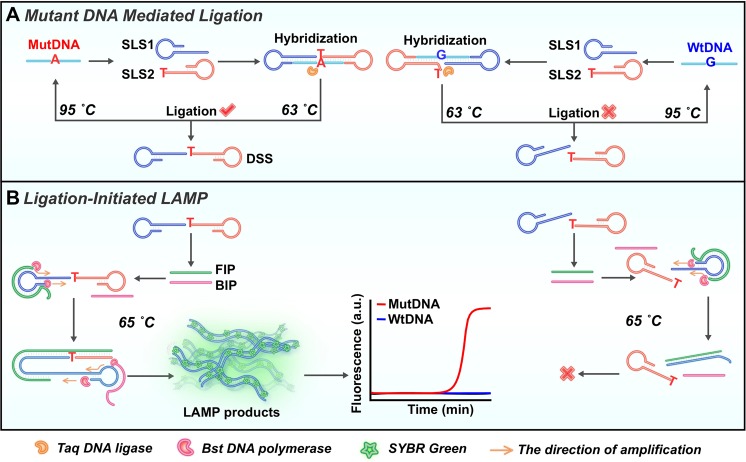
Herein, making use of the remarkable specificity of ligation reaction and highly amplified efficiency of LAMP, we developed a label-free ligation-initiated LAMP strategy for specific and sensitive detection of KRAS mutation. As shown in Fig. 1, the developed method mainly comprises two step reactions: the ligation reaction mediated by mutDNA and the ligation-initiated LAMP reaction. In the ligation reaction, two ligation substrates, stem-loop substrate 1 (SLS1) and stem-loop substrate 2 (SLS2), are designed for discriminating the single base between mutant KRAS DNA (mutDNA) and wild-type KRAS DNA (wtDNA) respectively. The SLS1 is completely complementary to mutDNA and wtDNA. While, compared with the mutDNA, there is one more position mismatch between wtDNA and SLS2 (Fig. S1, Supporting Information). In the presence of mutDNA, SLS1 and SLS2 can be ligated to form the dumbbell-shaped structure (DSS) by the catalysis of Taq DNA ligase at 63 °C. Meanwhile, the base T at the 3′-end of the SLS2 is not complementary to the base G in the wtDNA owing to the one more position mismatch. Thus, no ligation occurs in the presence of wtDNA. The DSS is just the starting template for LAMP amplification. Along with the gradient temperature cycles, more DSSs are produced in the presence of mutDNA. In the second step, forward inner primer (FIP) first hybridizes with the loop in the DSS for strand displacement DNA synthesis. Meanwhile, self-primed DNA synthesis is initiated from the 3′ to 5′ by using self-structure of DSS as the template. With high-efficiency strand displacement activity, the Bst DNA polymerase initiates the amplification reaction and creates more elongated structures along the DSS. Then, backward inner primer (BIP) hybridizes to the elongated structures to initiate subsequent rounds of amplification reaction, generating a large amount of double strand DNA (dsDNA). As a result, a significant fluorescence signal is achieved by the preferential binding between the fluorescence dye SYBR Green I and dsDNA. On the contrary, without formation of DSS, there is a negligible fluorescence background from the detection of wtDNA. The developed strategy kept a low background signal for the detection of KRAS mutation by adding a mismatch site. Moreover, the developed strategy can also be successfully applied for the detection of KRAS mutation from tissue samples of colorectal cancer patients, presenting a promising application for ultrasensitive detection of mutation in clinical diagnosis.
Figure 1.
The principle of ligation-initiated LAMP strategy. (A) The formation of dumbbell-shaped structure (DSS) by mutDNA mediated ligation reaction. (B) LAMP reaction initiated by the DSS.
Results and Discussion
The feasibility of the ligation-initiated LAMP
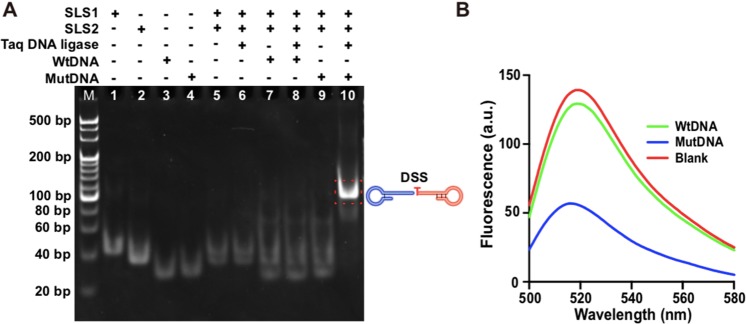
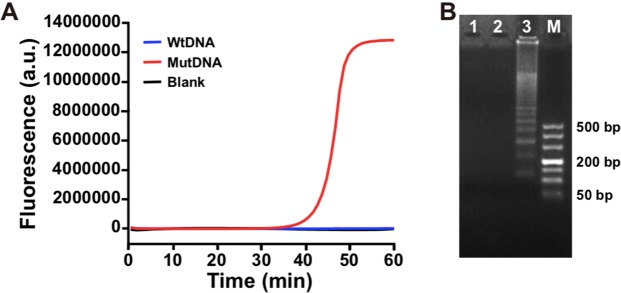
The feasibility of ligation reaction was firstly verified by 8% native PAGE. As shown in Fig. 2A, lanes 1–4 indicated the position of substrate SLS1, substrate SLS2, wtDNA and mutDNA, respectively. Lane 5 represented the mixture of SLS1 and SLS2. With addition of Taq DNA ligase to the mixture of SLS1 and SLS2, unchanged electrophoretic bands (lane 6) were achieved compared to lane 5, indicating the ligation reaction did not take place between SLS1 and SLS2 without the complementary DNA template. With addition of the wtDNA (or mutDNA) to the mixture of SLS1 and SLS2, a small band with lower electrophoresis migration was observed in the lanes 7 and 9. Then, with the addition of Taq DNA ligase (lane 8), there was no difference in the products of wtDNA mediated ligation compared to lane 7, while there was a distinct band with much lower mobility appeared in the mutDNA mediated ligation reaction (lane 10), indicating the formation of ligation products (DSS). Further, we labeled a fluorophore on the 5′-end of SLS1-1, and a quencher at the 3′-end of SLS2-1 to further confirm the mutDNA mediated ligation reaction. As shown in Fig. 2B, the highest fluorescence intensity was obtained in the wtDNA mediated ligation reaction, which is comparable to the fluorescence intensity of the blank (without mutDNA or wtDNA). However, owing to the short distance between fluorophore and quencher after mutDNA mediated ligation, a low fluorescence intensity was obtained. These results proved that the ligation could only be achieved in the presence of mutDNA. Further, the ligation products were amplified and quantified by real-time fluorescence monitored LAMP. As shown in Fig. 3A, the fluorescence intensity raised sharply with increase of the time after 35 min and then reached a plateau within 60 min in the presence of mutDNA mediated ligation product. However, no apparent fluorescence signal change was observed in the presence of wtDNA mediated ligation product or blank sample, due to no formation of DSS. Meanwhile, agarose electrophoresis was adopted to analyze the products of LAMP (Fig. 3B). A ladder-like pattern bands emerged from the LAMP product initiated by mutDNA mediated ligation (Fig. 3B, lane 3), demonstrating the successful amplification of the LAMP to produce long dsDNA product with cauliflower-like structures. In contrast, there were no apparent bands for the LAMP product initiated by wtDNA mediated ligation (Fig. 3B, lane 2) and blank sample (Fig. 3B, lane 1). These results were in good agreement with the real-time fluorescence measurements, further validating the feasibility of the ligation-initiated LAMP strategy for mutDNA detection.
Figure 2.
(A) PAGE analysis of the ligation products. 20 bp DNA marker, SLS1 (lane 1), SLS2 (lane 2), wtDNA (lane 3), mutDNA (lane 4), the mixture of SLS1 and SLS2 (lane 5), the mixture of SLS1 and SLS2 under ligation of Taq DNA ligase (lane 6), the mixture of SLS1, SLS2 and wtDNA (lane 7), the mixture of SLS1, SLS2 and wtDNA under ligation of Taq DNA ligase (lane 8), the mixture of SLS1, SLS2 and mutDNA (lane 9), the mixture of SLS1, SLS2 and mutDNA under ligation of Taq DNA ligase (lane 10). (B) Fluorescence spectra from the ligation reaction products mediated by blank, wtDNA and mutDNA, respectively.
Figure 3.
(A) Real-time fluorescence curves produced by 1 fM mutDNA (red line), 1 fM wtDNA (blue line) and blank (black line) with developed ligation-initiated LAMP strategy, respectively. (B) Gel electrophoresis analysis of 10 µL LAMP products: ligation products of blank (lane 1), wtDNA (lane 2), mutDNA (lane 3), (Lane M, DL500 DNA marker).
Optimization of the experimental conditions
In order to achieve the optimal analytical performance, several experimental conditions were investigated. According to the recent reports, inserting an extra mismatch at certain positions of the probes could change the Tm of duplex, which has been verified to affect the discrimination between wild and mutant gene37,38. Therefore, an additional mismatch at different sites (2nd, 3rd, 4th, 5th, 6th) within SLS2 was designed (Table S1) to increase the allele-specificity ligation. The ligation product of SLS1 and mismatched SLS2 mediated by mutDNA or wtDNA was added to real-time monitored LAMP reaction, separately. Meanwhile, the POI value (point of inflection, represents the time corresponding to the maximum slope of fluorescence curves) was employed to quantitatively evaluate the amplification efficiency. The lowest POIm POIw−1 ratio (POIm, POI of mutDNA; POIw, POI of wtDNA) could make the best distinction between mutDNA and wtDNA. As shown in Fig. S2, the lowest POIm POIw−1 ratio was obtained from the additional mismatch at the 3rd site. Thus, the 3rd site mismatch within SLS2 was the best optimal mismatch site. The other experimental parameters, including the ligation temperature, cycle numbers of ligation reaction, the concentration of Bst DNA polymerase and the temperature of LAMP reaction, were also investigated (Figs S3–S6). As a result, the optimal ligation temperature and cycle number were 63 °C and 30 cycles, respectively. The concentration of Bst DNA polymerase was optimized to be 0.4 U µL−1, and optimal temperature of ligation-initiated LAMP was 65 °C.
Analytical performance of the ligation-initiated LAMP strategy in synthetic mutDNA
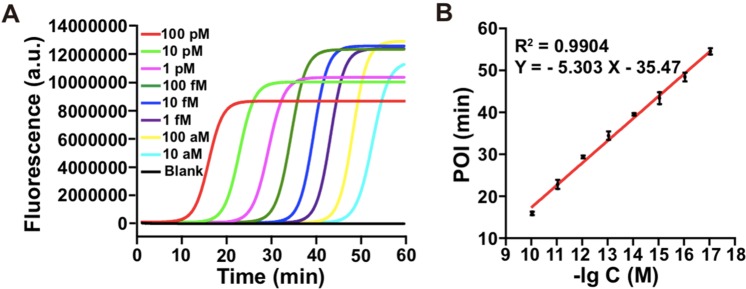
Under the optimal experimental conditions, the analytical performance of the developed ligation-initiated LAMP method was evaluated by real-time fluorescence monitor of different concentrations of synthetic mutDNA. As shown in Fig. 4A, the POI values were gradually shortened with the increasing concentration of mutDNA in the range from 10 aM to 100 pM. As illustrated in Fig. 4B, the POI value (Y) exhibited a good linear correlation with the logarithm of mutDNA concentrations (X) in the range from 10 aM to 100 pM, and the correlation equation was Y = −5.303X −35.47 with R2 = 0.9904. The developed strategy had a wide linear dynamic range over 7 orders of magnitude, and down to 10 aM target DNA could be detected in 60 min. Compared with the reported methods ligation-based strand displacement amplification and ligation-based rolling circle amplification (Supporting Information, Table S2), the ultra-sensitivity achieved for detection of mutDNA can be ascribed to the highly efficient signal amplification of LAMP, which generates upwards of 109 copies from less than 100 copies of target DNA within one hour39.
Figure 4.
(A) Real-time fluorescence measurement of different concentrations (100 pM, 10 pM, 1 pM, 100 fM, 10 fM, 1 fM, 100 aM, 10 aM and 0) of mutDNA based on ligation-initiated LAMP strategy. (B) The linear relationship between POI (Y) and the logarithm of mutDNA concentrations (X). Error bars show the standard deviation of three replicative tests.
Specificity of the ligation-initiated LAMP strategy in synthetic DNA strands
To test the specificity of the developed method for effective discrimination of trace mutDNA, the wtDNA and mutDNA were mixed with a total concentration of 10 fM, in which the ratio of mutDNA to the total DNA varying from 0% to 100%. Afterwards, the mixed samples were assayed, and the corresponding real-time fluorescence curves were monitored. As shown in Fig. 5A, along with the decreasing concentration of mutDNA, the POI values were significantly increased (Fig. 5B). As a result, even 0.1% mutDNA could be clearly discriminated from that of 10 fM wtDNA, which was much lower than those reported mutation detection strategies (Supporting Information, Table S2). These strategies mainly depend on the 3’ terminal nucleotide of a primer that corresponds to a specific mutation site to discriminate mutation, whereas it is not sufficient to achieve reliable discrimination between the alleles. Therefore, according to the combination of rational probe design and high amplification efficiency of LAMP, the strategy achieved a high specificity and superior sensitivity for mutation detection in DNA strand.
Figure 5.
Detection of KRAS mutation with different proportions between mutDNA and wtDNA. (A) Real-time fluorescence curves of ligation-initiated LAMP reaction produced by a mixture of target DNA with different ratios of mutDNA to the total DNA (allele frequency). The allele frequency was 0%, 0.1%, 1%, 10%, 50% and 100%, and the total DNA concentration was 10 fM. (B) The relationship between the POI values from (A) and allele frequency. The error bars represent standard deviation of three replicate measurements.
Detection of the KRAS mutation from tissue samples
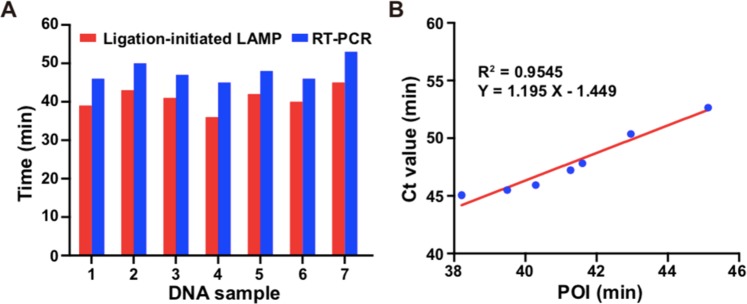
Finally, the genomic DNA extracted from tissue samples of ten colorectal cancer patients were assayed by the developed strategy to evaluate the potential application. As a result, 7 of 10 samples harbored the KRAS mutation, whereas the other 3 samples were negative. The result was consistent with the result obtained from the commercial RT-PCR qualitative kit approved by the China Food and Drug Administration (CFDA), indicating that the developed strategy could discriminate between negative and positive tissue samples. Figure 6A showed the mean POI value and corresponding time of cycle threshold (Ct, RT-PCR) of seven positive samples. The relationship between the results of RT-PCR and the ligation-initiated LAMP strategy was further investigated by regression analysis of the corresponding time of Ct and the mean value of POI. As shown in Fig. 6B, the regression equation was Y = 1.195X −1.449 with R2 = 0.9545, indicating there was a good correlation between the two methods. These results demonstrated the proposed strategy can be applied for the identification of KRAS mutation in genomic DNA from tissue samples, holding a great potential in the diagnosis of mutation-related human diseases.
Figure 6.
Detection of KRAS mutation in genomic DNA from tissue samples. (A) Histogram and (B) correlation plot of POI value of proposed strategy and Ct value of RT-PCR.
Conclusion
In summary, in combination with ligation and LAMP, a novel fluorescent sensing strategy has been developed for detection of DNA mutation. Owing to rational probe design by adding an extra mismatch to SLS2 and efficient amplification of LAMP, this ligation-initiated LAMP strategy not only achieves an ultrahigh sensitivity and specificity for detection of mutDNA at aM level but also can quantitate the rare mutation even in a large excess of coexisting wtDNA with a selectivity factor down to 0.1% in synthetic DNA strands. Moreover, the strategy has been successfully applied for the accurate detection of mutation from the tissue samples of colorectal cancer patients, and the results of our method are consistent with the that of RT-PCR. Therefore, the ligation-initiated LAMP strategy holds great potential application for the detection of DNA mutation, and also offers a new insight into disease diagnostics, personalized medicine and biomedical research.
Experiment Section
Materials and reagents
The DNA oligonucleotides were synthesized by Sangon Biotech Co., Ltd. (Shanghai, China) and purified by high-performance liquid chromatography. Taq DNA ligase (40 U μL−1), 10 × Taq DNA ligase reaction buffer (200 mM Tris-HCl, 250 mM KAc, 100 mM Mg(Ac)2, 100 mM DTT, 10 mM NAD, 1% Triton X-100, pH 7.6), Bst 2.0 DNA polymerase (8 U μL−1) and 10 × ThermoPol reaction buffer (200 mM Tris-HCl, 100 mM (NH4)2SO4, 500 mM KCl, 20 mM MgSO4, 1% Tween-20, pH 8.8) were all purchased from New England Biolabs (Beijing, China). SYBR Green I was obtained from Generay Biotech. Co., Ltd. (Shanghai, China). DNA marker, 6 × loading buffer and dNTPs were purchased from Takara (Dalian, China). TIANamp Genomic DNA Kit was purchased from Tiangen Biotech Co., Ltd. (Beijing, China). The nuclease-free water was purchased from Thermo Fisher Scientific Inc (Vilnius, Lithuania) and used in all ligation reaction and ligation-initiated LAMP assays. All the reagents were of analytical grade and used without further purification. All solutions were prepared with ultrapure water from Millipore Milli-Q water purification system (Millipore, USA). The detailed oligonucleotide sequences were listed in Table S1 (Supporting Information).
Ligation reaction
The ligation mixture contained 1 µL Taq DNA ligase reaction buffer, 1 U µL−1 Taq DNA ligase, 100 nM SLS1 and 100 nM SLS2. For the detection of different target, various concentrations of mutDNA, wtDNA or genomic DNA was added to the ligation mixture to yield a final volume of 10 µL. Then the reaction mixture was subjected to the following thermal cycling profile in a thermal cycler (Bio-Rad, USA): initial denaturation at 95 °C for 3 min, 30 cycles at 95 °C for 0.5 min and 63 °C for 1 min. Then, another denaturation at 95 °C for 5 min was carried out, followed by quickly cooled down to 4 °C. The ligation reaction products were subjected to LAMP reaction.
LAMP reaction and real-time fluorescence measurement
The LAMP reaction buffer contained 200 nM FIP, 200 nM BIP, and 0.25 mM dNTPs and 2 µL ThermoPol reaction buffer. For the detection of different ligation products, 1 µL mutDNA or wtDNA mediated ligation reaction products was added to the LAMP buffer with a total volume of 18 µL. Then the reaction mixture was heated to 95 °C for 5 min, followed by cooling at 4 °C for 5 min. After that, 1 µL SYBR Green I and 0.4 U µL−1 Bst 2.0 DNA polymerase were added. The reaction was performed at 65 °C in a StepOne Real-Time PCR instrument (Applied Biosystems, USA) for 60 min. The real-time fluorescence intensity was monitored at an interval of 1 min employing the SYBR Green channel.
Gel electrophoresis
The products of ligation reaction were analyzed by 8% native polyacrylamide gel electrophoresis (PAGE) in 1 × TBE buffer (89 mM Tris, 89 mM boric acid, 2 mM EDTA) at a 90 V constant voltage for 45 min. The products of LAMP reaction obtained were analyzed by 2% agarose gel in 1 × TBE buffer at 110 V for 35 min. After gold view (GV) staining, the gels were photographed with a gel imaging system (Bio-Rad, USA).
Genomic DNA extraction and purification from clinical samples
Samples and clinical data were collected from ten colorectal cancer patients at the first affiliated hospital of Chongqing Medical University with informed consent. Among which, seven samples were positive and three were negative. Genomic DNA from formalin-fixed, paraffin-embedded (FFPE) tissue samples was extracted by TIANamp Genomic DNA Kit according to the manufacture’s instruction, and stored at −80 °C until use, and the final concentration of genomic DNA was 0.4 ng µL−1. The DNA concentration was determined by NanoDrop One Microvolume UV-vis spectrophotometer (Thermo Scientific, USA).
Supplementary information
Detection of KRAS mutation via ligation-initiated LAMP reaction
Acknowledgements
This study was funded by the National Natural Science Foundation of China (81460317 and 31660241), and Special Financial Aid for Post-doctors in Chongqing City (Xm2017027).
Author Contributions
X.M. and S.D. coordinated the overall research. Y.F., X.D., J.H., L.H., L.Z. and W.C. conceived and conducted the experiments. Y.F. and X.D. analyzed experimental data, prepared all the Figures and wrote the main manuscript text. S.D. and X.M. contributed to the revision of the manuscript. All authors contributed to the discussion about the results. All authors reviewed the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yixin Fu and Xiaolei Duan contributed equally.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-42542-x.
References
- 1.Sachidanandam R, et al. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 2001;409:928–933. doi: 10.1038/35057149. [DOI] [PubMed] [Google Scholar]
- 2.Brookes AJ. The essence of SNPs. Gene. 1999;234:177–186. doi: 10.1016/S0378-1119(99)00219-X. [DOI] [PubMed] [Google Scholar]
- 3.Xu J, et al. Double-stem Hairpin Probe and Ultrasensitive Colorimetric Detection of Cancer-related Nucleic Acids. Theranostics. 2016;6:318–327. doi: 10.7150/thno.13533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bryant KL, Mancias JD, Kimmelman AC, Der CJ. KRAS: feeding pancreatic cancer proliferation. Trends Biochem Sci. 2014;39:91–100. doi: 10.1016/j.tibs.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barault L, et al. Mutations in the RAS-MAPK, PI(3)K (phosphatidylinositol-3-OH kinase) signaling network correlate with poor survival in a population-based series of colon cancers. Int J Cancer. 2008;122:2255–2259. doi: 10.1002/ijc.23388. [DOI] [PubMed] [Google Scholar]
- 6.De Roock W, De Vriendt V, Normanno N, Ciardiello F, Tejpar S. KRAS, BRAF, PIK3CA, and PTEN mutations: implications for targeted therapies in metastatic colorectal cancer. Lancet Oncol. 2011;12:594–603. doi: 10.1016/S1470-2045(10)70209-6. [DOI] [PubMed] [Google Scholar]
- 7.Patolsky F, Lichtenstein A, Willner I. Detection of single-base DNA mutations by enzyme-amplified electronic transduction. Nat Biotechnol. 2001;19:253–257. doi: 10.1038/85704. [DOI] [PubMed] [Google Scholar]
- 8.Xu H, et al. Label-free colorimetric detection of cancer related gene based on two-step amplification of molecular machine. Biosens Bioelectron. 2017;90:314–320. doi: 10.1016/j.bios.2016.12.003. [DOI] [PubMed] [Google Scholar]
- 9.Zhu K, et al. Mutations of KRAS and PIK3CA as independent predictors of distant metastases in colorectal cancer. Med Oncol. 2014;31:16. doi: 10.1007/s12032-014-0016-6. [DOI] [PubMed] [Google Scholar]
- 10.Sidransky D, et al. Identification of ras oncogene mutations in the stool of patients with curable colorectal tumors. Science. 1992;256:102–105. doi: 10.1126/science.1566048. [DOI] [PubMed] [Google Scholar]
- 11.Ahrendt SA, et al. Cigarette smoking is strongly associated with mutation of the K-ras gene in patients with primary adenocarcinoma of the lung. Cancer. 2001;92:1525–1530. doi: 10.1002/1097-0142(20010915)92:6<1525::AID-CNCR1478>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 12.Iacobuzio-Donahue CA. Genetic evolution of pancreatic cancer: lessons learnt from the pancreatic cancer genome sequencing project. Gut. 2012;61:1085–1094. doi: 10.1136/gut.2010.236026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Minamoto T, Ronai Z. Gene mutation as a target for early detection in cancer diagnosis. Crit Rev Oncol Hematol. 2001;40:195–213. doi: 10.1016/S1040-8428(01)00098-1. [DOI] [PubMed] [Google Scholar]
- 14.Lievre A, Blons H, Laurent-Puig P. Oncogenic mutations as predictive factors in colorectal cancer. Oncogene. 2010;29:3033–3043. doi: 10.1038/onc.2010.89. [DOI] [PubMed] [Google Scholar]
- 15.Meldrum C, Doyle MA, Tothill RW. Next-generation sequencing for cancer diagnostics: a practical perspective. Clin Biochem Rev. 2011;32:177–195. [PMC free article] [PubMed] [Google Scholar]
- 16.Li X, et al. Surface Enhanced Raman Spectroscopy (SERS) for the Multiplex Detection of Braf, Kras, and Pik3ca Mutations in Plasma of Colorectal Cancer Patients. Theranostics. 2018;8:1678–1689. doi: 10.7150/thno.22502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alvarez-Garcia V, et al. A simple and robust real-time qPCR method for the detection of PIK3CA mutations. Sci Rep. 2018;8:4290. doi: 10.1038/s41598-018-22473-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ho TH, et al. Extendable blocking probe in reverse transcription for analysis of RNA variants with superior selectivity. Nucleic Acids Res. 2015;43:e4. doi: 10.1093/nar/gku1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mehes G, et al. Activating BRAF V600E mutation in aggressive pediatric Langerhans cell histiocytosis: demonstration by allele-specific PCR/direct sequencing and immunohistochemistry. Am J Surg Pathol. 2014;38:1644–1648. doi: 10.1097/PAS.0000000000000304. [DOI] [PubMed] [Google Scholar]
- 20.Gibriel AA, Adel O. Advances in ligase chain reaction and ligation-based amplifications for genotyping assays: Detection and applications. Mutat Res. 2017;773:66–90. doi: 10.1016/j.mrrev.2017.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu Q, Huang SQ, Ma F, Tang B, Zhang CY. Controllable Mismatched Ligation for Bioluminescence Screening of Known and Unknown Mutations. Anal Chem. 2016;88:2431–2439. doi: 10.1021/acs.analchem.5b04540. [DOI] [PubMed] [Google Scholar]
- 22.Barany F. Genetic disease detection and DNA amplification using cloned thermostable ligase. Proc Natl Acad Sci USA. 1991;88:189–193. doi: 10.1073/pnas.88.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen W, Lim CL, Gao Z. A ferrofluid-based homogeneous assay for highly sensitive and selective detection of single-nucleotide polymorphisms. Chem Commun (Camb) 2013;49:8114–8116. doi: 10.1039/c3cc43281e. [DOI] [PubMed] [Google Scholar]
- 24.Zhou L, et al. DNAzyme based gap-LCR detection of single-nucleotide polymorphism. Biosens Bioelectron. 2013;45:141–147. doi: 10.1016/j.bios.2013.01.061. [DOI] [PubMed] [Google Scholar]
- 25.Chen X, Ying A, Gao Z. Highly sensitive and selective colorimetric genotyping of single-nucleotide polymorphisms based on enzyme-amplified ligation on magnetic beads. Biosens Bioelectron. 2012;36:89–94. doi: 10.1016/j.bios.2012.03.045. [DOI] [PubMed] [Google Scholar]
- 26.Krzywkowski T, Nilsson M. Fidelity of RNA templated end-joining by chlorella virus DNA ligase and a novel iLock assay with improved direct RNA detection accuracy. Nucleic Acids Res. 2017;45:e161. doi: 10.1093/nar/gkx708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wee EJ, Rauf S, Shiddiky MJ, Dobrovic A, Trau M. DNA ligase-based strategy for quantifying heterogeneous DNA methylation without sequencing. Clin Chem. 2015;61:163–171. doi: 10.1373/clinchem.2014.227546. [DOI] [PubMed] [Google Scholar]
- 28.Cheng Y, Zhao J, Jia H, Yuan Z, Li Z. Ligase chain reaction coupled with rolling circle amplification for high sensitivity detection of single nucleotide polymorphisms. Analyst. 2013;138:2958–2963. doi: 10.1039/c3an36920j. [DOI] [PubMed] [Google Scholar]
- 29.Kou HS, Wang CC. Molecular inversion probes equipped with discontinuous rolling cycle amplification for targeting nucleotide variants: Determining SMN1 and SMN2 genes in diagnosis of spinal muscular atrophy. Anal Chim Acta. 2017;977:65–73. doi: 10.1016/j.aca.2017.04.037. [DOI] [PubMed] [Google Scholar]
- 30.Xie S, Tang Y, Tang D. Converting pyrophosphate generated during loop mediated isothermal amplification to ATP: Application to electrochemical detection of Nosema bombycis genomic DNA PTP1. Biosens Bioelectron. 2018;102:518–524. doi: 10.1016/j.bios.2017.11.066. [DOI] [PubMed] [Google Scholar]
- 31.Seok Y, et al. A Paper-Based Device for Performing Loop-Mediated Isothermal Amplification with Real-Time Simultaneous Detection of Multiple DNA Targets. Theranostics. 2017;7:2220–2230. doi: 10.7150/thno.18675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu N, et al. Development of a multiplex loop-mediated isothermal amplification method for the simultaneous detection of Salmonella spp. and Vibrio parahaemolyticus. Sci Rep. 2017;7:45601. doi: 10.1038/srep45601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang X, Lowe SB, Gooding JJ. Brief review of monitoring methods for loop-mediated isothermal amplification (LAMP) Biosens Bioelectron. 2014;61:491–499. doi: 10.1016/j.bios.2014.05.039. [DOI] [PubMed] [Google Scholar]
- 34.Zhang LL, et al. Combination of Loop-Mediated Isothermal Ampli fi cation Assay and Nested PCR for Detection of Borrelia burgdorferi sensu lato in Human Serum Samples. Biomed Environ Sci. 2015;28:312–315. doi: 10.3967/bes2015.044. [DOI] [PubMed] [Google Scholar]
- 35.Li Z, et al. Detection of Novel duck reovirus (NDRV) using visual reverse transcription loop-mediated isothermal amplification (RT-LAMP) Sci Rep. 2018;8:14039. doi: 10.1038/s41598-018-32473-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Du W, Lv M, Li J, Yu R, Jiang J. A ligation-based loop-mediated isothermal amplification (ligation-LAMP) strategy for highly selective microRNA detection. Chem Commun (Camb) 2016;52:12721–12724. doi: 10.1039/c6cc06160e. [DOI] [PubMed] [Google Scholar]
- 37.Liu J, et al. An improved allele-specific PCR primer design method for SNP marker analysis and its application. Plant Methods. 2012;8:34. doi: 10.1186/1746-4811-8-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu T, Xiao X, Zhang Z, Zhao M. Enzyme-mediated single-nucleotide variation detection at room temperature with high discrimination factor. Chem Sci. 2015;6:1206–1211. doi: 10.1039/c4sc03375b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang YS, et al. Robust strand exchange reactions for the sequence-specific, real-time detection of nucleic acid amplicons. Anal Chem. 2015;87:3314–3320. doi: 10.1021/ac504387c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Detection of KRAS mutation via ligation-initiated LAMP reaction