Abstract
Lipid composition and macromolecular crowding are key external effectors of protein activity and stability whose role varies between different proteins. Therefore, it is imperative to study their effects on individual protein function. CYP2J2 is a membrane‐bound cytochrome P450 in the heart involved in the metabolism of fatty acids and xenobiotics. In order to facilitate this metabolism, cytochrome P450 reductase (CPR), transfers electrons to CYP2J2 from NADPH. Herein, we use nanodiscs to show that lipid composition of the membrane bilayer affects substrate metabolism of the CYP2J2‐CPR nanodisc (ND) system. Differential effects on both NADPH oxidation and substrate metabolism by CYP2J2‐CPR are dependent on the lipid composition. For instance, sphingomyelin containing nanodiscs produced more secondary substrate metabolites than discs of other lipid compositions, implying a possible conformational change leading to processive metabolism. Furthermore, we demonstrate that macromolecular crowding plays a role in the lipid‐solubilized CYP2J2‐CPR system by increasing the Km and decreasing the Vmax, and effect that is size‐dependent. Crowding also affects the CYP2J2‐CPR‐ND system by decreasing both the Km and Vmax for Dextran‐based macromolecular crowding agents, implying an increase in substrate affinity but a lack of metabolism. Finally, protein denaturation studies show that crowding agents destabilize CYP2J2, while the multidomain protein CPR is stabilized. Overall, these studies are the first report on the role of the surrounding lipid environment and macromolecular crowding in modulating enzymatic function of CYP2J2‐CPR membrane protein system.
Keywords: macromolecular crowding, CYP2J2, cytochrome P450, protein–protein interactions, lipid bilayer
Abbreviations
- CAR
carebastine
- Chol
cholesterol
- CPR
cytochrome P450 reductase
- CYP
cytochrome P450
- EBS
ebastine
- FRET
Forster resonance energy transfer
- HYD
hydroxyebastine
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- POPC
1‐palmitoyl‐2‐oleoyl‐sn‐glycero‐3‐phosphocholine
- POPS
1‐palmitoyl‐2‐oleoyl‐sn‐glycero‐3‐phospho‐L‐serine
- SM
sphingomyelin
Introduction
Membrane proteins are surrounded by lipids. Therefore, it is important to consider the consequence of membrane lipid composition on enzyme activity. Lipid composition is not uniform throughout different organelles within a cell. For instance, the endoplasmic reticulum (ER) in particular contains over 50% phosphatidylcholine (PC) and approximately 20% phosphatidylethanolamine (PE).1 This bears relevance to our investigations on metabolism and activity because the surrounding lipid environment can affect protein stability and activity.2, 3, 4, 5 Previous research has shown that GM2 activator protein is inhibited by high levels of cholesterol (CHOL) and sphingomyelin (SM) in the membrane, whereas sphingomyelinase is activated by high levels of CHOL.5, 6, 7 Membrane lipid composition has also been shown to affect protein structural conformations. For instance varying lipid compositions change the helicity of hIAPP.8 There have been reports that both CHOL and SM are enriched in lipid rafts and there have been indications that protein activity is different in lipid rafts.9, 10, 11 Herein, we use different lipid compositions, to best understand the effects of lipid type on a well‐characterized membrane protein system.
The cellular environment is also naturally crowded, with numerous proteins and small molecules. For instance, Escherichia coli contain between 300–400 mg/mL of assorted proteins and RNA, while mammalian cells are 20%–30% occupied by macromolecules.12, 13, 14 Such a complex environment is difficult to mimic in an in vitro experiment. Therefore, most biochemical studies are performed in buffers where enzymes and substrates are in dilute solutions. However, it has been noted that some enzymes function differently in a crowded environment than in dilute solution that makes it necessary to mimic a crowded environment in enzymatic studies.15, 16 Investigating enzymatic behavior in the presence of other cellular components reveals that enzymes have been evolutionarily optimized to function in crowded conditions.17 While a cell may be filled with macromolecules at a high overall concentration, individual proteins are far less concentrated.15 However, the presence of other macromolecules leaves much of the cellular volume unavailable, greatly increasing their effective concentration—often termed the excluded volume effect.12, 18, 19 Therefore, instead of floating uninhibited, molecules are intertwined in a delicate balance of steric hindrance and soft attractions.20, 21 It is commonly hypothesized that such a crowded environment promotes compact protein conformations, thus favoring substrate metabolism.22, 23 Previous studies on macromolecular crowding effects have mostly focused on protein folding, structure, and stability of soluble proteins.16, 24, 25, 26, 27, 28 For instance, it was shown that macromolecular crowding, could affect subunit association of the tetrameric protein GAPD.16 Other investigations revealed gains in secondary structure for the proteins flavodoxin and GGBP, as well as increased thermal stability.27 However, it has also been shown that macromolecular crowding contributes to protein aggregation.29, 30, 31, 32, 33, 34 There have been numerous previous studies on the enzymatic behavior in a crowded environment focusing on soluble proteins.17, 19, 30, 35 In some cases, crowding promotes increased catalytic efficiency. Poggi et al. showed that malate oxidation by malate dehydrogenase (MDH) increased in rate when performed in the presence of charged protein crowding agents.17 In contrast, other reports have shown that increased solution viscosity due to macromolecular crowding slows diffusion, which can alter enzyme kinetics if the rate limiting step is affected.19 For example, alcohol dehydrogenase (YADH) was found to have a lower Vmax and Km when crowded, meaning that although kinetics were slowed, the enzyme had higher substrate affinity.19 Enzymes with multiple distinct kinetic steps have even been found to have increased kinetics for one step, while another is decreased. Myosin ATPase activity increased by 30% when crowded with Ficoll 70, whereas dissociation and hydrolysis decreased by 45‐ and 3.5‐fold, respectively.30 Hence, while some research reports report a relation between crowding agent size and enzymatic activity,35 other groups do not find such correlation.17, 19
Herein, we study the role of both lipid composition and macromolecular crowding in controlling the function of membrane bound cytochrome P450 2 J2 (CYP2J2) and its redox partner cytochrome P450 reductase (CPR). CYP2J2 is an epoxygenase expressed mainly in human cardiomyocytes and brain. It oxidizes a variety of substrates including xenobiotics and fatty acids.36 It is anchored into the membrane by its N‐terminus hydrophobic residues and F‐G loop residues.37 CPR is an essential oxidoreductase for nearly all P450s and thus it is important to understand the protein–protein interactions between CYP2J2 and CPR. We use nanodiscs that are nanoscale lipid bilayers to incorporate CYP2J2‐CPR with controlled membrane compositions in order to evaluate the role of lipid composition on the catalytic activity of this system [Fig. 1(A)]. Previous studies have shown that lipid membrane composition can affect not only P450 substrate metabolism, but also localization.38 CYP1A2 has increased metabolic activity in the presence of cholesterol and sphingomyelin containing membranes and has been shown to localize to ordered domains within the membrane.39 CYP2E1, however, localizes to disordered domains, while CYP2B4 shows no specific localization.38 Also of importance is the effect of the membrane environment on redox potential and protein folding compared to that of a detergent or lipid‐solubilized system. It has previously been shown that while both soluble‐ and membrane‐bound CPR have similar structures, the redox potential of CPR in a lipid bilayer shifts in the positive direction.40 Herein, using the CYP2J2‐CPR‐ND system, we will be able to specifically tune lipid composition to determine the effects of lipids on metabolic activity and protein–protein interactions.
Figure 1.
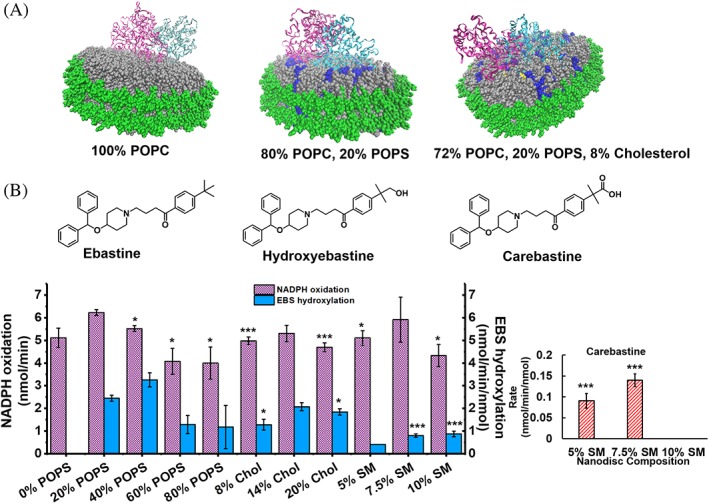
Variation of nanodisc lipid composition and its effects on CYP2J2 substrate metabolism. (A) CYP2J2 and CPR docked in nanodiscs of varying lipid compositions. Gray = POPC; blue = POPS; yellow = cholesterol; green = MSP. CYP2J2 is docked in the nanodisc via its N‐terminus and F‐G loop. CPR is added to solution separately and self‐associates with the lipid bilayer. (B) Structures of ebastine and its metabolites. Measurement of rate of NADPH oxidation and rate of metabolite formation from ebastine metabolism by CYP2J2‐CPR‐nanodiscs with varied lipid compositions. Inset shows carebastine production in SM containing nanodiscs. Rates are all calculated from within the initial, linear range. All metabolism data is compared to 20% POPS. * = P < 0.05; ** = P < 0.01; *** = P < 0.005. SEM is calculated from multiple measurements.
CYP2J2 and its redox partner CPR are membrane‐bound proteins, and there are no previous studies on the effects of crowding on membrane proteins. Most studies that show macromolecular crowding can affect the rate of substrate metabolism are performed with soluble proteins.17, 19, 30 Therefore, this is the first report on the effect of macromolecular crowding on CYP2J2‐CPR enzymatic activity. Cytochrome P450 activity is known to be affected by the presence of other P450s in CYP–CYP interactions which, while not an exact equivalent to the effects of a nonspecific macromolecule, can still be considered a form of macromolecular crowding. Most pertinent to our system would be the formation of P450–P450 complexes that affect the binding of CPR or catalytic efficiency.41 Such phenomena have been noted in literature for the P450s 1A2, 2B4, 2E1, 3A4, and others.42, 43, 44
In our study, we chose to compare the effects of the branched‐chain polymers—Ficoll and Dextran—on CYP2J2–CPR interactions in nanodiscs. Two sizes of each crowding agent will be used, one that is similar in size to the 57 kDa CYP2J2, as well as a larger size more than seven times the mass of the protein of interest, which may exacerbate the macromolecular crowding phenomena. Polymer crowding agents such as Ficoll are popular in macromolecular crowding investigations due to their low potential for interaction with the system of interest.
Overall, we determine the role of lipid composition on CYP2J2‐CPR‐ND mediated substrate metabolism. Then we further use macromolecular crowding agents to understand if they alter the CYP2J2‐CPR‐ND enzyme kinetics with respect to substrate metabolism.
Results
Assembly of CYP2J2‐CPR‐NDs with varying lipid composition
In order to study how the variation in lipid composition affects CYP2J2‐CPR‐ND activity, we prepared multiple sets of nanodiscs with different percentages of various lipids to mimic lipids composition of the cell membranes as described in materials and methods section. For our study, lipids were chosen to vary the amount of zwitterionic, anionic, and sphingolipids, as well as CHOL as these are all key components of cellular membranes. The corresponding chromatograms of the nanodiscs with different lipid compositions are provided in the Supporting Information Fig. S1. All the nanodiscs were prepared with POPC (1‐palmitoyl‐2‐oleoyl‐sn‐glycero‐3‐phosphocholine) as the base lipid, as the zwitterionic phosphatidyl choline is a principal component of most mammalian lipid bilayers.1, 45 POPS (1‐palmitoyl‐2‐oleoyl‐sn‐glycero‐3‐phospho‐L‐serine) was used to provide anionic lipid content and its percentage was varied from 0% to 80% in the nanodisc. Previously it was shown that these lipid compositions lead to the formation of stable nanodiscs.46 We then used the most optimum 20% POPS nanodiscs and changed the cholesterol composition from 0% to 20%. We separately prepared nanodiscs with 20% POPS and varying sphingomyelin concentration from 0% to 10%. Both sphingomyelin and cholesterol are important components of ER membranes where CYP2J2 and CPR are found inside the cells. All the CYP2J2‐CPR‐NDs with different lipid compositions formed uniform, homogenous nanodiscs according to size exclusion chromatography (Fig. S1). Both 14% and 20% CHOL‐containing discs were slightly smaller than nanodiscs with alternate compositions, as shown by the slightly later elution time [Supporting Information Fig. S1(g), S1(h)].
Characterization of ebastine metabolism by CYP2J2‐CPR‐NDs with varying lipid composition
In order to metabolize drugs, CYP2J2 requires electrons that are provided by NADPH via CPR. Therefore, we first measured NADPH oxidation by CYP2J2‐CPR‐ND in presence of ebastine. CYP2J2 is mainly responsible for the hydroxylation of the H1‐receptor antagonist ebastine in human liver and intestinal microsomes.47 We observed that NADPH oxidation was the greatest in 20% POPS discs, followed closely by 40% POPS discs [Fig. 1(B)]. As shown in Figure 1, nanodisc compositions containing greater than 60% and 80% POPS had lower NADPH oxidation rates compared to those at 20% POPS. Both the 8% and 20% CHOL‐containing discs produced slightly lower oxidation rates. The 7.5% SM‐containing nanodiscs had similar NADPH oxidation rates to 20% POPS discs.
In order to evaluate if the electrons from NADPH are productively used to metabolize ebastine, we next measured ebastine (EBS) hydroxylation by CYP2J2‐CPR‐NDs. We found that the production of EBS metabolites followed a similar trend to NADPH oxidation observed for the different nanodiscs [Fig. 1(B)]. Ebastine metabolism gives rise to two products—hydroxyebastine (HYD) and carebastine (CAR). Carebastine is formed when hydroxyebastine is further metabolized by CYPs. We developed an HPLC‐UV method to detect EBS, HYD, and CAR as mentioned in the materials and methods section. The highest amount of HYD was formed with the 20% POPS discs and the 40% POPS discs. Interestingly, 100% POPC NDs had much lower metabolism of EBS indicating a need for the presence of anionic lipids to facilitate CYP2J2 and CPR interactions, as has been seen reported in previous work from our laboratory and others.4, 48, 49 All the three CHOL‐containing disc compositions resulted in similar HYD production. All varieties of POPC/POPS discs and POPC/POPS/CHOL discs exhibited limited formation of carebastine (CAR). It is important to note that SM containing discs had a much higher percentage of CAR formation for the 5% and 7.5% SM discs, respectively [Fig. 1(B), inset], at the expense of HYD formation. This was interesting as this indicated that in SM containing nanodiscs, the HYD product did not leave the active site and was metabolized again to CAR.
Next, we evaluated the coupling efficiency, which is defined as the rate of EBS hydroxylation (HYD formation) divided by the rate of NADPH oxidation and is used as a measure of efficient use of electrons from NADPH to facilitate metabolism. While bacterial P450s such as cytochrome P450cam have high coupling efficiencies, this value is typically low for membrane‐bound CYPs.50, 51, 52, 53 For different nanodiscs containing only POPC and POPS, coupling efficiency increases from 0% to 40% POPS, indicating that having anionic lipids in the nanodisc increases the efficiency of electron transfer from CPR to CYP2J2 (Fig. S2). CHOL‐containing discs did not have significant coupling efficiency variation. For further macromolecular crowding studies, we chose to use the 20% POPS discs, as they have robust metabolite production and NADPH oxidation, as shown in Figure 1.
Kinetic effects of macromolecular crowding on the metabolism of ebastine by CYP2J2‐CPR‐NDs in comparison to a lipid‐solubilized system
To evaluate the role of the macromolecular crowding agents on the CYP2J2‐CPR mediated metabolism of EBS, we investigated the changes in kinetics of EBS hydroxylation in the presence of sucrose, Ficoll 70, and Ficoll 400 in a lipid‐solubilized system (LSS). There was a steady increase in Km with the addition of increasing sizes of crowding agents. Of note was the effect of Ficoll 70 on CYP2J2‐CPR‐LSS, which has a much lower Vmax (6.64 nmol∙min−1∙nmol−1) than the control or other crowding agents. The LSS was also used to examine the effects of different concentrations of macromolecular crowding agent. As seen in Figure 2, Ficoll 70 exhibited a decrease in NADPH oxidation with increasing crowding agent [Fig. 2(A)], and an increase both in EBS hydroxylation and coupling efficiency [Fig. 2(B,C)].
Figure 2.
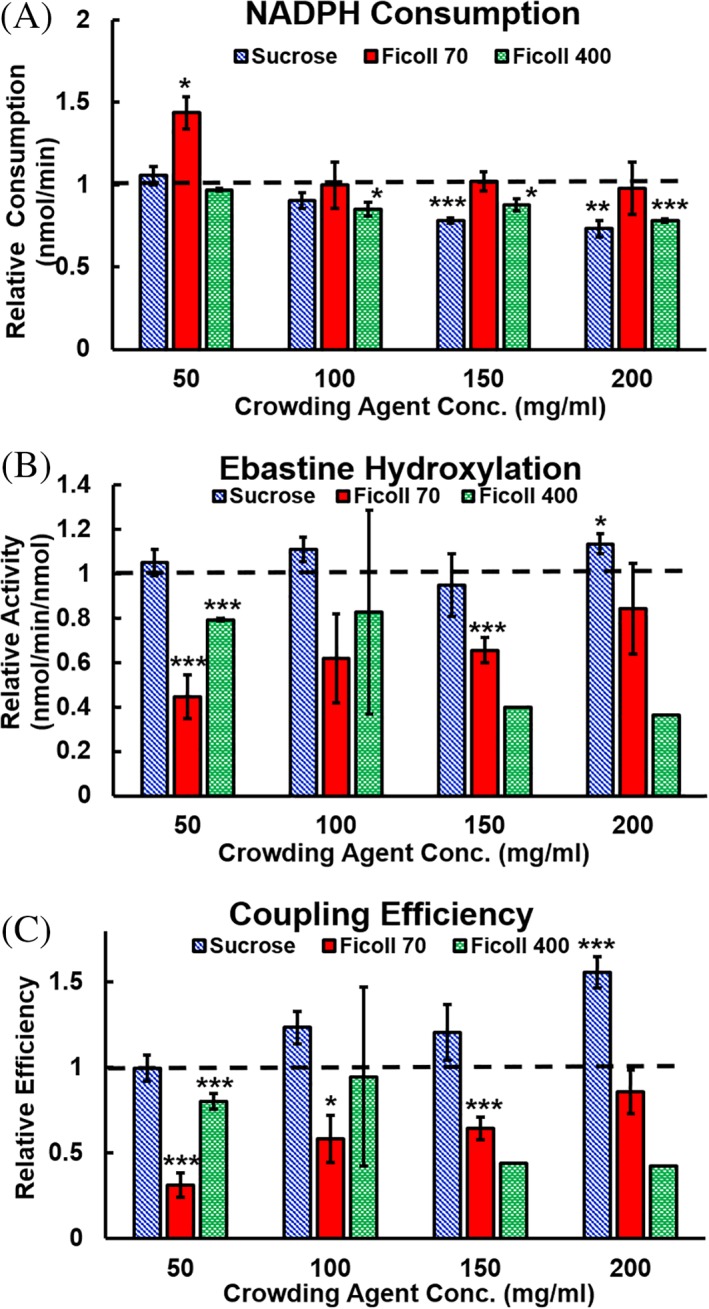
Relative kinetics for a lipid solubilized system with varying concentrations of macromolecular crowding agent. (A) NADPH consumption for the CYP2J2‐CPR‐LSS, (B) ebastine hydroxylation, and (C) coupling efficiency. The dashed line indicates the uncrowded control to which the crowding data is compared. Significant difference from uncrowded solution is indicated by asterisks: *P < 0.05, **P < 0.01, ***P < 0.005.
To examine the effect of crowded protein–protein interactions in a more membrane‐like environment we studied the CYP2J2‐CPR‐ND system with Dextran 75 and Dextran 500 in addition to Ficoll 70 and Ficoll 400 as macromolecular crowding agents. As shown in Figure 3(B), uncrowded 20% POPS nanodiscs exhibited a Vmax of 4.05 nmol∙min−1∙nmol−1 with a Km of 9.44 μM. Nanodiscs crowded by the monomer sucrose had a similar Vmax of 3.93 nmol∙min−1∙nmol−1 and a Km of 7.91 μM. Ficoll 70 crowded nanodiscs had no significant change in Vmax, but the Km did slightly decrease to 5.87 μM. Ficoll 400 crowded nanodiscs were the only group that showed an elevated Km (13.34 μM). Of note were the effects of Dextran crowding on the CYP2J2‐CPR‐ND system. While Dextran 500 decreased the Vmax to 2.72 nmol∙min−1∙nmol−1, it was Dextran 75 that had the greatest effect—decreasing the Vmax to 1.40 nmol∙min−1∙nmol−1 and the Km to 2.84 μM. Overall, macromolecular crowding exhibits a marked effect on the CYP2J2‐CPR‐ND system. This is the first account of measuring the role of crowding agents on membrane proteins. In order to understand why the crowded environment affects membrane protein function, we studied the effect of macromolecular crowding on the folding of the individual membrane proteins—CYP2J2 and CPR.
Figure 3.
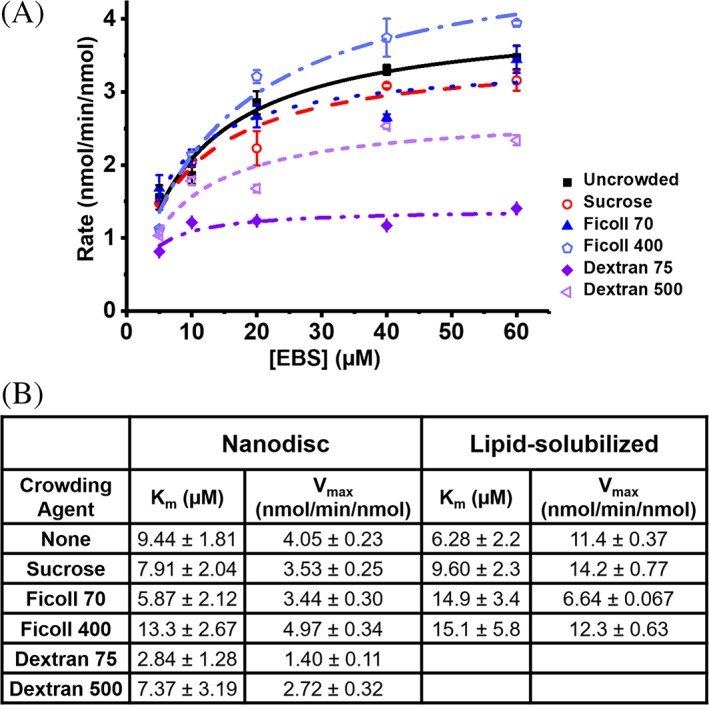
Kinetics for a nanodisc solubilized system and effects of macromolecular crowding on CYP2J2 and CPR stability. (A) Michaelis–Menten curves for 80% POPC/20% POPS nanodiscs crowded with 100 mg/mL of macromolecular crowding agent. (B) Kinetic constants for nanodisc and lipid‐solubilized CYP2J2 metabolism of EBS. Error shown is the standard error for all points at that concentration of EBS (separated by crowding agent).
Protein stability and ΔG unfolding
Macromolecular crowding usually affects protein folding as shown previously with soluble proteins.18, 24, 27, 54 Therefore, equilibrium protein unfolding of CYP2J2 and CPR was measured in the presence of increasing concentrations of the chaotropic agent guanidinium hydrochloride (Gu‐HCl), in the presence of different crowding agents. Tryptophan fluorescence was excited at 295 nm and measured at 321(λmax, folded) and 360 nm (λmax, unfolded) as described in the material and methods section. The ratio of A360/A321 was used to determine the fraction of unfolded proteins. The equation ΔG = −RT∙ln(fD/[1‐fD])55 was used to calculate ΔG, with fD representing the fraction of denatured protein. From this ΔG0 was calculated using ΔG = m[Gu‐HCl] + ΔG0. As shown in Figure 4, CYP2J2 denaturation was destabilized by both Ficoll 70 and sucrose. The ΔG0 value decreased from 4.32 kJ/mol for uncrowded protein to 3.44 and 2.89 kJ/mol for sucrose and Ficoll 70 crowded protein, respectively [Fig. 4(A), 4(C)]. In contrast, for CPR sucrose stabilized CPR while there was no effect from Ficoll 70. ΔG0 values increased from 4.56 kJ/mol for uncrowded CPR, to 5.82 kJ/mol for sucrose crowded protein while remaining at 4.60 kJ/mol for Ficoll 70 crowded protein [Fig. 4(B, C)]. However, in comparison to CYP2J2, CPR is less stable and unfolds in the presence of lower concentrations of denaturant, reaching maximum denaturation between 3 and 4 M Gu‐HCl, while CYP2J2 continued to denature up to 6–7 M Gu‐HCl [Fig. 4(A,B)].
Figure 4.
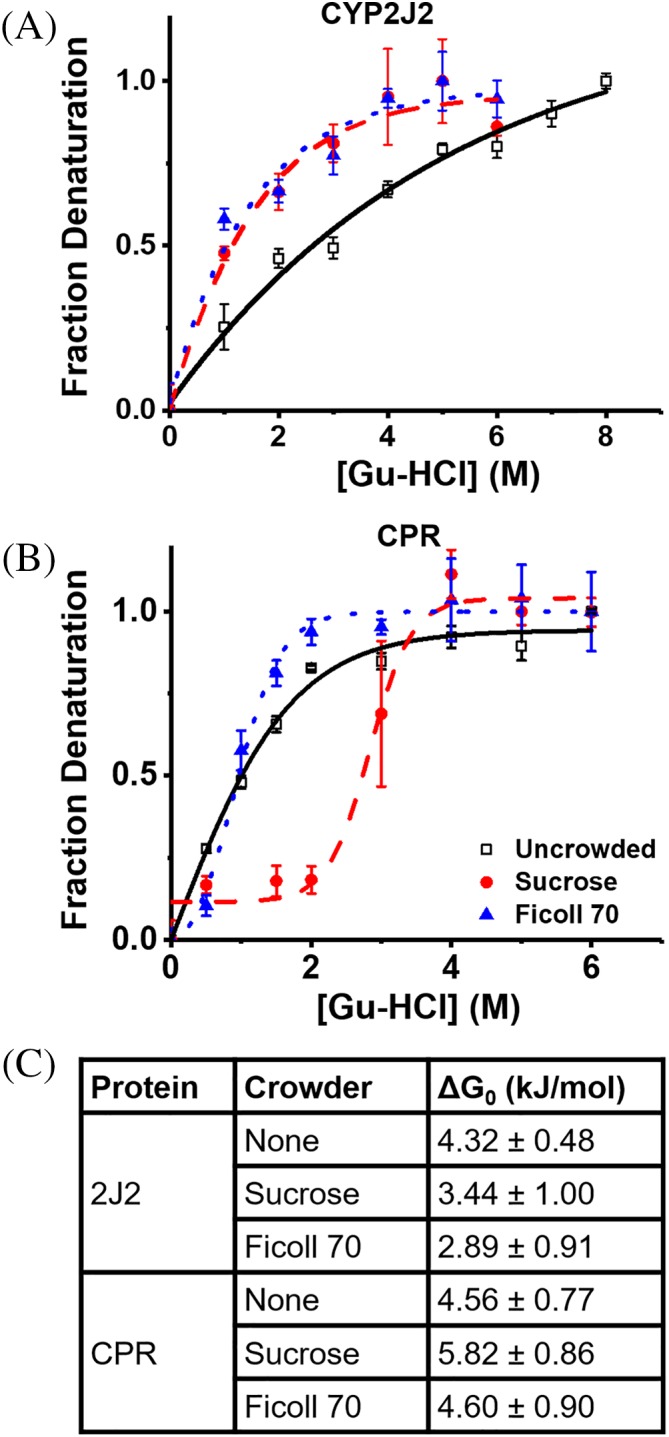
Effects of macromolecular crowding on CYP2J2 and CPR stability. (A) Denaturation curves for CYP2J2 in the presence of water, sucrose, and Ficoll 70. (B) Denaturation curves for CPR under the same conditions. (C) Calculated ΔG0 values for CPR and CYP2J2 in crowded and uncrowded solution. Higher ΔG0 values indicate a higher degree of stability.
Discussion
The importance of lipid composition
CYP2J2 epoxygenase and its redox partner CPR were assembled into NDs with controlled ratios of lipids as described in the materials and methods section (Table 1). Lipids were chosen to test a variety of key ER membrane lipids at a wide range of concentrations and their effects on CYP2J2‐CPR enzymatic activity as shown previously with other P450s. The ER is a major site of lipid and CHOL biosynthesis and consists of the zwitterionic lipids phosphatidylcholine (PC) and phosphatidylethanolamine (PE), as well as a smaller percentage of the anionic lipids phosphatidylinositol (PI) and phosphatidylserine (PS).56 CHOL and sphingolipids, both of which are synthesized in the ER and then transported to other organelles, are also present in low concentrations.39 CYP2J2 metabolizes drugs such as EBS and converts it to HYD and CAR [Fig. 1(B)]. In order to facilitate the metabolism of drugs by CYP2J2, CPR shuttles electrons from NADPH to the CYP2J2 heme active site.57, 58 Therefore, we measured the NAPDH oxidation rates which are indicative of how fast CPR transfers electrons gained from NADPH oxidation to CYP2J2. These electrons are used by CYP2J2 either in the productive pathway to generate ebastine metabolites, or an unproductive pathway to generate reactive oxygen species and water. NADPH oxidation data showed the highest activity in 20% POPS nanodiscs implying that anionic lipids aid favorable interactions between CYP2J2 and CPR that lead to efficient electron transfer [Fig. 1(B)]. CHOL and SM‐containing nanodiscs had much less variation in NADPH oxidation and substrate metabolism between the concentrations tested, as mentioned previously. This is expected, as the percentages of CHOL and SM were varied more narrowly than POPS, and remain closer to physiological membrane composition. CHOL‐containing discs have an overall lower rate of metabolism and NADPH oxidation compared to 20% POPS discs. There is a possibility that CHOL may directly bind to the protein active site as it does for CYP3A4, thus inhibiting enzymatic function.59 However such binding to CYP2J2 was not observed when CYP2J2‐CPR‐NDs were incubated with cholesterol. For all nanodisc types, NADPH oxidation rates were higher than rates of substrate metabolism and the electrons from NADPH and in such cases were shunted for the production of excess water and hydrogen peroxide, leading to poor coupling efficiencies.60, 61 This is common in mammalian P450s which show low coupling efficiency in comparison to bacterial P450s.62 Prokaryotic P450s are known to orient the A‐propionate of the heme to the distal side, while in eukaryotic P450s it is proximal. This distal orientation in prokaryotes is due to a stabilizing residue upstream from the heme‐binding residue that closes the substrate access channel and may increase catalytic efficiency.62
Table 1.
Lipid composition of all nanodisc varieties used in this article
| Nanodiscs | % POPC | % POPS | % Chol | % SM |
|---|---|---|---|---|
| 1 | 100 | 0 | 0 | 0 |
| 2 | 80 | 20 | 0 | 0 |
| 3 | 60 | 40 | 0 | 0 |
| 4 | 40 | 60 | 0 | 0 |
| 5 | 20 | 80 | 0 | 0 |
| 6 | 72 | 20 | 8 | 0 |
| 7 | 66 | 20 | 14 | 0 |
| 8 | 60 | 20 | 20 | 0 |
| 9 | 75 | 20 | 0 | 5 |
| 10 | 72.5 | 20 | 0 | 7.5 |
| 11 | 70 | 20 | 0 | 10 |
Ebastine metabolism gives rise to two products—hydroxyebastine (HYD) and carebastine (CAR). While all the different types of CYP2J2‐CPR‐NDs produced HYD from ebastine except for 100% POPC discs (just below quantifiable range), few lipid compositions lead to the production of any measurable CAR. SM‐containing discs were the only disc type to produce quantifiable CAR, the secondary metabolite of CYP2J2. The increased CAR production in SM nanodiscs may be due to substrate retention in the binding pocket of CYP2J2, which would promote the direct conversion of HYD to CAR as the product release is delayed. This is not unprecedented, as several other P450s have exhibited processive metabolism of substrates, such as the metabolism of dialkylnitrosamines by CYP2A6.63 This indicates that in the presence of SM, the CYP2J2 adopts a conformation where the HYD is unable to exit the active site and is hydroxylated again to form CAR. SM is known to have a high phase transition temperature compared to other lipids, which could lead to more stable CYP2J2‐CPR‐ND interactions.64 Another possibility is allosteric modulation of the CYP‐CPR complex by SM. While SM has not yet been implicated as an allosteric regulator, other membrane components have been shown to have allosteric effects on membrane proteins such as the GPCRs β2AR and A2AR.65, 66 Some CYPs are also known to be allosterically regulated, though not by membrane lipids.67, 68 However, multiple P450s and CPR have been shown to localize to ordered domains enriched with SM and CHOL.38 Nanodiscs formed from native membrane lipids also show similar enrichment.69 Proving allosteric regulation by SM is outside the scope of this article, but it is an important point to consider for future investigations. Overall the EBS metabolism by CYP2J2‐CPR‐ND system shows that the SM nanodiscs have lower coupling efficiency [Supporting Information Fig. S2] and lead to the formation of the secondary metabolite CAR, though the decreased coupling efficiency may also be attributed to the increased production of CAR.
Decreased coupling efficiency was also seen in discs with a high percentage of POPS, and can be attributed to changes in electrostatic repulsion, as well as differential metabolite formation. It has been previously shown that anionic phospholipids can be beneficial to P450 activity in NDs.70 However, anionic phospholipids normally comprise less than 30% of the membrane.71 Excess anionic phospholipids will experience electrostatic repulsion from one another, destabilizing the lipid bilayer. Conversely, anionic phospholipids have been shown to stabilize some CYPs and shift the high‐spin equilibrium.72 There is existing evidence that repulsion between POPC and the positively charged rhodopsin has a negative impact on arrestin‐1 binding, which is ameliorated with the addition of anionic POPS.73 There is also literature evidence of phosphatidylserine (PS) and phosphatidylethanolamine (PE) working in concert to provide opportunities for protein‐membrane binding that are not possible with lipids like POPC.74
Enzyme kinetics in a crowded system
There have been few investigations centered on the role of macromolecular crowding in membrane‐bound proteins or protein systems. We first used CYP2J2‐CPR in a lipid‐solubilized environment to study for how macromolecular crowding affects protein–protein interactions in a membrane‐bound protein system in the absence of lipid bilayers. We then refined this using CYP2J2‐CPR‐NDs. Soluble proteins are constantly affected by the contents of the cytosol, whereas membrane proteins have inherent protection from the cytosolic contents as they are embedded in a membrane. The lipid bilayer, while also crowding the proteins it contains, can simultaneously prevent extraneous interactions with cellular components. Therefore we first examined the effects of macromolecular crowding on CYP2J2‐CPR kinetics using a LSS before transitioning to the CYP2J2‐CPR‐ND system to determine if the membrane‐like environment of the nanodiscs would serve as a protective buffer for CYP2J2 and CPR. Outside the protection of the nanodisc environment, macromolecular crowding agents exert notable effects on NADPH oxidation, substrate metabolism, and coupling efficiency (Fig. 2). Of note are the effects exerted by Ficoll 70. A Ficoll 70 crowded LSS produces less EBS metabolites compared to the uncrowded system, which could be due to size effects, as Ficoll 70 is similar in molecular weight to CYP2J2 and CPR. It has been shown in literature that crowding agents of similar size to the proteins of interest have the greatest effect, but we must also consider the effect of crowding agent concentration. Because the greatest effects of Ficoll are seen at the lowest concentrations of crowding agent, there is a possibility that soft interactions are contributing to the change in kinetics. Literature shows that although Ficoll is widely considered to be neutral, it has a weak negative charge at the pH used for our experiments, likely due to random oxidation of end groups on its branched chain structure.75 Thus, weak electrostatic interactions are the main contributor to crowding effects on NADPH oxidation and EBS hydroxylation at low concentrations of macromolecular crowding agent. In the CYP2J2‐CPR‐LSS, both Ficoll 70 and Ficoll 400 have increased Km values [Fig. 3(B)], and Ficoll 70 has a decreased Vmax—showing that Ficoll 70 affects the kinetics of CYP2J2 substrate metabolism. Potential variations in structure and folding must also be considered, as the CYP2J2‐CPR‐LSS is outside the realm of a protective membrane bilayer. Solution NMR experiments on cytochrome b 5 and c interactions have shown that while a soluble system has faster micro‐rates, the productive complex as a whole is more stable in a membrane bilayer.76
Within CYP2J2‐CPR‐NDs, Dextran 75 and Dextran 500 exert the greatest effects [Fig. 3(B)]. When ensconced in a nanodisc, CYP2J2 and CPR no longer experience macromolecular crowding as independent entities, but as a larger complex. Thus one might assume any meaningful steric interactions would require a crowding agent of similar size35, 77 to the protein‐ND complex in order to effectively exclude volume. However, the Vmax and Km of EBS hydroxylation are approximately 3‐fold lower in Dextran 75 crowded solution compared to uncrowded, and over 2‐fold lower compared to Ficoll 70. This indicates that the effects of macromolecular crowding are not solely dependent on the molecular mass. While the Ficoll and Dextran crowding agents used were of similar masses, Ficoll is much more compressible than Dextran, and has been shown to adsorb monovalent cations such as K+. Additionally, at concentrations above 2.5% (wt/vol), relative excluded volume by Ficoll is reduced, likely due to compression.75, 78, 79 Dextran has a larger Stokes‐Einstein radius and therefore can exclude greater volume and more effectively crowd a given solution. It may also be that various crowding agents alter the complex formation of CYP2J2 and CPR. Previous literature on CYP2B4 and CPR shows that the anionic loop of the FMN domain interacts with cationic CYP surface to form a productive complex with the two prosthetic groups in range for electron transfer.80 The presence of larger volume Dextran molecules could force the CYP‐CPR complex away from an optimal position for electron transfer, thus affecting the kinetics. Therefore, we can conclude that crowding exerts effects on the nanodisc solubilized system dependent on the molecular volume and crowding agent identity.
The stabilizing effects of macromolecular crowding agents
It has been shown previously that the presence of crowding agents increases the stability of soluble globular proteins.19 Herein, we see different effects of the crowding on the two membrane protein CYP2J2 and CPR. Interestingly, CYP2J2 is destabilized by macromolecular crowding agents indicating changes in the hydration shell of the protein.81 Biological macromolecules are normally surrounded by a constrained layer of water that does not behave as free‐flowing liquid and can have pronounced effects on molecular dynamics.82 Due to the disruption caused by crowding agents, a compromised hydration shell has led to CYP2J2 denaturant susceptibility. Previous experiments have shown the gradual loss of heme via chaotropic denaturation and support our findings that uncrowded CYP2J2 is stable.37 However, it is clear that macromolecular crowding agents perturb the stability. On the contrary, CPR is stabilized in the presence of sucrose, causing the protein to adopt a more compact state. It is known that CPR has two flavin domains that come together for electron transfer. While we speculated that the presence of macromolecular crowders would aid in bringing the two domains together, thus giving it a more stable conformation,83, 84, 85 there is no evidence that this occurs in the presence of Ficoll 70. The stabilizing effects of sucrose may also be due to favorable changes in the hydration shell. Literature shows that as guanidinum ions induce changes in the hydration shell, the affected protein becomes more flexible.86 While this would be undesirable in CYP2J2, as it would eventually lead to heme loss, in CPR it could cause domain movement leading to a more compact, stable state.
Conclusions
Taken together, we have found that membrane lipid composition plays a key role in CYP2J2‐CPR interactions and ebastine metabolism. We show that a certain percentage of anionic lipid increases rate of metabolism and coupling efficiency. We also observe that including sphingomyelin along with 20% POPS nanodisc leads to the formation of the CAR, implying that this particular lipid composition is either an allosteric regulator or induces conformational change that disfavors HYD product release and promotes processive metabolism to form the CAR product.
We further probe the role of macromolecular crowding on the CYP2J2‐CPR‐LSS and CYP2J2‐CPR‐ND system. In the CYP2J2‐CPR‐LSS system, Ficoll 70 decreases the Vmax and increases the Km, implying that substrate affinity decreases. In CYP2J2‐CPR‐ND system, Ficoll has no effect on the kinetics of ebastine metabolism. However, Dextran decreases Km and Vmax showing that the substrate is tightly bound in the active site and does not get metabolized. Separately, macromolecular crowding agents have differential effects on the stability of both CYP2J2 and CPR that depend on a complex combination of macromolecular hydration, flexibility, and the identity of the protein. CPR is clearly stabilized by the presence of crowders, while CYP2J2 is destabilized.
Overall, these studies present the interesting observation that depending on the environment of the enzyme (lipid composition or macromolecular crowding), the affinity for the substrate and rate of metabolism changes.
Materials and Methods
Materials
Human CYP2J2 cDNA was obtained from OriGene (Catalog No. SC321730) and N‐terminally modified.60 Ampicillin, arabinose, chloramphenicol, isopropyl β‐D‐1‐thiogalactopyranoside (IPTG), and Ni‐NTA resin were obtained from Gold Biotechnology and Sigma. δ‐aminolevulinic acid (δ‐ALA) was obtained from Frontier Scientific. NADPH and NADP+ were obtained from Research Products International. POPC and POPS were purchased from Avanti Polar Lipids, Inc. Ficoll 70 and Ficoll 400 were purchased from Sigma. Sucrose and CHOL were purchased from Fisher Scientific. POPC, POPS, and SM were obtained from Avanti Polar Lipids. Guanidine hydrochloride was also purchased from Sigma.
Protein expression and purification
Truncated M2D34G‐CYP2J2 was first expressed in E. coli and purified via Ni‐NTA chromatography as previously published.60, 87 This construct of CYP2J2 contains an N‐terminal modification in which residues 3–35 have been deleted, as well as an L2A mutation. Full length Rattus norvegicus Cytochrome P450 Reductase (RnCPR) was expressed in E. coli and thereafter purified via ADP‐agarose chromatography following previously published protocols.88
Lipid solubilized system preparation
A mixture of 80% POPC and 20% POPS was dried on a rotary evaporator, followed by overnight drying in a vacuum desiccator. The lipids were then solubilized to 10 mM in ddH2O via sonication and were stored at −20°C until needed.
Nanodisc incorporation
Purified CYP2J2 was incorporated into nanodiscs containing POPC, POPS, CHOL, and SM according to the concentrations listed in Table 1. Lipids solubilized in CHCl3 were aliquoted to the desired mol% and dried on a rotary evaporator, then in a vacuum desiccator overnight. After resolubilizing the lipids to 50 mM in 200 mM cholate, membrane scaffold protein (MSP) was added and incubated at 4°C with rocking for at least 15 minutes. CYP2J2 was then added to the mixture and incubated again at 4°C with rocking for at least 45 minutes. Spontaneous assembly of CYP2J2‐NDs was then initiated through removal of the detergent by addition and incubation of BioBeads (8 hours, 4°C). All batches of nanodiscs were prepared according to a 130:1 lipids:MSP ratio and a 1:10 CYP2J2:MSP ratio. CYP2J2 NDs were purified by use of high performance liquid chromatography (HPLC) on a Superdex 200 10/300 GL size exclusion column in 0.1 M potassium phosphate buffer (pH 7.4) following established protocols.89 Construction and stability of nanodiscs has been previously verified.90, 91, 92, 93 Size has been previously verified using water soluble protein standards on a size‐exclusion column and by small‐angle x‐ray scattering.60, 92
NADPH oxidation assay
NADPH oxidation by CYP2J2 and CPR was completed following a previously published method on the same CYP2J2 ND and RnCPR constructs as mentioned earlier by monitoring the decline in NADPH absorption.60 NADPH absorption was measured at 340 nm by use of a Cary 300 UV–vis spectrophotometer in kinetics mode. Reaction incubations were completed in a quartz cuvette. All reactions were carried out by equilibrating 0.2 μM CYP2J2‐ND, 0.6 μM rnCPR, and 40 μM EBS (solubilized in DMSO) in a buffered solution of 100 mM potassium phosphate (pH = 7.4) and crowding agent at 37°C for 5 minutes. Following the incubation, the reaction was initiated at t = 0 min by the addition of 200 μM NADPH (solubilized in ddH2O) to reach a final reaction volume of 250 μL. All concentrations listed are final. The decline in NADPH absorbance at 340 nm was then recorded over the next 10 minutes. At t = 10 min the reaction was quenched with 250 μL ethyl acetate, after which it was frozen at −20°C until extraction of the metabolites.
Crowding agents were added to the initial equilibration buffer in concentrations of either 50 mg/mL, 100 mg/mL, 150 mg/mL, or 200 mg/mL from a 250 mg/mL stock. All stocks were solubilized in ddH2O. All kinetics experiments shown in Figure 3 were performed at a crowding agent concentration of 100 mg/mL. Molar concentration ranges are as follows: Sucrose, 146–584 mM; Ficoll 70, 0.714–2.857 mM; Ficoll 400, 0.125–0.500 mM; Dextran 75, 1.333 mM only; Dextran 500, 0.200 mM only.
Michaelis–Menten kinetic assay
Michaelis–Menten assays were carried out using the same basic procedure as the NADPH oxidation assay, with the amount of substrate varied. EBS concentration was varied from 5 to 60 μM for the majority of experiments. Actual reactions were carried out in test tubes and incubated in an aluminum heat block. These samples are separate from NADPH oxidation samples. After quenching, the samples were subjected to metabolite extraction so EBS and its metabolites could be analyzed by HPLC and LC–MS/MS to determine the rate of EBS hydroxylation.
Metabolite extraction
EBS and its metabolites were extracted from quenched NADPH oxidation and Michaelis–Menten samples via phase separation. Glacial acetic acid (100 μL) and 4 M NaCl were added to quenched, room temperature samples to facilitate clean layer separation. Samples were vortexed for 1 minute, followed by centrifugation for 5 minutes at 3000 rpm, 4°C. The organic layer (top) was then removed to a clean tube and the remaining substrates were extracted from the aqueous layer twice more, for a total of three extractions. The second and third extractions were facilitated by the readdition of 250 μL ethyl acetate, followed by the same process of vortexing and centrifugation. Combined extractions were dried on a rotary evaporator and stored at −20°C until analysis.
HPLC metabolism analysis
HPLC was performed using an Alliance 2695 analytical separation module (Waters, Milford, MA) and a Waters 996 photodiode diode array detector (Waters). Samples were resolubilized in 100 μL mobile phase A and filtered before injection onto a 250 mm x 4.6 mm cyano column (5 μm beads, Alltech) Mobile phase A was composed of 0.012 M ammonium acetate buffer, methanol, and acetonitrile in a 48:30:20 ratio, respectively. Acetate buffer and mobile phase A were prepared according to Matsuda and colleagues.94 A linear gradient was run over 30 minutes from 100% mobile phase A to a 50/50 ratio of A and B (100% acetonitrile) at 40°C. EBS and its metabolites HYD and CAR were detected at 254 nm. Peak integration was performed using MassLynx and Origin software, and data was quantified via an EBS standard curve.
LC–MS/MS metabolism analysis
EBS and HYD for the nanodisc solubilized Michaelis–Menten experiments were quantified using a 5500 QTRAP LC/MS/MS system (Sciex, Framingham, MA) in the Metabolomics Lab of the Roy J. Carver Biotechnology Center, University of Illinois Urbana‐Champaign, as previously described.95 A linear gradient was run at a 0.4 mL/min flow rate as follows: 0–2 min, 90% A; 8 min, 55% A; 13–25 min, 40% A; 30 min, 30% A; 35 min, 25% A; 40 min, 20% A; 45–47 min, 15% A; 48–54 min, 90% A. mobile phase A was composed of 0.1% formic acid in water; mobile phase B was composed of 0.1% formic acid in acetonitrile. The autosampler was set at 15°C, with a 1 μL injection volume. Mass spectra were acquired under positive electrospray ionization (ESI) with ion spray voltage at 5500 V with 450°C source temperature. The curtain gas, ion source gas 1, and ion source gas 2 were 32, 50, and 65 psi, respectively. Multiple reaction monitoring (MRM) was used for quantitation: Ebastine m/z 470.4 → m/z 302.3; Hydroxyebastine m/z 486.4 → m/z 318.3; internal standard terfenadine m/z 472.4 → m/z 262.1.
ΔG unfolding
For chemical denaturation experiments, CYP2J2 (2.5 μM) was combined with 50 μM lipids (80% POPC, 20% POPS), 50 mg/mL crowding agent (146 mM sucrose, 0.714 mM Ficoll 70), and a gradient of guanidinium hydrochloride (Gu‐HCl) from 0 to 8 M (100 μL total volume). After incubating for two hours at room temperature, endpoint measurement readings were taken. Fluorescence was excited at 295 nm and measured at 321 (λmax, folded) and 360 nm (λmax, unfolded). All readings were taken with a SpectraMax Gemini EM at room temperature, using a black, half‐volume 96‐well plate. The ratio of A360/A321 was used to determine the fraction of unfolded proteins. The equation ΔG = −RT∙ln(fD/[1‐fD])55 was used to calculate ΔG, with fD representing the fraction of denatured protein. The two negative controls consisted of protein that not been exposed to denaturant and protein that had neither been exposed to denaturant nor incubated. The same procedure was used for CPR. Ficoll 400 was not used as it was insoluble at the high concentrations necessary for this assay (400 mg/mL was brought to 50 mg/mL final).
Statistical analysis
All data were produced from multiple repeats. Statistical significance was determined based on a two‐tailed t‐test with a confidence level of P < 0.05. All curve fittings were preformed using OriginPro 2017 (Origin Laboratories Inc., Northampton, MA).
Author Contributions
HCH designed and performed the experiments, analyzed data and wrote the manuscript. DM performed the unfolding experiments and made figures. AD designed the experiments and wrote the manuscript. All authors approved the final version of the manuscript.
Conflict of Interest
The authors declare no competing financial interests.
Supporting information
Figure S1 HPLC spectra of nanodisc library. Main 13 nm disc peaks are eluted between 28–29 minutes and can be confirmed by monitoring elution at 280 nm (MSP and 2 J2) and 417 nm (2 J2). 280 nm peaks are shown. (a) 100% POPC (b) 80% POPC, 20% POPS (c) 60% POPC, 40% POPS (d) 40% POPC, 60% POPS (e) 20% POPC, 80% POPS (f) 72% POPC, 20% POPS, 8% CHOL (g) 66% POPC, 20% POPS, 14% CHOL (h) 60% POPC, 20% POPS, 20% CHOL (i) 75% POPC, 20% POPS, 5% SM (j) 72.5% POPC, 20% POPS, 7.5% SM (k) 70% POPC, 20% POPS, 10% SM.
Figure S2: Coupling efficiency by CYP2J2 and CPR in nanodiscs of varying lipid compositions in uncrowded solution. Asterisks indicate a significant difference between 80% POPC/20% POPS nanodiscs and all other discs as determined by t‐test; n ≥ 3: *P < 0.05, **P < 0.01, ***P < 0.005.
Acknowledgments
The authors would like to thank the Sligar Lab for use of their fermenter to grow MSP for nanodiscs and Dr. Li Zhong of the UIUC Roy J. Carver Metabolomics Center for the LC–MS/MS analyses. This research is supported by the American Heart Association Scientist Development Grant 15SDG25760064 (A.D.) and in part by the National Institutes of Health Grants R01 GM1155884 and R03 DA 04236502 (A.D.).
Statement of importance: This work is of interest to researchers studying macromolecular crowding, protein‐lipid interactions, and cytochrome P450s. This work provides key insights into the kinetic effects of lipid composition as well as macromolecular crowding, a constant force within any cell. We also determine the effects of a crowded environment on membrane protein stability. As CYPs are important drug metabolizers, such data are pertinent to current and future studies.
References
- 1. van Meer G, de Kroon AIPM (2011) Lipid map of the mammalian cell. J Cell Sci 124:5–8. [DOI] [PubMed] [Google Scholar]
- 2. Das A, Varma SS, Mularczyk C, Meling DD (2014) Functional investigations of thromboxane synthase (CYP5A1) in lipid bilayers of nanodiscs. Chembiochem 15:892–899. [DOI] [PubMed] [Google Scholar]
- 3. Ingólfsson HI, Melo MN, van Eerden FJ, Arnarez C, Lopez CA, Wassenaar TA, Periole X, de Vries AH, Tieleman DP, Marrink SJ (2014) Lipid organization of the plasma membrane. J Am Chem Soc 136:14554–14559. [DOI] [PubMed] [Google Scholar]
- 4. Roy J, Pondenis H, Fan TM, Das A (2015) Direct capture of functional proteins from mammalian plasma membranes into nanodiscs. Biochemistry 54:6299–6302. [DOI] [PubMed] [Google Scholar]
- 5. Sanders CR, Mittendorf KF (2011) Tolerance to changes in membrane lipid composition as a selected trait of membrane proteins. Biochemistry 50:7858–7867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Anheuser S, Breiden B, Schwarzmann G, Sandhoff K (2015) Membrane lipids regulate ganglioside GM2 catabolism and GM2 activator protein activity. J Lipid Res 56:1747–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Oninla VO, Breiden B, Babalola JO, Sandhoff K (2014) Acid sphingomyelinase activity is regulated by membrane lipids and facilitates cholesterol transfer by NPC2. J Lipid Res 55:2606–2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dignon GL, Zerze GH, Mittal J (2017) Interplay between membrane composition and structural stability of membrane‐bound hIAPP. J Phys Chem B 121:8661–8668. [DOI] [PubMed] [Google Scholar]
- 9. Lei L, Lu S, Wang Y, Kim T, Mehta D, Wang Y (2014) The role of mechanical tension on lipid raft dependent PDGF‐induced TRPC6 activation. Biomaterials 35:2868–2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lucero HA, Robbins PW (2004) Lipid rafts–protein association and the regulation of protein activity. Arch Biochem Biophys 426:208–224. [DOI] [PubMed] [Google Scholar]
- 11. Wang L, Murphy‐Ullrich JE, Song Y (2014) Molecular insight into the effects of lipid bilayer environments on thrombospondin‐1 and calreticulin interactions. Biochemistry 53:6309–6322. [DOI] [PubMed] [Google Scholar]
- 12. Ellis RJ (2001) Macromolecular crowding: obvious but underappreciated. Trends Biochem Sci 26:597–604. [DOI] [PubMed] [Google Scholar]
- 13. Ellis RJ (2001) Macromolecular crowding: an important but neglected aspect of the intracellular environment. Curr Opin Struct Biol 11:114–119. [DOI] [PubMed] [Google Scholar]
- 14. Zeiger AS, Loe FC, Li R, Raghunath M, Van Vliet KJ (2012) Macromolecular crowding directs extracellular matrix organization and mesenchymal stem cell behavior. PLoS ONE 7:e37904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kuznetsova IM, Turoverov KK, Uversky VN (2014) What macromolecular crowding can do to a protein. Intl J Mol Sci 15:23090–23140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Minton AP, Wilf J (1981) Effect of macromolecular crowding upon the structure and function of an enzyme: glyceraldehyde‐3‐phosphate dehydrogenase. Biochemistry 20:4821–4826. [DOI] [PubMed] [Google Scholar]
- 17. Poggi CG, Slade KM (2015) Macromolecular crowding and the steady‐state kinetics of malate dehydrogenase. Biochemistry 54:260–267. [DOI] [PubMed] [Google Scholar]
- 18. Christiansen A, Wittung‐Stafshede P (2013) Quantification of excluded volume effects on the folding landscape of Pseudomonas aeruginosa apoazurin In Vitro . Biophys J 105:1689–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schneider SH, Lockwood SP, Hargreaves DI, Slade DJ, LoConte MA, Logan BE, McLaughlin EE, Conroy MJ, Slade KM (2015) Slowed diffusion and excluded volume both contribute to the effects of macromolecular crowding on alcohol dehydrogenase steady‐state kinetics. Biochemistry 54:5898–5906. [DOI] [PubMed] [Google Scholar]
- 20. Minton AP (2013) Quantitative assessment of the relative contributions of steric repulsion and chemical interactions to macromolecular crowding. Biopolymers 99:239–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhou H‐X (2013) Polymer crowders and protein crowders act similarly on protein folding stability. FEBS Lett 587:394–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Paudel BP, Rueda D (2014) Molecular crowding accelerates ribozyme docking and catalysis. J Am Chem Soci 136:16700–16703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kuznetsova IM, Zaslavsky BY, Breydo L, Turoverov KK, Uversky VN (2015) Beyond the excluded volume effects: mechanistic complexity of the crowded milieu. Molecules 20:1377–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Benton LA, Smith AE, Young GB, Pielak GJ (2012) Unexpected effects of macromolecular crowding on protein stability. Biochemistry 51:9773–9775. [DOI] [PubMed] [Google Scholar]
- 25. Fonin AV, Silonov SA, Sitdikova AK, Kuznetsova IM, Uversky VN, Turoverov KK (2017) Structure and conformational properties of D‐glucose/D‐galactose‐binding protein in crowded milieu. Molecules 22:E244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Somkuti J, Török Z, Pfalzfraf F, Smeller L (2017) Low crowding agent concentration destabilizes against pressure unfolding. Biophys Chem 231:125–134. [DOI] [PubMed] [Google Scholar]
- 27. Stagg L, Zhang S‐Q, Cheung MS, Wittung‐Stafshede P (2007) Molecular crowding enhances native structure and stability of α/β protein flavodoxin. Proc Natl Acad Sci U S A 104:18976–18981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang Y, Sarkar M, Smith AE, Krois AS, Pielak GJ (2012) Macromolecular crowding and protein stability. J Am Chem Soc 134:16614–16618. [DOI] [PubMed] [Google Scholar]
- 29. Gao M, Winter R (2015) The effects of lipid membranes, crowding and osmolytes on the aggregation, and fibrillation propensity of human IAPP. J Diabetes Res 2015:849017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ge J, Bouriyaphone SD, Serebrennikova TA, Astashkin AV, Nesmelov YE (2016) Macromolecular crowding modulates actomyosin kinetics. Biophys J 111:178–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lee CF, Bird S, Shaw M, Jean L, Vaux DJ (2012) Combined effects of agitation, macromolecular crowding, and interfaces on amyloidogenesis. J Biol Chem 287:38006–38019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Munishkina LA, Ahmad A, Fink AL, Uversky VN (2009) Guiding protein aggregation with macromolecular crowding. Biochemistry 47:8993–9006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Munishkina LA, Cooper EM, Uversky VN, Fink AL (2004) The effect of macromolecular crowding on protein aggregation and amyloid fibril formation. J Mol Recogn 17:456–464. [DOI] [PubMed] [Google Scholar]
- 34. Siddiqui GA, Naeem A (2018) Aggregation of globular protein as a consequences of macromolecular crowding: a time and concentration dependent study. Intl J Biol Macromol 108:360–366. [DOI] [PubMed] [Google Scholar]
- 35. Balcells C, Pastor I, Vilaseca E, Madurga S, Cascante M, Mas F (2014) Macromolecular crowding effect upon in vitro enzyme kinetics: mixed activation‐diffusion control of the oxidation of NADH by pyruvate catalyzed by lactate dehydrogenase. J Phys Chem B 118:4062–4068. [DOI] [PubMed] [Google Scholar]
- 36. Zelasko S, Arnold WR, Das A (2015) Endocannabinoid metabolism by cytochrome P450 monooxygenases. Prostagland Other Lipid Mediat 116‐117:112–123. [DOI] [PubMed] [Google Scholar]
- 37. McDougle DR, Baylon JL, Meling DD, Kambalyal A, Grinkova YV, Hammernik J, Tajkhorshid E, Das A (2015) Incorporation of charged residues in the CYP2J2 F‐G loop disrupts CYP2J2‐lipid bilayer interactions. Biochim Biophys Acta Biomembr 1848:2460–2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Park JW, Reed JR, Brignac‐Huber LM, Backes WL (2014) Cytochrome P450 system proteins reside in different regions of the endoplasmic reticulum. Biochem J 464:241–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Brignac‐Huber L, Reed JR, Backes WL (2011) Organization of NADPH‐cytochrome P450 reductase and CYP1A2 in the endoplasmic reticulum—microdomain localization affects monooxygenase function. Mol Pharm 79:549–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Das A, Sligar SG (2009) Modulation of the cytochrome P450 reductase redox potential by the phospholipid bilayer. Biochemistry 48:12104–12112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Reed JR, Backes WL (2012) Formation of P450 P450 complexes and their effect on P450 function. Pharm Therapeut 133:299–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Davydov DR, Petushkova NA, Bobrovnikova EV, Knyushko TV, Dansette P (2001) Association of cytochromes P450 1A2 and 2B4: are the interactions between different p450 species involved in the control of the monooxygenase activity and coupling? Advan Experim Med Biol 500:335–338. [DOI] [PubMed] [Google Scholar]
- 43. Kelley RW, Cheng D, Backes WL (2006) Heteromeric complex formation between CYP2E1 and CYP1A2: evidence for the involvement of electrostatic interactions. Biochemistry 45:15807–15816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yamazaki H, Gillam EMJ, Dong M‐S, Johnson WW, Guengerich FP (1997) Reconstitution of recombinant cytochrome P450 2C10(2C9) and comparison with cytochrome P450 3A4 and other forms: effects of cytochrome P450‐P450 and cytochrome P450‐b5 interactions. Arch Biochem Biophys 342:329–337. [DOI] [PubMed] [Google Scholar]
- 45. Spector AA, Yorek MA (1985) Membrane lipid composition and cellular function. J Lipid Res 26:1015–1035. [PubMed] [Google Scholar]
- 46. Shaw AW, Pureza VS, Sligar SG, Morrissey JH (2006) The local phospholipid environment modulates the activation of blood clotting. J Biol Chem 282:6556–6563. [DOI] [PubMed] [Google Scholar]
- 47. Kang W, Elitzer S, Noh K, Bednarek T, Weiss M (2011) Myocardial pharmacokinetics of ebastine, a substrate for cytochrome P450 2J, in rat isolated heart. Brit J Pharm 163:1733–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Klein N, Hellmann N, Schneider D (2015) Anionic lipids modulate the activity of the aquaglyceroporin GlpF. Biophys J 109:722–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vorobyov I, Allen TW (2011) On the role of anionic lipids in charged protein interactions with membranes. Biochim Biophys Acta Biomembr 1808:1673–1683. [DOI] [PubMed] [Google Scholar]
- 50. Colthart AM, Tietz DR, Ni Y, Friedman JL, Dang M, Pochapsky TC (2016) Detection of substrate‐dependent conformational changes in the P450 fold by nuclear magnetic resonance. Sci Rep 6:22035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Denisov IG, Baas BJ, Grinkova YV, Sligar SG (2007) Cooperativity in cytochrome P450 3A4. J Biol Chem 282:7066–7076. [DOI] [PubMed] [Google Scholar]
- 52. Sevrioukova IF, Poulos TL (2011) Structural biology of redox partner interactions in P450cam monooxygenase: a fresh look at an old system. Arch Biochem Biophys 507:66–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yang W, Bell SG, Wang H, Zhou W, Hoskins N, Dale A, Bartlam M, Wong L‐L, Rao Z (2010) Molecular characterization of a class I P450 electron transfer system from Novosphingobium aromaticivorans DSM12444. J Biol Chem 285:27272–27384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jia J, Peng X, Qi W, Su R, He Z (2017) Effects of macromolecular crowding on alkaline phosphatase unfolding, conformation and stability. Intl J Biol Macromol 101:373–382. [DOI] [PubMed] [Google Scholar]
- 55. Pace CN (1986) Determination and analysis of urea and guanidine hydrochloride denaturation curves. Methods Enzymol 131:266–280. [DOI] [PubMed] [Google Scholar]
- 56. van Meer G, Voelker DR, Feigenson GW (2008) Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol 9:112–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kandel SE, Lampe JN (2014) Role of protein‐protein interactions in cytochrome P450‐mediated drug metabolism and toxicity. Chem Res Toxicol 27:1474–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Xia C, Hamdane D, Shen AL, Choi V, Kasper CB, Pearl NM, Zhang H, Im S‐C, Waskell L, Kim J‐JP (2011) Conformational changes of NADPH‐cytochrome P450 oxidoreductase are essential for catalysis and cofactor binding. J Biol Chem 286:16246–16260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Shinkyo R, Guengerich FP (2011) Inhibition of human cytochrome P450 3A4 by cholesterol. J Biol Chem 286:18426–18433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. McDougle DR, Palaria A, Magnetta E, Meling DD, Das A (2013) Functional studies of N‐terminally modified CYP2J2 epoxygenase in model lipid bilayers. Protein Sci 22:964–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Reed JR, Cawley GF, Backes WL (2013) Interactions between cytochromes P450 2B4 (CYP2B4) and 1A2 (CYP1A2) lead to alterations in toluene disposition and P450 uncoupling. Biochemistry 52:4003–4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Denisov IG, Shih AY, Sligar SG (2012) Structural differences between soluble and membrane bound cytochromes P450. J Inorgan Biochem 108:150–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Guengerich FP, Sohl CD, Chowdhury G (2011) Multi‐step oxidations catalyzed by cytochrome P450 enzymes: Processive vs. distributive kinetics and the issue of carbonyl oxidation in chemical mechanisms. Arch Biochem Biophys 507:126–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Barenholz Y, Thompsion TE (1999) Sphingomyelin: biophysical aspects. Chem Phys Lipids 102:29–34. [DOI] [PubMed] [Google Scholar]
- 65. Bruzzese A, Gil C, Dalton JAR, Giraldo J (2018) Structural insights into positive and negative allosteric regulation of a G protein‐coupled receptor through protein‐lipid interactions. Sci Rep 8:4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Guixà‐González R, Albasanz JL, Rodriguez‐Espigares I, Pastor M, Sanz F, Martí‐Solano M, Manna M, Martinz‐Seara H, Hildebrand PW, Martín M, Selent J (2017) Membrane cholesterol access into a G‐protein‐coupled receptor. Nat Commun 8:14505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Polic V, Auclair K (2017) Allosteric activation of cytochrome P450 3A4 via progesterone bioconjugation. Bioconjug Chem 28:885–889. [DOI] [PubMed] [Google Scholar]
- 68. Anderson KW, Mast N, Hudgens JW, Lin JB, Turko IV, Pikuleva IA (2016) Mapping of the allosteric site in cholesterol hydroxylase CYP46A1 for Efavirenz, a drug that stimulates enzyme activity. J Biol Chem 291:11876–11886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Barnaba C, Sahoo BR, Ravula T, Medina‐Meza IG, Im S‐C, Anantharamaiah GM, Waskell L, Ramamoorthy A (2018) Cytochrome‐P450‐induced ordering of microsomal membranes modulates affinity for drugs. Angew Chem Intl Ed 57:3391–3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Grinkova YV, Denisov IG, McLean MA, Sligar SG (2013) Oxidase uncoupling in heme monoxygenases: human cytochroome P450 CYP3A4 in Nanodiscs. Biochem Biophys Res Commun 430:1223–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Dowhan W (1997) Molecular basis for membrane phospholipid diversity: why are there so many lipids? Annu Rev Biochem 66:199–232. [DOI] [PubMed] [Google Scholar]
- 72. Ravula T, Barnaba C, Mahajan M, Anantharamaiah GM, Im S‐C, Waskell L, Ramamoorthy A (2017) Membrane environment drives cytochrome P450's spin transition and its interaction with cytochrome b 5 . Chem Commun 53:12798–12801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Bayburt TH, Vishnivetskiy SA, McLean MA, Morizumi T, C‐c H, Tesmer JJG, Ernst OP, Sligar SG, Gurevich VV (2011) Monomeric rhodopsin is sufficient for normal rhodopsin kinase (GRK1) phosphorylation and arrestin‐1 binding. J Biol Chem 286:1420–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Tavoosi N, Davis‐Harrison RL, Pogorelov TV, Ohkubo YZ, Arcario MJ, Clay MC, Rienstra CM, Tajkhorshid E, Morrissey JH (2011) Molecular determinants of phospholipid synergy in blood clotting. J Biol Chem 286:23247–23253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wang Y, Dubin PL (1998) Observation of Ficoll charge using size‐exclusion chromatography. J Chromatog A 800:181–185. [Google Scholar]
- 76. Gentry KA, Prade E, Barnaba C, Zhang M, Mahajan M, Im S‐C, Anantharamaiah GM, Nagao S, Waskell L, Ramamoorthy A (2017) Kinetic and structural characterization of the effects of membrane on the complex of cytochrome b 5 and cytochrome c . Sci Rep 7:7793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Sharp KA (2015) Analysis of the size dependence of macromolecular crowding shows that smaller is better. Proc Natl Acad Sci U S A 112:7990–7995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Shah G, Dubin PL (1995) Adsorptive interaction of Ficoll standards with porous glass size‐exclusion chromatography columns. J Chromatog A 693:197–203. [Google Scholar]
- 79. Venturoli D, Rippe B (2005) Ficoll and dextran vs. globular proteins as probes for testing glomerular permselectivity: effects of molecular size, shape, charge, and deformability. Am J Physiol Renal Physiol 288:F605–F613. [DOI] [PubMed] [Google Scholar]
- 80. Prade E, Mahajan M, Im S‐C, Zhang M, Gentry KA, Anantharamaiah GM, Waskell L, Ramamoorthy A (2018) A minimal functional complex of cytochrome P450 and FBD of cytochrome P450 reductase in nanodiscs. Angew Chem Int Ed 57:8458–8462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Woodbury CP. Introduction to macromolecular binding equilibria. Boca Raton: CRC Press, 2008. [Google Scholar]
- 82. King JT, Arthur EJ, Brooks CLI, Kubarych KJ (2014) Crowding induced collective hydration of biological macromolecules over extended distances. J Am Chem Soc 136:188–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Ellis J, Gutierrez A, Barsukov IL, Huang WC, Grossmann JG, Roberts GCK (2009) Domain motion in cytochrome P450 reductase conformational equilibria revealed by NMR and small‐angle X‐ray scattering. J Biol Chem 284:36628–36637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Wang M, Roberts DL, Paschke R, Shea TM, Masters BSS, Kim JJP (1997) Three‐dimensional structure of NADPH‐cytochrome P450 reductase: prototype for FMN‐ and FAD‐containing enzymes. Proc Natl Acad Sci U S A 94:8411–8416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Kim JJP, Wang M, Roberts D, Paschke R, Shea T, Masters BSS (1997) Three‐dimensional structure of NADPH‐cytochrome P450 reductase: a prototype for FMN‐ and FAD‐containing enzymes. Faseb Journal 11(9):A781–A781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Tripathy J, Mueller JJ, Shepherd NC, Beck WF (2013) Dynamic solvation and coupling of the hydration Shell of ZnII‐substituted cytochrome c in the presence of Guanidinium ions. Journal of Physical Chemistry B 117:14589–14598. [DOI] [PubMed] [Google Scholar]
- 87. Zelasko S, Palaria A, Das A (2013) Optimizations to achieve high‐level expression of cytochrome P450 proteins using Escherichia coli expression systems. Prot Expr Purif 92:77–87. [DOI] [PubMed] [Google Scholar]
- 88. Shen AL, Porter TD, Wilson TE, Kasper CB (1989) Structural analysis of the FMN binding domain of NADPH‐cytochrome P‐450 oxidoreductase by site‐directed mutagenesis. J Biol Chem 264:7584–7589. [PubMed] [Google Scholar]
- 89. McDougle DR, Kambalyal A, Meling DD, Das A (2014) Endocannabinoids anandamide and 2‐arachidonoylglycerol are substrates for human CYP2J2 epoxygenase. J Pharm Exper Therapeut 351:616–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Bayburt TH, Sligar SG (2003) Self‐assembly of single integral membrane proteins into soluble nanoscale phospholipid bilayers. Protein Sci 12:2476–2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Civjan NR, Bayburt TH, Schuler MA, Sligar SG (2003) Direct solubilization of heterologously expressed membrane proteins by incorporation into nanoscale lipid bilayers. Biotechniques 35:556–563. [DOI] [PubMed] [Google Scholar]
- 92. Denisov IG, McLean MA, Shaw AW, Grinkova YV, Sligar SG (2005) Thermotropic phase transition in soluble nanoscale lipid bilayers. J Phys Chem B 109:15580–15588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Shaw AW, McLean MA, Sligar SG (2004) Phospholipid phase transitions in homogeneous nanometer scale bilayer discs. FEBS Lett 556:260–264. [DOI] [PubMed] [Google Scholar]
- 94. Matsuda M, Mizuki Y, Terauchi Y (2001) Simultaneous determination of the histamine H1‐antagonist ebastine and its two metabolites, carebastine and hydroxyebastine, in human plasma using high‐performance liquid chromatography. J Chromatog B 757:173–179. [DOI] [PubMed] [Google Scholar]
- 95. Arnold WR, Baylon JL, Tajkhorshid E, Das A (2016) Asymmetric binding and metabolism of polyunsaturated fatty acids (PUFAs) by CYP2J2 epoxygenase. Biochemistry 55:6969–6980. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 HPLC spectra of nanodisc library. Main 13 nm disc peaks are eluted between 28–29 minutes and can be confirmed by monitoring elution at 280 nm (MSP and 2 J2) and 417 nm (2 J2). 280 nm peaks are shown. (a) 100% POPC (b) 80% POPC, 20% POPS (c) 60% POPC, 40% POPS (d) 40% POPC, 60% POPS (e) 20% POPC, 80% POPS (f) 72% POPC, 20% POPS, 8% CHOL (g) 66% POPC, 20% POPS, 14% CHOL (h) 60% POPC, 20% POPS, 20% CHOL (i) 75% POPC, 20% POPS, 5% SM (j) 72.5% POPC, 20% POPS, 7.5% SM (k) 70% POPC, 20% POPS, 10% SM.
Figure S2: Coupling efficiency by CYP2J2 and CPR in nanodiscs of varying lipid compositions in uncrowded solution. Asterisks indicate a significant difference between 80% POPC/20% POPS nanodiscs and all other discs as determined by t‐test; n ≥ 3: *P < 0.05, **P < 0.01, ***P < 0.005.


