Abstract
Protein‐based biological drugs and many industrial enzymes are unstable, making them prohibitively expensive. Some can be stabilized by formulation with excipients, but most still require low temperature storage. In search of new, more robust excipients, we turned to the tardigrade, a microscopic animal that synthesizes cytosolic abundant heat soluble (CAHS) proteins to protect its cellular components during desiccation. We find that CAHS proteins protect the test enzymes lactate dehydrogenase and lipoprotein lipase against desiccation‐, freezing‐, and lyophilization‐induced deactivation. Our data also show that a variety of globular and disordered protein controls, with no known link to desiccation tolerance, protect our test enzymes. Protection of lactate dehydrogenase correlates, albeit imperfectly, with the charge density of the protein additive, suggesting an approach to tune protection by modifying charge. Our results support the potential use of CAHS proteins as stabilizing excipients in formulations and suggest that other proteins may have similar potential.
Keywords: biologics, desiccation tolerance, enzymes, enzyme protection, excipients, intrinsically disordered proteins, tardigrades
Abbreviations
- BSA
bovine serum albumin
- CAHS
cytosolic abundant heat soluble protein
- dextran 20
20 kDa dextran
- Ficoll 70
70 kDa Ficoll®
- Hphob
Kyte and Doolittle hydrophobicity
- IDP
intrinsically disordered protein
- LB
Lennox broth
- LDH
lactate dehydrogenase
- LEA
late embryogenesis abundant
- LPL
lipoprotein lipase
- NAD+
oxidized nicotinamide adenine dinucleotide
- NADH
reduced nicotinamide adenine dinucleotide
- PBS
phosphate buffered saline
- PEG 3350
3.35 kDa polyethylene glycol
- pI
isoelectric point
- SH3
the T22G variant of the N‐terminal SH3 domain of Drosophila signal transduction protein drk
Introduction
Biologics, protein‐based drugs derived from living organisms, are among the most effective therapeutic treatments in the market. However, they are unstable, have short half‐lives, and require low temperature storage, which makes them expensive. Environmentally friendly industrial enzymes often confront similar challenges. Manufacturers increase the half‐life of biologics and enzymes by adding excipients, molecules that stabilize the active ingredient.1
Trehalose is a common excipient and osmolyte that is synthesized by many desiccation tolerant organisms.2, 3, 4, 5 This nonreducing disaccharide can increase the mid‐point denaturation temperature of a protein by as much as 18°C and its modified standard‐state Gibbs free energy of denaturation by almost 5 kcal/mol at physiologically relevant temperatures.6 However, trehalose also enhances autophagy,7, 8 and can interfere with drug efficacy in treatments to repress autophagy in autoimmune diseases like lupus.9 Albumedix produces a protein‐based excipient, recombinant human serum albumin, which is FDA approved in five biologics including the M‐M‐R®II vaccine, the type II diabetes drug Tanzeum®, and the hemophilia treatment Idelvion®. Nevertheless, biologics still require storage at temperatures as low as −80°C, even after formulation.10 Discovery of better excipients would make these products more affordable and accessible.
We turned to tardigrades as a potential source of novel excipients. These microscopic animals survive conditions ranging from −274°C to 151°C, from vacuum to 6000 atm, 1000‐times more radiation than the average organism, 10 days exposed to outer space, 30 years frozen, and up to 10 years of desiccation.11, 12, 13, 14, 15 Genomic and transcriptomic data suggest that, unlike many desiccation tolerant organisms, some species of tardigrades lack trehalose phosphatase,16, 17, 18, 19, 20, 21 and therefore cannot synthesize this sugar. Nevertheless, tardigrades produce several families of intrinsically disordered proteins (IDPs), including the cytosolic abundant heat soluble (CAHS) family.22 IDPs have many functions and are thought to play a role in stress tolerance across all kingdoms of life,23, 24, 25, 26, 27 and in vitro the intrinsically disordered late embryogenesis abundant (LEA) proteins protect enzymes against environmental stress.23, 24, 28, 29, 30 These observations led us to investigate whether CAHS proteins could perform a similar function. We showed that CAHS proteins allow tardigrades to survive desiccation and have been detected only in these organisms.16 Furthermore, recombinant expression of CAHS proteins in yeast and bacteria enhances the desiccation tolerance of both organisms by over 100‐fold.16
We studied the protection of two enzymes by CAHS proteins. Lactate dehydrogenase (LDH, Scheme 1), is a 150 kDa tetramer with an isoelectric point (pI) of 6.0 that is frequently studied in stress tolerance of in vitro samples.16, 24, 28, 29, 30, 31, 32, 33, 34 Here, we expand our initial results16 by quantifying the protective properties of osmolytes, sugars, synthetic polymers, globular proteins, disordered proteins, and CAHS proteins at room temperature and 95°C. We also studied protection of the unstable enzyme lipoprotein lipase (LPL, Scheme 2), a 110 kDa dimer with a pI of 8.5 that possesses therapeutic potential for treating hypertriglyceridemia.35, 36, 37
Scheme 1.
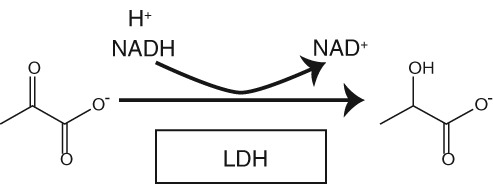
LDH reaction. A hydride from NADH and a proton are transferred to pyruvate, forming the reduced product, lactate, and the oxidized cofactor NAD+.
Scheme 2.
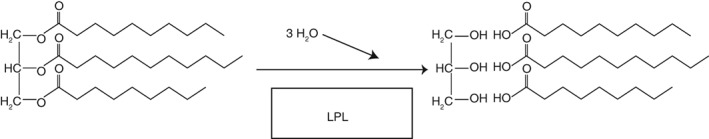
LPL reaction. Triglycerides are hydrolyzed to fatty acids and glycerol.
Results
Protecting LDH against desiccation induced inactivation
LDH is commercially available and sensitive to H2O loss from vacuum‐drying.16, 30, 32, 38 The enzyme was desiccated in the presence of osmolytes, sugars, synthetic polymers, globular proteins, disordered proteins, and CAHS proteins (Fig. 1), with some occupying multiple categories (Fig. 2), using a Genevac® EZ‐2 Personal Evaporator. Protection was quantified as the percent activity of the desiccated samples compared to unstressed controls (Fig. 3).
Figure 1.
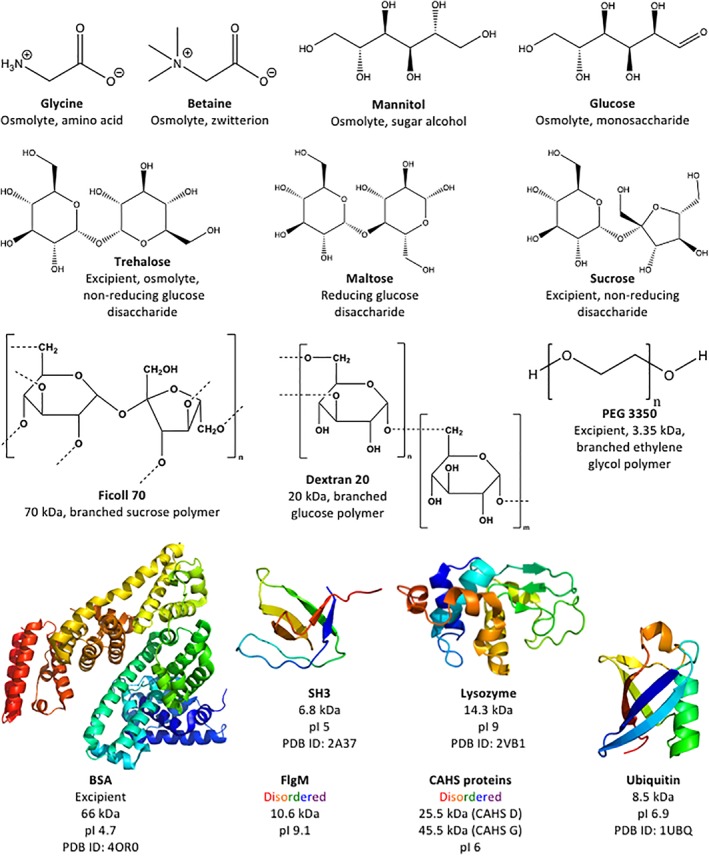
Additives tested as potential protectants.
Figure 2.
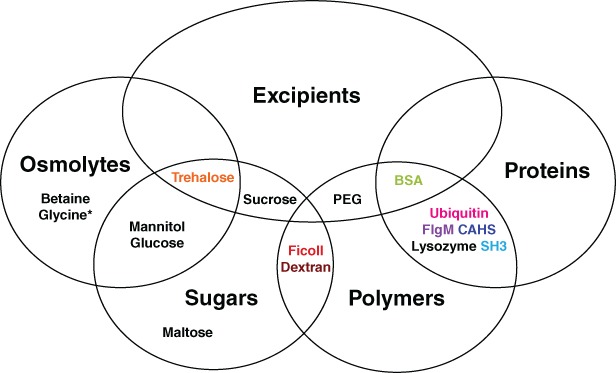
Venn diagram of additives tested as potential protectants and their categorization. Most additives fall into multiple categories. *Glycine is an amino acid and osmolyte.
Figure 3.
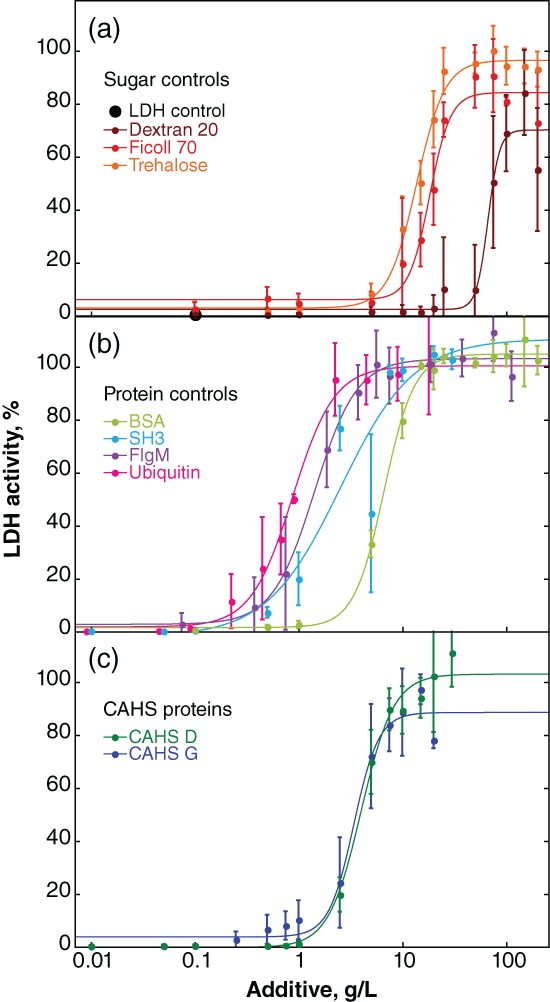
Protection of LDH activity against desiccation‐induced inactivation. Buffered LDH (0.10 g/L) was desiccated and rehydrated by itself and with additives. Percent activity was determined by comparison to a sample of the same solution stored at 4°C. Uncertainties are the standard deviations of the mean from triplicate measurements. Data for the less effective additives sucrose, maltose, and mannitol are given in Supporting Information Figure S1. “LDH control” represents the activity in the absence of additive or in the presence of glucose, glycine, betaine or PEG 3350 (i.e., these additives are ineffective). Sigmoidal curves were added as a visual guide and used to calculate the concentration of additive required for 50% protection (Fig. 4). Data for trehalose and BSA have been published.16 Data for CAHS D and G are slightly different from our previous report16 because, as described in the Materials and Methods section, we improved the purification protocol.
Trehalose, a nonreducing glucose disaccharide, is the most protective sugar, most protective osmolyte, and most protective small molecule tested [Fig. 3(a)]. Ficoll 70, a branched 70 kDa sucrose polymer containing nonreducing linkages, and dextran 20, a 20 kDa complex branched glucose polymer containing reducing linkages, are also protective. Sucrose, the nonreducing disaccharide and the monomer of Ficoll, protects only up to 70% of activity (Supporting Information Fig. S1). Maltose, the reducing glucose disaccharide comprising dextran, protects up to 30% of activity at concentrations between 10 g/L and 50 g/L, but is not effective at higher concentrations. The sugar alcohol and osmolyte mannitol protects no more than 10% of LDH activity. The monosaccharide glucose, the osmolytes glycine and betaine, and the polymer polyethylene glycol (PEG) 3350 are ineffective (data not shown).
All proteins tested outperform trehalose [Fig. 3(a,b)]. Bovine serum albumin (BSA), a homolog of the excipient human serum albumin, is the least protective protein and ubiquitin the most protective on a g/L basis. The disordered protein flgM39 and the stabilized SH3 domain, SH3 T22G (SH3),40 are less protective than ubiquitin but more protective than BSA. Lysozyme protects LDH activity at low concentrations, but at high concentrations it inactivates LDH in the unstressed controls (data not shown).
The g/L‐concentration and mole ratio of additive required for 50% protection (Fig. 4) were then calculated. The CAHS homologs tested fall between SH3 and BSA [Figs. 3(c) and 4(a)] on a g/L basis. When these data are converted from g/L to mole ratio, CAHS G is the most protective with a confidence interval of at least 67% [Fig. 4(b)].
Figure 4.
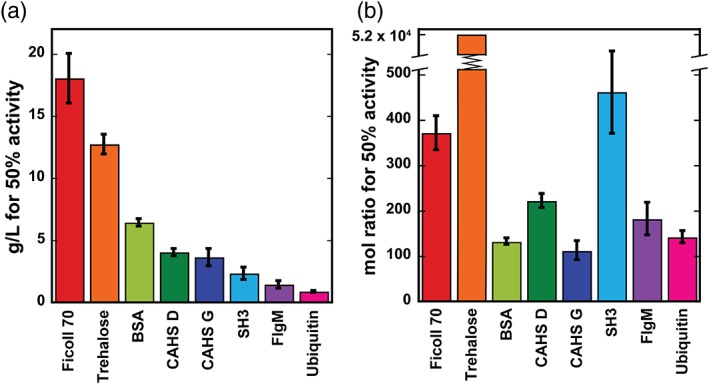
Additive concentrations affording 50% LDH protection from desiccation‐induced inactivation as determined from the sigmoidal fits of the data in Figure 3. (a) g/L concentrations. Uncertainties are the standard deviations of the mean from triplicate measurements. (b) Mole ratios of additive to LDH. The upper and lower uncertainties for trehalose are 55,000 and 49,000.
Protecting desiccated LDH against heat inactivation
Next, we tested the ability of trehalose, BSA, flgM, ubiquitin, CAHS D, and CAHS G to protect the desiccated enzyme at 95°C (Fig. 5). We chose a common additive concentration of 20 g/L because all additives tested protect LDH at this concentration (Fig. 3). Trehalose has no protective ability above 80°C and at 95°C almost all activity is lost in 5 min (Fig. S2). All proteins show some ability to protect the desiccated enzyme at 95°C (Fig. 5). BSA is the least protective with activity disappearing in about 4 h. Ubiquitin is the most protective, preserving activity for 2 h at 95°C before a decline. The potency of CAHS D and G fall between flgM and BSA, similar to their behavior at room temperature (Fig. 3).
Figure 5.
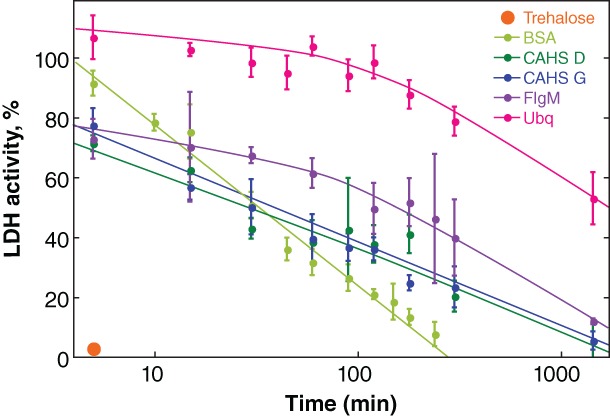
Protecting dried LDH from heat‐inactivation. Buffered LDH (0.10 g/L) was desiccated in the presence of 20 g/L protectant before exposure to 95°C. After rehydration, the percent activity was determined by comparison to a control comprising the same solution stored at 4°C. Uncertainties are the standard deviations of the mean from triplicate measurements. Curves were added as a visual guide but have no theoretical significance.
Protecting LPL against lyophilization‐induced inactivation and long‐term storage
We also investigated the ability of a limited selection of additives to protect bovine LPL, an enzyme that normally requires storage at −80°C (Fig. 6). LPL was frozen or lyophilized in solutions of trehalose, BSA, and CAHS D. Protection was measured as percent activity compared to unstressed controls. Trehalose does not protect the enzyme against inactivation by either freeze/thaw stress or lyophilization. BSA and CAHS D protect slightly better against freeze/thaw‐inactivation than lyophilization induced‐inactivation, but CAHS D is more protective against both processes.
Figure 6.
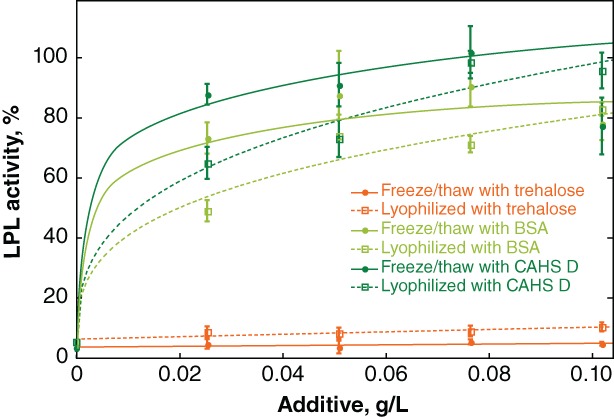
Protecting LPL against freeze/thaw and lyophilization‐induced inactivation. Buffered LPL (0.28 mg/L) was formulated with various concentrations of additives before flash freezing. Freeze/thawe samples were stored at −80°C. Other samples were lyophilized. Normalized activity was determined by comparison to an LPL solution stored at −80°C without freeze/thaw stress. Curves were added as a visual guide but have no theoretical significance.
LPL was then lyophilized from solutions of trehalose, BSA, or CAHS D and its activity monitored at 4°C as a function of time (Fig. 7). Consistent with the observations described earlier, trehalose is not protective at 4°C whether the samples are lyophilized or in solution. In solution, BSA and CAHS D were both slightly protective, but activity diminished over a week. BSA and CAHS D provided longer‐term stabilization for lyophilized LPL stored at 4°C, with activity gradually declining over 2 months. In both cases, CAHS D outperforms BSA.
Figure 7.
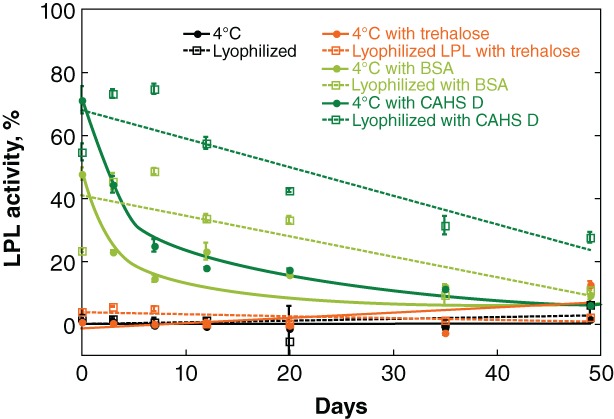
Protecting LPL against lyophilization‐induced inactivation. Buffered LPL (0.28 mg/L) was formulated with 0.026 g/L additive. Samples were stored at either 4°C or flash frozen and lyophilized before storage at 4°C. Normalized activity was determined by comparison to a control comprising the same solution stored at −80°C. Curves were added as a visual guide but have no theoretical significance.
Discussion
Trehalose outperforms other sugars, polymers, and osmolytes as a protectant of LDH
The nonreducing sugars trehalose and the sucrose polymer Ficoll 70 protect LDH better than the reducing sugars maltose, glucose, and the glucose polymer dextran 20. Nonreducing sugars may be more effective protectants because they lack the reactive carbonyl group of reducing sugars. However, this idea does not explain why the reducing sugar dextran 20 is more protective than the nonreducing sugar sucrose. Additionally, sugar size does not have a consistent effect. Both sugar polymers outperform the disaccharides sucrose and maltose but not the disaccharide trehalose. These observations may explain why trehalose is one of the most widely used excipients. The sugar alcohol and osmolyte mannitol protects poorly against desiccation‐induced LDH inactivation. The nonsugar osmolyte betaine, despite its ability to stabilize proteins in vitro and in cells,41, 42, 43 does not protect LDH against desiccation‐induced inactivation. The same is true for the amino acid and osmolyte glycine.42 Similarly, despite its use as an excipient and its ability to protect LDH against freeze/thaw and freeze/dry stress,31 the synthetic polymer PEG 3350 does not protect LDH against desiccation‐induced inactivation. With the exception of glucose, all sugars have some ability to protect LDH against desiccation‐induced inactivation, but we find no other clear patterns among sugars, polymers, and osmolytes.
Proteins outperform trehalose
We chose the proteins ubiquitin, flgM, SH3, and BSA because they are easy to produce and store, and they represent two protein classes: globular and disordered. These proteins protect LDH against desiccation‐induced inactivation at room temperature and 95°C more effectively than trehalose and other sugars, polymers, and osmolytes (Figs. 3, 4, 5). BSA also protects LPL against freeze/thaw and lyophilization‐induced inactivation (Figs. 6 and 7). By contrast, trehalose does not protect LDH from these stresses.
Protein protection and physical properties
We sought to explain the protective properties of proteins (Table 1). With the exception of SH3, their protective ability, measured in g/L, directly correlates to the total charge at pH 7 divided by the number of amino acid residues, which we call the sequence charge density [Fig. 8(a)]. With two exceptions (discussed later), the more positive its sequence charge density, the better the protein protects negatively charged LDH (pI 5 to 6, depending on the isozyme).
Table 1.
Ability of Additives to Protect LDH and Some of Their Physical Properties
| Additive | g/L for 50% activity | Mole ratio for 50% activity | pI | MW (kDa) | # residues | Hphoba | Hphoba density | Charge at pH 7 | Sequence charge density, pH 7 | Surface hydrophobicity59 |
|---|---|---|---|---|---|---|---|---|---|---|
| BSA | 6.4 | 130 | 4.8 | 66 | 583 | −279.2 | −0.48 | −13.9 | −0.024 | 0.545 |
| CAHS D | 4.0 | 220 | 6.0 | 25.5 | 227 | −223.5 | −0.99 | −3.1 | −0.014 | n/db |
| CAHS G | 3.6 | 110 | 5.9 | 45.5 | 414 | −341.5 | −0.82 | −5.1 | −0.012 | n/db |
| SH3 | 2.3 | 460 | 4.8 | 6.8 | 59 | −85.8 | −0.95 | −5.6 | −0.095 | 0.596 |
| FlgM | 1.4 | 180 | 9.1 | 10.6 | 97 | −59.8 | −0.62 | 1.9 | 0.020 | n/db |
| Ubiquitinc | 0.84 | 140 | 8.5 | 10.4 | 90 | −85.8 | −0.95 | 2.4 | 0.027 | 0.491 |
| Lysozyme | n/dd | n/dd | 9.0 | 14.3 | 129 | −60.9 | −0.47 | 7.9 | 0.061 | 0.471 |
| LDH – 1e | n/af | n/af | 6.0 | 147 | 1336 | −186.6 | −0.002 | −18 | −0.013 | n/af |
| LDH – 2e | n/af | n/af | 6.5 | 146 | 1334 | −347.4 | 0.011 | −9 | −0.007 | n/af |
| LPL | n/af | n/af | 8.5 | 107 | 956 | −499.8 | −0.15 | 11 | 0.012 | n/ab , f |
Hphob, Kyte, and Doolittle hydrophobicity.60
n/d, not determined because there is no PDB.
n/d, not determined because lysozyme inactivates LDH in solution.
Isozymes LDH – 1 and LDH – 2 are both present in commercial LDH.
n/a, not applicable because these are test enzymes, not additives.
Figure 8.
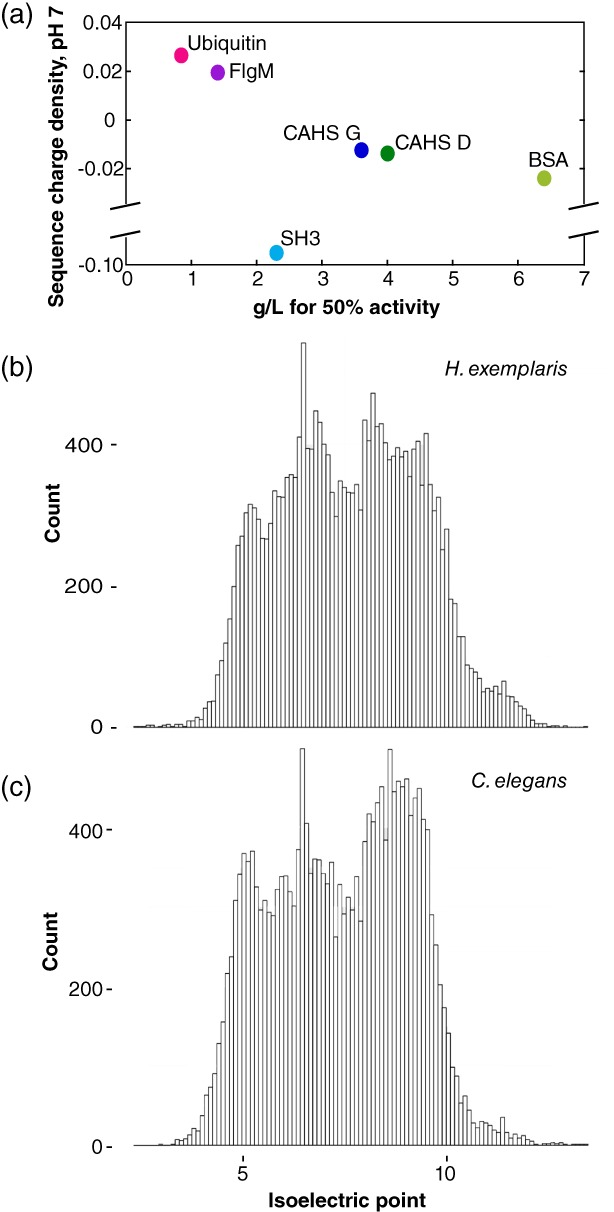
(a) Charge at pH 7 divided by the number of residues in each protein plotted against the concentration for 50% protection of LDH activity. (b) Histogram of pI values at 0.1 intervals for predicted open reading frames in the genome of Hypsibius exemplaris.44, 45 (c) Histogram of pI values at 0.1 intervals for predicted open reading frames in the genome of Caenorhabditis elegans.46, 47
SH3 deviates from this pattern, possibly because of its small size or its large surface hydrophobicity (Table 1). Lysozyme is the other exception. Its protective ability cannot be quantified because it inactivates LDH even in unstressed control solutions, consistent with its generally destabilizing effect.48, 49, 50, 51 In solution, test proteins are stabilized by crowders of the same charge and destabilized by crowders of the opposite charge.41, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58 If negatively charged CAHS proteins function by protecting more positively charged proteins against desiccation, then it is likely that the mechanism of desiccation tolerance differs from the mechanism that stabilizes test proteins under crowded conditions in solution.
We suggest that CAHS proteins (pI ~6) are best at protecting proteins with isoelectric points greater than 6. This idea holds for LDH and LPL, and perhaps tardigrades. In the species Hypsibius exemplaris, from which CAHS D and G are derived, 80% of proteins in predicted open reading frames have a higher pI than CAHS proteins [Fig. 8(b)], consistent with our suggestion. This trend is also true for LEA proteins (pI ~5), an intrinsically disordered protein family required for desiccation tolerance in Caenorhabditis elegans, 62, 63 which also protects enzymes against environmental stress.24, 28, 30, 64, 65 Ninety‐one percent of proteins in predicted open reading frames of C. elegans have a higher isoelectric point than LEA proteins [Fig. 8(c)].
CAHS proteins protect against multiple stresses
CAHS D and G protect LDH against desiccation‐induced inactivation and heat‐induced inactivation of the desiccated enzyme but do not outperform the control proteins ubiquitin, flgM, and SH3 on a g/L basis. CAHS proteins and BSA also protect LPL against freeze/thaw. Most noticeably, CAHS proteins outperform BSA in stabilizing LPL after lyophilization and storage at 4°C. Tardigrades require CAHS proteins to survive desiccation, but not freeze/thaw stress,16 suggesting redundant strategies to protect their cellular components against freeze/thaw stress. CAHS proteins may arrest formation of ice crystals, or they may protect enzyme activity orthogonally to antifreeze proteins.66 Either way, CAHS proteins may be valuable for organ preservation.67, 68, 69, 70 Other proteins with no known link to freeze/thaw or desiccation tolerance may also have these protective properties.
Conclusions
Intrinsically disordered CAHS proteins from tardigrades can protect test enzymes against desiccation‐, freezing‐, and lyophilization‐induced deactivation, consistent with the idea that protection of enzymes and other globular proteins is part of their native function. Our data also show that a variety of globular and disordered protein controls with no known link to desiccation tolerance can protect test enzymes. LDH protection correlates, albeit imperfectly, with the sequence charge density of the protein additive, suggesting a method to tune the protective ability of CAHS proteins or other protective proteins by modifying their charge.
CAHS proteins may also be optimized in other ways to confer desiccation tolerance. For instance, they may be more effective at long‐term protection against temperature fluctuations, as would be experienced with the changing of seasons. CAHS proteins may protect membranes in addition to proteins. CAHS proteins could also be inert to essential biochemical pathways compared to other proteins. Additionally, their intrinsic disorder and the disorder of other proteins linked to desiccation tolerance may facilitate degradation after rehydration. Additional studies are needed to explore these possibilities. Nonetheless our results support the potential use of CAHS proteins as a stabilizing excipient in biologic formulations.
Materials and Methods
Commercial additives
Betaine and Ficoll 70 were purchased from Sigma Life Science, St. Louis, MO, USA. PEG 3350 and sucrose were purchased from Sigma Chemical, St. Louis, MO, USA. Bovine serum albumin and lysozyme were purchased from Sigma, St. Louis, MO, USA. D‐Glucose was purchased from MP Biomedicals, Santa Ana, CA, USA. D‐Maltose was purchased from Fluka Biochemika, Morris Plains, NJ, USA. D‐Mannitol was purchased from Acros Organics, Geel, Belgium. D‐Trehalose was purchased from Aldrich Chemical, St. Louis, MO, USA and Acros Organics, Geel, Belgium. Dextran 20 was purchased from Alfa Aesar, Heysham, Lancashire, UK. Glycine was purchased from Fisher Chemical, Fairlawn, NJ, USA. H2O with a conductivity of 17 MΩcm−1 was used for protein purification and LDH assays.
CAHS D purification
Plasmids for CAHS D, also known as CAHS 94063, were engineered and transformed into BL21star(DE3) cells as described.16 A single colony was used to inoculate 10 mL of Lennox broth (LB, 10 g/L tryptone, 5 g/L yeast extract, 5 g/L NaCl) supplemented with the antibiotic kanamycin to a final concentration of 60 μg/mL. The culture was shaken at 37°C overnight (New Brunswick Scientific I26 incubator, 225 rpm). This 10 mL culture was used to inoculate 1 L of kanamycin supplemented LB.
One‐L cultures were shaken at 37°C until the optical density at 600 nm was greater than 0.4 but less than 0.8 (BioRad SmartSpec Plus spectrophotometer). Isopropyl β‐D‐1‐thiogalactopyranoisde (1 mM final concentration) was added to induce expression. After 4 h, cells were pelleted at 1000 g at 10°C for 30 min (Sorvall Instruments RC‐3B, Fairlawn, NJ, USA). Pellets were stored at −20°C.
Three cell pellets were resuspended in 15 mL of 25 mM Tris/HCl (pH 7.0) supplemented with 50 μL protease inhibitors (Sigma‐Aldrich P2714 containing 40 mM AEBSF, 6 μM aprotinin, 2.3 mM bestatin, 280 μM E‐64, 20 μM leupeptin, and 20 mM EDTA) per cell pellet and then heat shocked at 95°C for 15 min. The heat‐insoluble fraction was removed by centrifugation at 20,000 g for 30 min (Du Pont RC‐5B) at room temperature. The supernatant was mixed with two volumes of 8 M urea containing 50 mM sodium acetate (pH 4.0), which had been passed through a 0.45 μm filter (Millipore SLGVM33RS, Burlington, MA, USA). This and all other urea solutions were deionized with 5 g/L Dowex® MB Mixed Ion Exchange Resin (Sigma) before filtering out the resin with a 0.22 μm filter (Corning Inc. 431161, Corning, NY, USA) and adding buffer salts.
Cation exchange chromatography was performed at room temperature (GE AKTA Start, 5 mL GE HiTrap SP HP) in 8 M urea, 50 mM sodium acetate (pH 4.0) with a 28‐column‐volume gradient of 0% to 50% 1 M NaCl. SDS‐PAGE (Bio‐Rad 4–20% Criterion™ TGX™ Gels) was used to identify fractions containing pure CAHS D. The fractions were pooled, filtered (0.45 μm), and transferred to 10,000 MWCO dialysis tubing (Fisher 68100). Samples were dialyzed against 2 M urea, 20 mM Tris/HCl (pH 7.5) for a minimum of 3 h followed by four changes of 20 mM Tris/HCl (pH 7.5) and one change of H2O for at least 3 h each. The samples were flash frozen in a CO2(s)/ethanol bath and lyophilized for 48 h (Labconco FreeZone) before storage at room temperature.
The purity of CAHS D was confirmed using a ThermoScientific Q Exactive HF‐X mass spectrometer. Lyophilized CAHS D was resuspended at a concentration of ~1 g/L protein in autoclaved H2O and filtered (0.20 μm, Millipore SLHVM33RS). The solution was diluted 1:1 with acetonitrile before injection.
CAHS G purification
CAHS G, also known as CAHS 89226,16 was expressed in BL21star(DE3) cells as described earlier. Two cell pellets were resuspended in 20 mL of 25 mM Tris/HCl (pH 7.0) supplemented with protease inhibitors and heat shocked as described earlier. The supernatant was mixed with two volumes of 3 M urea, 20 mM Tris/HCl (pH 9.0) that had been passed through a 0.45 μm filter.
Anion exchange chromatography was performed at room temperature on the AKTA Start (5 mL GE HiTrap SP Q) in 3 M urea, 20 mM Tris/HCl (pH 9.0) with a 24‐column‐volume gradient of 0% to 100% 150 mM NaCl. SDS‐PAGE was used to identify the fractions containing pure CAHS G. These fractions were pooled, filtered (0.45 μm), and transferred to 10,000 MWCO dialysis tubing. Samples were dialyzed against 20 mM Tris/HCl (pH 7.5) for a minimum of 4 h followed by five changes of H2O for at least 3 h each. The samples were flash frozen, lyophilized, and stored as described for CAHS D. Purity was determined only by SDS PAGE because CAHS G is insoluble under the conditions used for mass spectrometry.
FlgM
A plasmid harboring the FlgM gene71 was used to express the protein in Escherichia coli BL21star(DE3) cells using the protocol earlier, except that antibiotic ampicillin was used for selection (100 μg/mL final concentration). Three pellets were resuspended in 15 mL of 50 mM Tris/HCl (pH 7.5) supplemented with protease inhibitors. The heat‐soluble fraction was collected as described earlier.
Anion exchange chromatography was performed at 4°C (GE Healthcare, Chicago, IL, USA, AKTA FPLC, GE Q Sepharose column, 1.6 cm × 10 cm) in 50 mM Tris/HCl (pH 7.5) using a gradient of 2.5% to 50% 2 M NaCl. FlgM does not bind. The flgM in the flow through was further purified by size‐exclusion chromatography (GE Superdex 75 column, 1.6 cm x 600 cm eluted with 50 mM Na2HPO4, 20 mM KH2PO4, 9 mM NaCl, pH 7.4). Purified protein was dialyzed against H2O for 4 h at room temperature or overnight at 4°C. Dialyzed samples were flash frozen in a CO2(s)/ethanol bath and lyophilized.
SH3
The stabilized T22G mutant40 was produced by site‐directed mutagenesis of the pET11d plasmid containing the drkN SH3 gene using the Agilent QuikChange kit (Santa Clara, CA, USA) and the following primers: forward 5’ GAC GAC GAG CTG AGT TTT CGC AAA GGT CAG ATT CTA AAG ATA TTA AAT ATG G 3′ and reverse 5’ C CAT ATT TAA TAT CTT TAG AAT CTG ACC TTT GCG AAA ACT CAG CTC GTC GTC 3′. The plasmid was used to express the protein in E. coli BL21star(DE3) cells using the protocol for CAHS expression except that ampicillin was used for selection (100 μg/mL final concentration). Six pellets were combined in 10 mL of 50 mM Tris/HCl (pH 7.5) supplemented with protease inhibitors and lysed by sonication (10% amplitude, 4 s on, 2 s off for 15 min). Anion exchange was performed as described for flgM. SH3 does bind to anion exchange resin. SH3 containing fractions identified by SDS PAGE were further purified by size exclusion chromatography as described for flgM. Purified protein was dialyzed and lyophilized a described for flgM.
Ubiquitin
A plasmid harboring the gene encoding histidine‐tagged ubiquitin58, 61 was used to express the protein in E. coli BL21star(DE3) cells using the protocol described for the CAHS proteins except ampicillin (100 μg/mL final concentration) was used for selection. Three pellets were resuspended in 15 mL of 50 mM Na2HPO4, 500 mM NaCl, 15 mM imidazole (pH 7.6) supplemented with protease inhibitors and purified as described.58
Lactate dehydrogenase assays
The assay is based on published protocols.16, 30 L‐LDH from rabbit muscle (Roche, Basel, Switzerland) was diluted to 0.1 g/L in 100 μL of 25 mM Tris/HCl (pH 7.0) containing various concentrations of CAHS D, CAHS G, ubiquitin, SH3, flgM, or other additives. Concentrations of CAHS D, CAHS G, ubiquitin, and SH3 were determined using the Bradford assay.72 Before addition to LDH, CAHS proteins were resuspended at high concentration by heating at 95°C. FlgM does not react in a linear fashion with the Bradford reagent, so we determined the concentration spectrophotometrically73 (ε280 = 1,490 M−1 cm−1) despite its lack of tryptophans. BSA concentration was also quantified spectrophotometrically (ε280 = 43,824 M−1 cm−1 according to supplier). Concentrations of lysozyme were based on mass.
Half of each sample was stored at 4°C. The other half was dehydrated in a Genevac® EZ‐2 Personal Evaporator using the aqueous setting (time to final stage: 16 h; final stage: 0 h) without added heat.
Protection assay samples were immediately rehydrated with 250 μL of H2O. Heat tolerance assay samples containing protein additives were exposed to 95°C (Fisherbrand™ Isotemp™ heat block, Pittsburgh, PA, USA) for various times, cooled on ice, and rehydrated with 250 μL of H2O. Heat tolerance assay samples containing trehalose were exposed to a fixed temperature for 5 min before cooling to 4°C (Eppendorf® Mastercycler Personal, Hambrug, Germany) and resuspension in 250 μL of H2O. Control samples were diluted with 200 μL of H2O.
If necessary for resuspension, samples were shaken (New Brunswick Scientific, Edison, NJ, USA, I26 incubator, 225 rpm) at 37°C for 10 min. Otherwise, samples were kept on ice until assayed. Enzyme activity was determined as described.16 Experiments were performed in triplicate.
Lipoprotein lipase assays
Lipoprotein lipase was purified from fresh, unpasteurized cow's milk as described.74 Samples containing 27.5 ng of LPL (100 μL of 2.5 nM LPL) were mixed with additive. Excipients were resuspended in phosphate‐buffered saline (PBS, 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 1.8 mM KH2PO4, pH 7.4) and mixed with LPL prior to flash freezing and lyophilization. Samples were brought to equal volumes with PBS, flash frozen, and lyophilized in a Savant Speed Vac attached to a lyophilizer. Positive controls were flash frozen but not lyophilized and stored at −80°C until lyophilization was complete. Following lyophilization, positive controls were adjusted to the same condition as their lyophilized counterpart.
Samples were resuspended in 100 μL LPL assay buffer, which has a final concentration of 20 mM Tris–HCl pH 8.0, 150 mM NaCl, 0.2% fatty‐acid free BSA (Sigma), and 1 mM sodium deoxycholate (Sigma). Immediately prior to measuring activity, 10 μM of the fluorescent substrate 1,2‐di‐O‐lauryl‐rac‐glycero‐3‐(glutaric acid 6‐methylresofurin ester) (DGGR, Sigma) in 0.01% Anzergent 3–16 (Anatrace, Maumee, OH, USA) was added.
Activity was measured as described.35 Assays were conducted at 37°C in triplicate. The activities were corrected by subtracting a buffer control comprising DGGR and the appropriate concentration of CAHS, BSA, or trehalose, but without LPL. Data were normalized to unstressed LPL.
Histograms of pI values for predicted open reading frames
The C. elegans (UP000001940) proteome was obtained from Uniprot.org. The H. exemplaris (formerly H. dujardini) proteome (Hypsibius_dujardini_nHd.3.1.5.proteins.fa) was obtained from tardigrades.org. Isoelectric points were calculated using ipc‐1.0.75
Supporting information
Supplemental Figure S1: Protecting LDH with sugars.
Supplemental Figure S2: Trehalose protects dry LDH from heat inactivation.
Acknowledgments
Our research is supported by the National Science Foundation (MCB 1410854 and CHE 1607359 to GJP), the National Institutes of Health (NIGMS 5T32GM008570‐20 to UNC, R01HL12564 to SBN, and R01GM127291 to GJP), the National Aeronautics and Space Administration (NNX15AB44G to TCB), the Life Sciences Research Foundation (Fellowship to TCB), the UNC Office of Graduate Education (Dissertation Completion Fellowship to SP), the UNC Office of Undergraduate Research (Summer Undergraduate Research Fellowship to AM), and the WinSPIRE Summer High School Research Program (Fellowship to AP). GJP thanks the K.C. Wong Education Foundation for travel support. We thank the University of North Carolina's Department of Chemistry Mass Spectrometry Core Laboratory, especially Brandie M Ehrmann, for assistance with mass spectrometry. The core laboratory is supported by the National Science Foundation (CHE 1726291). We thank Eric Brustad for use of the UV–Visible spectrophotometer, as well as Elizabeth Pielak and Dennis Piszkiewicz for comments on the manuscript. GJP, TCB, and SP are inventors on two provisional patent applications pertinent to this work (62/585,740 and 62/375,238).
Impact statement: Protein‐based drugs called biologics are among the most effective therapeutic treatments in the market. However, these drugs are unstable and require refrigerated storage, which makes them expensive. Manufacturers increase the shelf‐life of biologics by adding excipients (molecules that protect active ingredients) but most still require refrigeration. Discovering better excipients would make these products more affordable and accessible, particularly for poor and remote populations without access to refrigeration. Furthermore, some potential biologics may never reach the market because of instability. Better methods to protect and stabilize biologics could provide new treatment options for numerous diseases. Here we describe a family of proteins from tardigrades that increase the shelf stability of two enzymes. This family has potential as an excipient for biologics and for formulating industrially useful enzymes.
References
- 1. Mensink MA, Frijlink HW, van der Voort Maarschalk K, Hinrichs WL (2017) How sugars protect proteins in the solid state and during drying (review): mechanisms of stabilization in relation to stress conditions. Eur J Pharm Biopharm 114:288–295. [DOI] [PubMed] [Google Scholar]
- 2. Liu K, Dong Y, Huang Y, Rasgon JL, Agre P (2013) Impact of trehalose transporter knockdown on Anopheles gambiae stress adaptation and susceptibility to Plasmodium falciparum infection. Proc Natl Acad Sci U S A 110:17504–17509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Erkut C, Penkov S, Khesbak H, Vorkel D, Verbavatz JM, Fahmy K, Kurzchalia TV (2011) Trehalose renders the dauer larva of Caenorhabditis elegans resistant to extreme desiccation. Curr Biol 21:1331–1336. [DOI] [PubMed] [Google Scholar]
- 4. Sakurai M, Furuki T, Akao K, Tanaka D, Nakahara Y, Kikawada T, Watanabe M, Okuda T (2008) Vitrification is essential for anhydrobiosis in an African chironomid, Polypedilum vanderplanki . Proc Natl Acad Sci U S A 105:5093–5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tapia H, Koshland DE (2014) Trehalose is a versatile and long‐lived chaperone for desiccation tolerance. Curr Biol 24:2758–2766. [DOI] [PubMed] [Google Scholar]
- 6. Kaushik JK, Bhat R (2003) Why is trehalose an exceptional protein stabilizer? An analysis of the thermal stability of proteins in the presence of the compatible osmolyte trehalose. J Biol Chem 278:26458–26465. [DOI] [PubMed] [Google Scholar]
- 7. Sarkar S, Davies JE, Huang Z, Tunnacliffe A, Rubinsztein DC (2007) Trehalose, a novel mTOR‐independent autophagy enhancer, accelerates the clearance of mutant huntingtin and α‐synuclein. J Biol Chem 282:5641–5652. [DOI] [PubMed] [Google Scholar]
- 8. Aguib Y, Heiseke A, Gilch S, Riemer C, Baier M, Schatzl HM, Ertmer A (2009) Autophagy induction by trehalose counteracts cellular prion infection. Autophagy 5:361–369. [DOI] [PubMed] [Google Scholar]
- 9. Muller S, Wallace DJ (2014) The importance of implementing proper selection of excipients in lupus clinical trials. Lupus 23:609–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mazzeo A, Carpenter P. Stability studies for biologics In: Huynh‐Ba K, Ed. (2009) Handbook of stability testing in pharmaceutical development: regulations, methodologies, and best practices. New York, NY: Springer New York; p. 353–369. [Google Scholar]
- 11. Becquerel P (1950) La suspension de la vie au‐dessous de 1/20‐degrees‐K absolu par demagnetisation adiabatique de lalun de fer dans le vide le plus eleve. Cr Hebd Acad Sci 231:261–263. [Google Scholar]
- 12. Guidetti R, Jonsson KI (2002) Long‐term anhydrobiotic survival in semi‐terrestrial micrometazoans. J Zool 257:181–187. [Google Scholar]
- 13. Horikawa DD, Sakashita T, Katagiri C, Watanabe M, Kikawada T, Nakahara Y, Hamada N, Wada S, Funayama T, Higashi S, Kobayashi Y, Okuda T, Kuwabara M (2006) Radiation tolerance in the tardigrade Milnesium tardigradum . Int J Radiat Biol 82:843–848. [DOI] [PubMed] [Google Scholar]
- 14. Jonsson KI, Rabbow E, Schill RO, Harms‐Ringdahl M, Rettberg P (2008) Tardigrades survive exposure to space in low earth orbit. Curr Biol 18:R729–R731. [DOI] [PubMed] [Google Scholar]
- 15. Tsujimoto M, Imura S, Kanda H (2016) Recovery and reproduction of an Antarctic tardigrade retrieved from a moss sample frozen for over 30 years. Cryobiology 72:78–81. [DOI] [PubMed] [Google Scholar]
- 16. Boothby TC, Tapia H, Brozena AH, Piszkiewicz S, Smith AE, Giovannini I, Rebecchi L, Pielak GJ, Koshland D, Goldstein B (2017) Tardigrades use intrinsically disordered proteins to survive desiccation. Mol Cell 65:975–984 e975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Boothby TC, Tenlen JR, Smith FW, Wang JR, Patanella KA, Nishimura EO, Tintori SC, Li Q, Jones CD, Yandell M, Messina DN, Glasscock J, Goldstein B (2015) Evidence for extensive horizontal gene transfer from the draft genome of a tardigrade. Proc Natl Acad Sci U S A 112:15976–15981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Koutsovoulos G, Kumar S, Laetsch DR, Stevens L, Daub J, Conlon C, Maroon H, Thomas F, Aboobaker AA, Blaxter M (2016) No evidence for extensive horizontal gene transfer in the genome of the tardigrade Hypsibius dujardini . Proc Natl Acad Sci U S A 113:5053–5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bemm F, Weiss CL, Schultz J, Forster F (2016) Genome of a tardigrade: horizontal gene transfer or bacterial contamination? Proc Natl Acad Sci U S A 113:E3054–E3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Levin M, Anavy L, Cole AG, Winter E, Mostov N, Khair S, Senderovich N, Kovalev E, Silver DH, Feder M, Fernandez‐Valverde SL, Nakanishi N, Simmons D, Simakov O, Larsson T, Liu SY, Jerafi‐Vider A, Yaniv K, Ryan JF, Martindale MQ, Rink JC, Arendt D, Degnan SM, Degnan BM, Hashimshony T, Yanai I (2016) The mid‐developmental transition and the evolution of animal body plans. Nature 531:637–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mali B, Grohme MA, Forster F, Dandekar T, Schnolzer M, Reuter D, Welnicz W, Schill RO, Frohme M (2010) Transcriptome survey of the anhydrobiotic tardigrade Milnesium tardigradum in comparison with Hypsibius dujardini and Richtersius coronifer . BMC Genomics 11:168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yamaguchi A, Tanaka S, Yamaguchi S, Kuwahara H, Takamura C, Imajoh‐Ohmi S, Horikawa DD, Toyoda A, Katayama T, Arakawa K, Fujiyama A, Kubo T, Kunieda T (2012) Two novel heat‐soluble protein families abundantly expressed in an anhydrobiotic tardigrade. PloS One 7:e44209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chakrabortee S, Tripathi R, Watson M, Schierle GS, Kurniawan DP, Kaminski CF, Wise MJ, Tunnacliffe A (2012) Intrinsically disordered proteins as molecular shields. Mol Biosyst 8:210–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dang NX, Popova AV, Hundertmark M, Hincha DK (2014) Functional characterization of selected LEA proteins from Arabidopsis thaliana in yeast and in vitro . Planta 240:325–336. [DOI] [PubMed] [Google Scholar]
- 25. Garay‐Arroyo A, Colmenero‐Flores JM, Garciarrubio A, Covarrubias AA (2000) Highly hydrophilic proteins in prokaryotes and eukaryotes are common during conditions of water deficit. J Biol Chem 275:5668–5674. [DOI] [PubMed] [Google Scholar]
- 26. Boothby TC, Pielak GJ (2017) Intrinsically disordered proteins and desiccation tolerance: elucidating functional and mechanistic underpinnings of anhydrobiosis. Bioessays 39:1700119. [DOI] [PubMed] [Google Scholar]
- 27. Theillet FX, Binolfi A, Frembgen‐Kesner T, Hingorani K, Sarkar M, Kyne C, Li C, Crowley PB, Gierasch L, Pielak GJ, Elcock AH, Gershenson A, Selenko P (2014) Physicochemical properties of cells and their effects on intrinsically disordered proteins (IDPs). Chem Rev 114:6661–6714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Popova AV, Rausch S, Hundertmark M, Gibon Y, Hincha DK (2015) The intrinsically disordered protein LEA7 from Arabidopsis thaliana protects the isolated enzyme lactate dehydrogenase and enzymes in a soluble leaf proteome during freezing and drying. Biochim Biophys Acta 1854:1517–1525. [DOI] [PubMed] [Google Scholar]
- 29. Hughes S, Graether SP (2011) Cryoprotective mechanism of a small intrinsically disordered dehydrin protein. Protein Sci 20:42–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Goyal K, Walton LJ, Tunnacliffe A (2005) LEA proteins prevent protein aggregation due to water stress. Biochem J 388:151–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Izutsu K, Yoshioka S, Terao T (1994) Stabilizing effect of amphiphilic excipients on the freeze‐thawing and freeze‐drying of lactate dehydrogenase. Biotechnol Bioeng 43:1102–1107. [DOI] [PubMed] [Google Scholar]
- 32. Anchordoquy TJ, Carpenter JF (1996) Polymers protect lactate dehydrogenase during freeze‐drying by inhibiting dissociation in the frozen state. Arch Biochem Biophys 332:231–238. [DOI] [PubMed] [Google Scholar]
- 33. Pyne A, Chatterjee K, Suryanarayanan R (2003) Solute crystallization in mannitol‐glycine systems‐‐implications on protein stabilization in freeze‐dried formulations. J Pharm Sci 92:2272–2283. [DOI] [PubMed] [Google Scholar]
- 34. Tamiya T, Okahashi N, Sakuma R, Aoyama T, Akahane T, Matsumoto JJ (1985) Freeze denaturation of enzymes and its prevention with additives. Cryobiology 22:446–456. [DOI] [PubMed] [Google Scholar]
- 35. Lafferty MJ, Bradford KC, Erie DA, Neher SB (2013) Angiopoietin‐like protein 4 inhibition of lipoprotein lipase: evidence for reversible complex formation. J Biol Chem 288:28524–28534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Osborne JC Jr, Bengtsson‐Olivecrona G, Lee NS, Olivecrona T (1985) Studies on inactivation of lipoprotein lipase: role of the dimer to monomer dissociation. Biochemistry 24:5606–5611. [DOI] [PubMed] [Google Scholar]
- 37. Pillarisetti S, Paka L, Sasaki A, Vanni‐Reyes T, Yin B, Parthasarathy N, Wagner WD, Goldberg IJ (1997) Endothelial cell heparanase modulation of lipoprotein lipase activity. Evidence that heparan sulfate oligosaccharide is an extracellular chaperone. J Biol Chem 272:15753–15759. [DOI] [PubMed] [Google Scholar]
- 38. Miller DP, Anderson RE, de Pablo JJ (1998) Stabilization of lactate dehydrogenase following freeze thawing and vacuum‐drying in the presence of trehalose and borate. Pharm Res 15:1215–1221. [DOI] [PubMed] [Google Scholar]
- 39. Daughdrill GW, Chadsey MS, Karlinsey JE, Hughes KT, Dahlquist FW (1997) The C‐terminal half of the anti‐sigma factor, FlgM, becomes structured when bound to its target, sigma 28. Nat Struct Biol 4:285–291. [DOI] [PubMed] [Google Scholar]
- 40. Mok YK, Elisseeva EL, Davidson AR, Forman‐Kay JD (2001) Dramatic stabilization of an SH3 domain by a single substitution: roles of the folded and unfolded states. J Mol Biol 307:913–928. [DOI] [PubMed] [Google Scholar]
- 41. Stadmiller SS, Gorensek‐Benitez AH, Guseman AJ, Pielak GJ (2017) Osmotic shock induced protein destabilization in living cells and its reversal by glycine betaine. J Mol Biol 429:1155–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Santoro MM, Liu Y, Khan SM, Hou LX, Bolen DW (1992) Increased thermal stability of proteins in the presence of naturally occurring osmolytes. Biochemistry 31:5278–5283. [DOI] [PubMed] [Google Scholar]
- 43. Rydeen AE, Brustad EM, Pielak GJ (2018) Osmolytes and protein‐protein interactions. J Am Chem Soc 140:7441–7444. [DOI] [PubMed] [Google Scholar]
- 44. Arakawa K, Yoshida Y, Tomita M (2016) Genome sequencing of a single tardigrade Hypsibius dujardini individual. Sci Data 3:160063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yoshida Y, Koutsovoulos G, Laetsch DR, Stevens L, Kumar S, Horikawa DD, Ishino K, Komine S, Kunieda T, Tomita M, Blaxter M, Arakawa K (2017) Comparative genomics of the tardigrades Hypsibius dujardini and Ramazzottius varieornatus . PLoS Biol 15:e2002266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Okimoto R, Macfarlane JL, Clary DO, Wolstenholme DR (1992) The mitochondrial genomes of two nematodes, Caenorhabditis elegans and Ascaris suum . Genetics 130:471–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Consortium CeS (1998) Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282:2012–2018. [DOI] [PubMed] [Google Scholar]
- 48. Sarkar M, Smith AE, Pielak GJ (2013) Impact of reconstituted cytosol on protein stability. Proc Natl Acad Sci U S A 110:19342–19347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Smith AE, Zhou LZ, Gorensek AH, Senske M, Pielak GJ (2016) In‐cell thermodynamics and a new role for protein surfaces. Proc Natl Acad Sci U S A 113:1725–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Monteith WB, Pielak GJ (2014) Residue level quantification of protein stability in living cells. Proc Natl Acad Sci U S A 111:11335–11340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Miklos AC, Sarkar M, Wang Y, Pielak GJ (2011) Protein crowding tunes protein stability. J Am Chem Soc 133:7116–7120. [DOI] [PubMed] [Google Scholar]
- 52. Danielsson J, Mu X, Lang L, Wang H, Binolfi A, Theillet FX, Bekei B, Logan DT, Selenko P, Wennerstrom H, Oliveberg M (2015) Thermodynamics of protein destabilization in live cells. Proc Natl Acad Sci U S A 112:12402–12407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ignatova Z, Krishnan B, Bombardier JP, Marcelino AM, Hong J, Gierasch LM (2007) From the test tube to the cell: exploring the folding and aggregation of a beta‐clam protein. Biopolymers 88:157–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Guseman AJ, Pielak GJ (2017) Cosolute and crowding effects on a side‐by‐side protein dimer. Biochemistry 56:971–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sarkar M, Lu J, Pielak GJ (2014) Protein crowder charge and protein stability. Biochemistry 53:1601–1606. [DOI] [PubMed] [Google Scholar]
- 56. Sarkar M, Pielak GJ (2014) An osmolyte mitigates the destabilizing effect of protein crowding. Protein Sci 23:1161–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Davis‐Searles PR, Saunders AJ, Erie DA, Winzor DJ, Pielak GJ (2001) Interpreting the effects of small uncharged solutes on protein‐folding equilibria. Annu Rev Biophys Biomol Struct 30:271–306. [DOI] [PubMed] [Google Scholar]
- 58. Wang Y, Sarkar M, Smith AE, Krois AS, Pielak GJ (2012) Macromolecular crowding and protein stability. J Am Chem Soc 134:16614–16618. [DOI] [PubMed] [Google Scholar]
- 59. Mahn A, Lienqueo ME, Asenjo JA (2004) Effect of surface hydrophobicity distribution on retention of ribonucleases in hydrophobic interaction chromatography. J Chromatogr A 1043:47–55. [DOI] [PubMed] [Google Scholar]
- 60. Kyte J, Doolittle RF (1982) A simple method for displaying the hydropathic character of a protein. J Mol Biol 157:105–132. [DOI] [PubMed] [Google Scholar]
- 61. Barnes CO, Monteith WB, Pielak GJ (2011) Internal and global protein motion assessed with a fusion construct and in‐cell NMR spectroscopy. Chembiochem 12:390–391. [DOI] [PubMed] [Google Scholar]
- 62. Erkut C, Vasilj A, Boland S, Habermann B, Shevchenko A, Kurzchalia TV (2013) Molecular strategies of the Caenorhabditis elegans dauer larva to survive extreme desiccation. PloS One 8:e82473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Gal TZ, Glazer I, Koltai H (2004) An LEA group 3 family member is involved in survival of C. elegans during exposure to stress. FEBS Lett 577:21–26. [DOI] [PubMed] [Google Scholar]
- 64. Hincha DK, Thalhammer A (2012) LEA proteins: IDPs with versatile functions in cellular dehydration tolerance. Biochem Soc Trans 40:1000–1003. [DOI] [PubMed] [Google Scholar]
- 65. Liu Y, Chakrabortee S, Li R, Zheng Y, Tunnacliffe A (2011) Both plant and animal LEA proteins act as kinetic stabilisers of polyglutamine‐dependent protein aggregation. FEBS Lett 585:630–634. [DOI] [PubMed] [Google Scholar]
- 66. Chapsky L, Rubinsky B (1997) Kinetics of antifreeze protein‐induced ice growth inhibition. FEBS Lett 412:241–244. [DOI] [PubMed] [Google Scholar]
- 67. Rubinsky B, Arav A, Hong JS, Lee CY (1994) Freezing of mammalian livers with glycerol and antifreeze proteins. Biochem Biophys Res Commun 200:732–741. [DOI] [PubMed] [Google Scholar]
- 68. Soltys KA, Batta AK, Koneru B (2001) Successful nonfreezing, subzero preservation of rat liver with 2,3‐butanediol and type I antifreeze protein. J Surg Res 96:30–34. [DOI] [PubMed] [Google Scholar]
- 69. Amir G, Horowitz L, Rubinsky B, Yousif BS, Lavee J, Smolinsky AK (2004) Subzero nonfreezing cryopresevation of rat hearts using antifreeze protein I and antifreeze protein III. Cryobiology 48:273–282. [DOI] [PubMed] [Google Scholar]
- 70. Amir G, Rubinsky B, Horowitz L, Miller L, Leor J, Kassif Y, Mishaly D, Smolinsky AK, Lavee J (2004) Prolonged 24‐hour subzero preservation of heterotopically transplanted rat hearts using antifreeze proteins derived from arctic fish. Ann Thorac Surg 77:1648–1655. [DOI] [PubMed] [Google Scholar]
- 71. Dedmon MM, Patel CN, Young GB, Pielak GJ (2002) FlgM gains structure in living cells. Proc Natl Acad Sci U S A 99:12681–12684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein‐dye binding. Anal Biochem 72:248–254. [DOI] [PubMed] [Google Scholar]
- 73. Gill SC, von Hippel PH (1989) Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem 182:319–326. [DOI] [PubMed] [Google Scholar]
- 74. Bengtsson‐Olivecrona G, Olivecrona T (1991) Phospholipase activity of milk lipoprotein lipase. Methods Enzymol 197:345–356. [DOI] [PubMed] [Google Scholar]
- 75. Kozlowski LP (2016) IPC ‐ isoelectric point calculator. Biol Direct 11:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure S1: Protecting LDH with sugars.
Supplemental Figure S2: Trehalose protects dry LDH from heat inactivation.


