Abstract
We previously described structural and functional characterization of the first ubiquitin variant (UbV), UbV.v27.1, engineered by phage display to bind with high affinity to a specific ubiquitin interacting motif (UIM). We identified two substitutions relative to ubiquitin (Gly10Val/His68Tyr) that were critical for enhancing binding affinity but could only rationalize the mechanism of action of the Tyr68 substitution. Here, we extend our characterization and uncover the mechanism by which the Val10 substitution enhances binding affinity. We show that Val10 in UbV.v27.1 drives UbV dimerization through an intermolecular β‐strand exchange. Dimerization serves to increase the contact surface between the UIM and UbV and also affords direct contacts between two UIMs through an overall 2:2 binding stoichiometry. Our identification of the role of Val10 in UbV dimerization suggests a general means for the development of dimeric UbVs with improved affinity and specificity relative to their monomeric UbV counterparts. Statement: Previously, we used phage display to engineer a UbV that bound tightly and specifically to a UIM. Here, we discovered that tight binding is partly due to the dimerization of the UbV, which increases the contact surface between the UbV and UIM. We show that UbV dimerization is dependent on the Gly10Val substitution, and posit that dimerization may provide a general means for engineering UbVs with improved binding properties.
Keywords: ubiquitin, phage display, protein engineering, ubiquitin interacting motif, dimer
Short abstract
PDB Code(s): 6NJG
Abbreviations
- SEC
size exclusion chromatography
- Ub.wt
ubiquitin wild type
- UbV
ubiquitin variant
- UIM
ubiquitin interacting motif
- Vps27
Vacuolar protein sorting‐associated protein 27
Introduction
We previously reported a ubiquitin (Ub) variant (UbV), UbV.v27.1 [see Fig. 1(A) for sequence of UbV.v27.1] that we developed by phage display. UbV.v27.1 was engineered to bind the first Ub interacting motif (UIM) of the yeast protein Vacuolar protein sorting‐associated protein 27 (Vps27), referred to here as yUIM‐1 [see Fig. 1(B) for yUIM‐1 sequence]. Impressively, UbV.v27.1 was found to bind to yUIM‐1 with almost 500‐fold (439 times) higher affinity than Ub.wt.1
Figure 1.
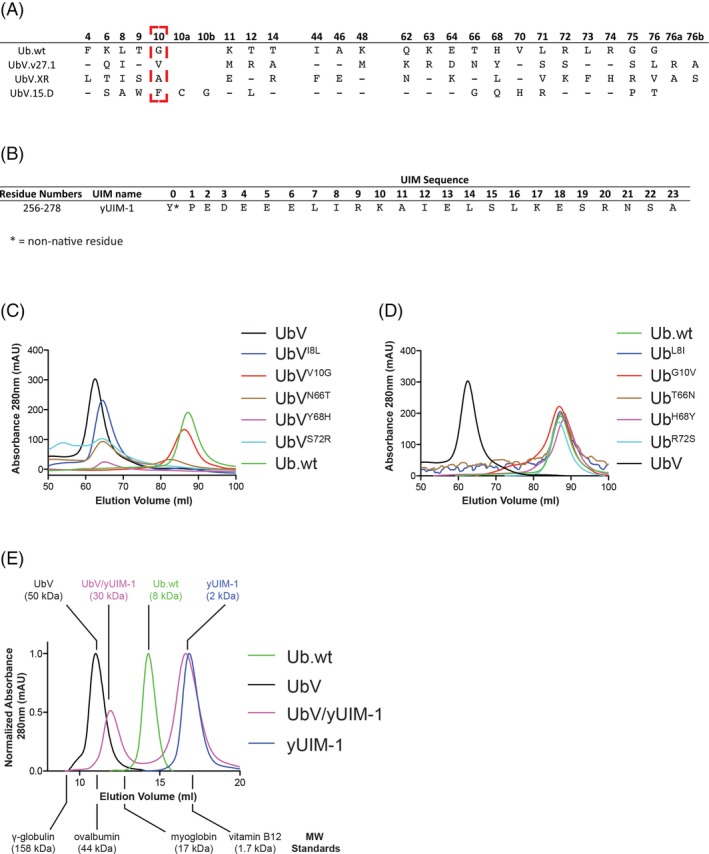
UbV.v27.1 is an oligomeric protein in solution. (A) Sequence alignment of Ub.wt, UbV.v27.1, UbV.XR, and UbV.15.D. Only positions that differ between Ub.wt and one of the UbVs are depicted. Positions that are conserved with Ub.wt are shown as dashes. UbV.15.D contains an insert of two amino acids designated as positions 10a and 10b. (B) Sequence of the yUIM‐1 peptide. The non‐native tyrosine residue at position 0 was included in the synthesis to aid concentration determination by absorbance at 280 nm. (C,D) Preparative scale (120‐ml column volume) size exclusion chromatography elution profiles of UbV.v27.1 with the indicated back substitutions or Ub.wt with the indicated forward substitutions, respectively. The profiles of UbV.v27.1 and Ub.wt are shown in each plot as references. (E) Analytical scale (Bio‐Rad Laboratories Enrich SEC 70 10 × 300 mm column) size exclusion chromatography elution profiles of Ub.wt, UbV.v27.1, yUIM‐1, and a UbV.v27.1/yUIM‐1 complex. The elution volumes for molecular weight standards are indicated along with the inferred molecular weights of Ub.wt, UbV.v27.1, yUIM‐1, and the UbV.v27.1/yUIM‐1 complex. As the molar absorptivity of yUIM‐1 is much larger than UbV.v27.1 and Ub, the absorbance profiles for the presented plots were normalized to the largest peak height.
We determined the crystal structure of yUIM‐1 in complex with UbV.v27.1, which revealed a 1:1 binding stoichiometry that was very similar to the canonical binding mode of Ub.wt to UIMs. Mutational and functional analysis identified two substitutions (Gly10Val/His68Tyr) in UbV.v27.1 relative to Ub.wt that were responsible in large part for the observed binding affinity of the UbV.v27.1 for yUIM‐1. From the structure, we could rationalize the importance of the Tyr68 substitution in its specific contribution of a ~13‐fold increase in affinity when introduced in Ub.wt, as the tyrosine side chain made extensive favorable hydrophobic contacts directly with yUIM‐1. However, we could not rationalize why the Val10 substitution when introduced on its own into Ub.wt was responsible for an ~8‐fold increase in binding affinity, as the Val side chain did not participate in the interface with yUIM‐1. Indeed, Val10 was >7 Å away from the closest residue in yUIM‐1 in our original structure. Here, we have revisited our structural and functional analysis of the UbV–UIM complex, discovering that the Gly10Val substitution in UbV.v27.1 contributes to the enhanced affinity of the UbV for yUIM‐1 through its unexpected ability to induce a dimer state.
Results
UbV.v27.1 is a dimer
In the process of purifying Ub, UbV.v27.1, and a series of intermediate UbVs used to probe the necessity and sufficiency of specific substitutions toward the enhancement of affinity for yUIM‐1, we observed that Ub.wt eluted at a volume consistent with a smaller size than UbV.v27.1 [Fig. 1(C)]. We initially reasoned that the larger apparent size of UbV.v27.1 in solution reflected nonspecific aggregation at high protein concentrations due to the numerous (18 in total) substitutions relative to Ub.wt.
Variants of UbV.v27.1 with individual back substitutions to the respective residue in Ub.wt (Ile8Leu, Val10Gly, Asn66Thr, Tyr68His, or Ser72Arg) also eluted as apparent oligomers with the exception of the Val10Gly back mutant. This mutant eluted at the same volume as Ub.wt [Fig. 1(C)], indicating that the Val10Gly substitution was essential for maintaining an oligomeric state. However, variants of Ub.wt with single‐site substitutions to the respective residues found in UbV.v27.1 (Leu8Ile, Gly10Val, Thr66Asn, His68Tyr, or Arg72Ser) all eluted as monodispersed monomers, indicating that no single substitution in the Ub.wt background was sufficient to induce an oligomeric state [Fig. 1(D)]. In sum, these results suggested that the Gly10Val substitution is necessary but not sufficient to drive the oligomerization of UbV.v27.1, providing a first clue of how Val10 might contribute to the enhanced binding affinity of UbV.v27.1 for yUIM‐1.
To gain insight into how the Gly10Val substitution might contribute to the oligomeric state of UbV.v27.1, we reanalyzed the crystal structure of UbV.v27.1 in complex with yUIM‐1. In the published structure (PDB ID 5UCL), Val10 was immediately adjacent to Thr9, which was left unmodeled due to ambiguity in interpreting the connectivity of the β1–β2 loop [Fig. 2(A,B) top panel]. With a realization that UbV.v27.1 forms an oligomer in solution, we hypothesized that the difficulty in modeling Thr9 may be due to its participation in an unexpected strand exchange mechanism between two crystallographic symmetry‐related protomers rather than a conventional β1–β2 loop observed in the Ub.wt monomer [Fig. 2(B)]. In the strand exchange configuration, residues 8–10 of UbV.v27.1 adopt a linear conformation that seamlessly links strand β1 from one protomer to strand β2 of the second protomer. We termed this extended β‐strand, β1’. The correctness of the strand exchange model was assessed in comparison to the original Thr9 delete model and a model in which Thr9 was included in the form of a conventional loop linking strands β1 and β2 in cis (termed the β1‐β2 loop model). Examination of 2Fo‐Fc and Fo‐Fc electron density maps and final refinement statistics after application of identical refinement protocols and resolution cutoffs to our models confirmed that the β1 loops of two UbV.v27.1 monomers undergo a strand exchange configuration, resulting in the formation of a dimer [Fig. 2(B); Supporting Information Fig. S1, Table S1].
Figure 2.
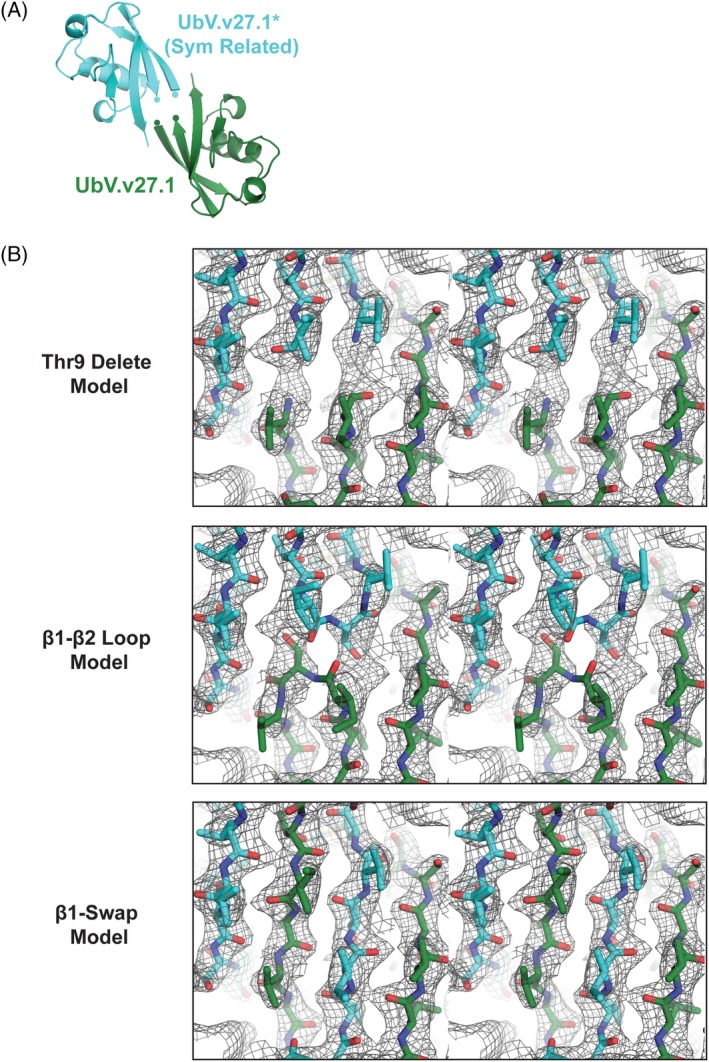
UbV.v27.1 is a β‐strand exchanged dimer in the crystal structure. (A) Architecture of UbV.v27.1 (UbV) and a crystal symmetry related UbV* (PDB entry: 5UCL). C‐terminus of β1 strand and N‐terminus of β2 strand are indicated by circles. (B) Comparison of Thr9 delete, β1–β2 loop, and β1‐swap models for the UbV.v27.1 structure. The panels depict the respective 2Fo‐Fc electron density maps contoured at 1.0 σ for the indicated models shown in stereo view. Models were refined in the same manner using identical data cutoffs.
To investigate the actual size of the UbV.v27.1 oligomer in solution, we reanalyzed UbV.v27.1 using an analytical scale size exclusion column calibrated with molecular weight markers (Supporting Information Fig. S2). Surprisingly, UbV.v27.1 eluted at a volume corresponding to a molecular weight considerably larger (50 kDa) than the theoretical molecular weight of a dimer (19.4 kDa) [Fig. 1(E)]. We reasoned that this discrepancy might be due to further oligomerization of UbV.v27.1 in the absence of a yUIM‐1 binding partner. Indeed, when UbV.v27.1 was mixed with a molar excess of yUIM‐1, its apparent molecular weight downshifted to 30 kDa [Fig. 1(E)], which closely coincided with the expected molecular weight of the dimeric UbV.v27.1:yUIM‐1 complex (26 kDa for a 2:2 binding stoichiometry) observed in the crystal structure. These observations supported the notion that UbV.v27.1 was in fact a functional dimer both in solution and in the crystal environment.
Revised binding mode and stoichiometry of UbV.v27.1 to yUIM‐1
The strand exchange structure of UbV.v27.1 resulted in an unexpected 2:2 binding stoichiometry for yUIM‐1, unlike the 1:1 binding stoichiometry of the previous model [Fig. 3(A,B)]. Notably, with the UbV.v27.1 dimer, each UIM now makes both canonical contacts (buried surface on yUIM‐1 = 537 Å2) typical of Ub–UIM complexes,2 and noncanonical contacts (buried surface area on yUIM‐1 = 297 Å2) not previously observed in Ub–UIM complexes. We previously overlooked the latter contacts as reflecting irrelevant crystal packing interactions. The canonical contacts involving each UIM are essentially as described previously, with the notable exception that the contact surface on the UbV is now composed of sequence elements from both UbV protomers in the strand exchange dimer [Fig. 3(C,D)]. The noncanonical contacts involving each UIM are also composed of sequence elements from both protomers in the UbV dimer. Included are hydrophobic interactions between Leu7 of yUIM‐1 and Ile8 of the first UbV protomer and Val10* of the second UbV* protomer, and hydrophilic interactions between Asp3 of yUIM‐1 and Gln6 of the first UbV protomer and Tyr68* of the second UbV* protomer [Fig. 3(E)].
Figure 3.
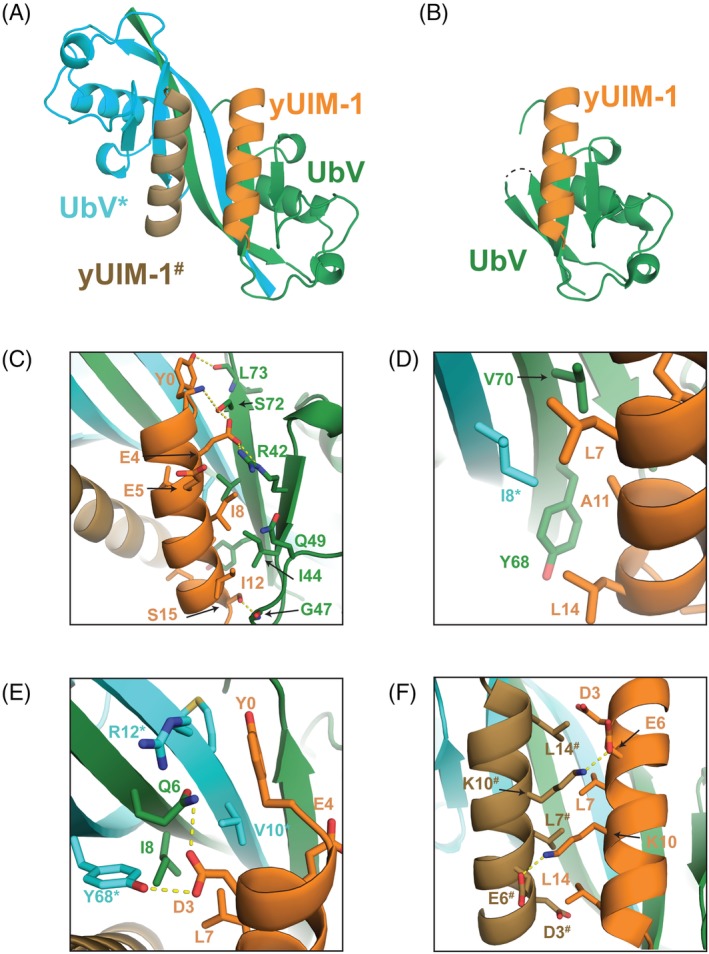
Crystal structure of UbV.v27.1 bound to yUIM‐1. (A) Overview of the dimeric UbV.v27.1‐yUIM‐1 structure. “*” or “#” denotes the symmetry related UbV or yUIM‐1, respectively. (B) Overview of the monomeric UbV.v27.1‐yUIM‐1 structure. (C,D) Details of the molecular interactions on the canonical interface. An asterisk “*” indicates residues from UbV*. (E) Details of the molecular interactions on the noncanonical interface. (F) Details of the molecular interactions on the noncanonical interface. A hash sign “#” indicates residues from yUIM‐1#.
Interestingly, the 2:2 binding interaction between yUIM‐1 and the UbV.v27.1 dimer juxtaposes the two UIMs in an anti‐parallel orientation with a buried surface area on each UIM of 276 Å2 [Fig. 3(A)]. Contacts include hydrophobic interactions between Leu residues at positions 7 and 14 of one protomer and the equivalent position of the second protomer [Fig. 3(F)] and reciprocal salt bridges between Glu and Lys residues at positions 6 and 10, respectively [Fig. 3(F)]. We hypothesized that these inter‐UIM contacts might also contribute to the enhanced affinity of yUIM‐1 binding to UbV.v27.1. In this regard, the salt bridge between Glu6 and Lys10# provided a useful test case as neither residue participates in direct contacts with UbV.v27.1. However, the individual substitutions Glu6Lys or Lys10Glu, which were expected to abrogate the salt interaction and the double substitution Glu6Lys/Lys10Glu, which was expected to restore the salt interaction with opposite polarity, did not appreciably change the affinity of yUIM‐1 for UbV.v27.1 [Fig. 4(A)]. Since all other contacting residues between the two UIMs also participate in direct contacts with the UbV, we could not explore this issue further unambiguously.
Figure 4.
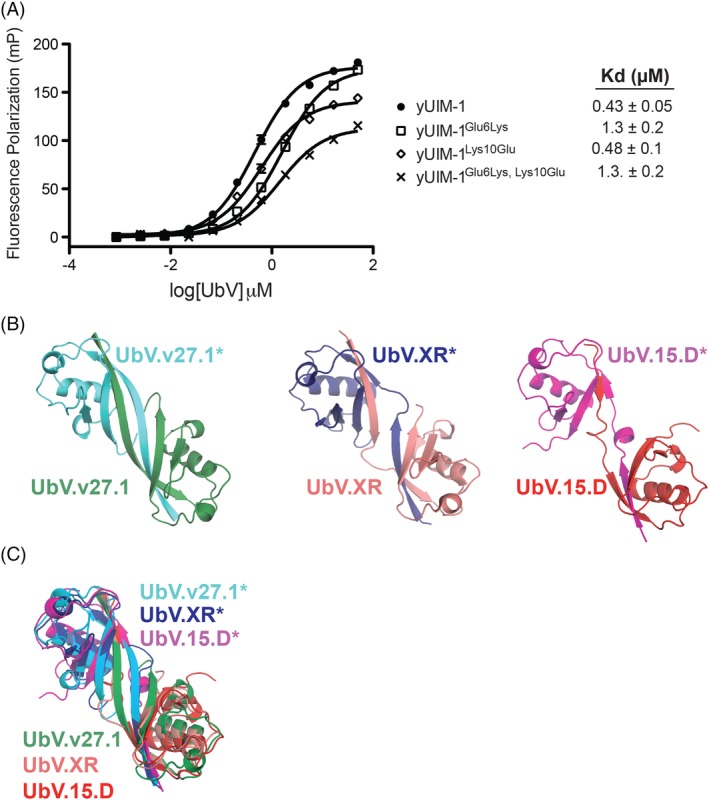
Mutational and comparative analysis of the yUIM‐1/UbV.v27.1 complex. (A) Effects of substitutions on yUIM‐1 binding to UbV.v27.1. Fluorescence polarization binding experiments are shown for the binding of UbV.v.27.1 to yUIM‐1 peptides harboring the indicated substitutions. Values represent mean of readings done in triplicate ± SD. (B,C) Comparison of dimeric UbVs. (B) Side by side view of the dimeric UbVs, UbV.v27.1, UbV.XR (PDB:5O6T), and UbV.15.D (PDB:6DJ9). (C) Superposition of UbV.v27.1. UbV.XR, and UbV.15.D.
Discussion
We have discovered that UbV.v27.1 forms a strand exchange dimer allowing it to bind yUIM‐1 with 2:2 stoichiometry. Interestingly, in the absence of yUIM‐1, UbV.v27.1 self associates as an oligomer with an apparent molecular weight of 50 kDa. This is consistent with either a tetramer (38.8 kDa), a pentamer (48.5 kDa), or a hexamer (58.2 kDa) of UbV.v27.1. We deem it most likely that this species is a tetramer or hexamer, representing further oligomerization of dimeric UbV.v27.1. It is unclear how yUIM‐1 binding drives UbV.v27.1 to its dimeric form but we speculate that oligomerization of the uncomplexed UbV.v27.1 dimer involves the UbV‐UIM contact surface. This allows y‐UIM1 binding to UbV.v27.1 to disrupt the formation of the larger sized oligomer.
We found that oligomerization of UbV.v27.1 in solution is critically dependent on the Gly10Val substitution, as the back substitution to Gly converts UbV.v27.1 to a monomer. How precisely does Val10 help to promote the dimer state? We surmised Val10 might act by destabilizing the monomer by preventing formation of the tight β‐turn between strands β1 and β2. While Val10 in the strand exchange dimer adopts different Ramachandran Phi and Psi values than Gly10 in Ub.wt monomer (77°, 17° versus −113°, 123°, respectively, PDB entry 1UBQ), the latter conformation is fully accessible to Val (i.e. it is not a disallowed conformation in the Ramachandran plot), suggesting that this may not be the case. Our finding that the Gly10Val single‐site substitution in Ub.wt behaves as a monomer in size exclusion chromatography (SEC) analysis proves that Valine at position 10 can support the tight β‐turn promoting conformation required in the monomer state. Thus, the exact reason why Val at position 10 promotes dimerization remains an open question.
The finding that the Gly10Val substitution is not sufficient to drive dimerization indicates that other substitutions in UbV.v27.1 relative to the observed positions in Ub.wt help to promote the stand exchange mechanism. Other substituted positions in UbV.v27.1 that may collaborate with the Gly10Val substitution include residues Gln6, Ile8, Met11, Arg12, and Ala14, which lie in close proximity to the strand exchange junction, and Leu76, which participates in UbV.27.1 dimer contacts elsewhere in the structure [see Fig. 1(A) for sequence comparison between Ub.wt and UbV.v27.1]. Resolving how precisely Val10 helps to promote UbV dimerization may require an understanding of how these substitutions cooperate.
The structure of UbV.v27.1 bound to yUIM‐1 allows for a more comprehensive rationalization of our previous mutational studies. The position of Tyr68 on the canonical contact surface between UbV.v27.1 and yUIM‐1 readily explains its strong contribution to binding. As described above, it creates favorable interactions with yUIM‐1, which directly enhance affinity. The contribution of the substitution Gly10Val, which was difficult to rationalize previously, is now also readily apparent. First, Val10 participates in direct favorable hydrophobic interactions with yUIM‐1 through its position on the noncanonical UIM binding surface. Second, by promoting strand exchange dimerization, Val10 indirectly supports the formation of the noncanonical contact surface with yUIM‐1 that affords additional opportunities for favorable interactions. Third, by enabling a 2:2 binding mode with yUIM‐1, Val10 also provides opportunities for favorable contacts between the two UIMs. We note that in addition to contributing to the enhanced binding affinity of UbV.v27.1 for yUIM‐1, the noncanonical contacts between UbV and yUIM‐1, and possibly between the two UIMs, could in principle contribute to the binding specificity of UbV.v27.1, which we previously showed was highly specific for yUIM‐1 over a panel of 11 other yeast UIMs.
Strand swaps have been observed for many other proteins including C‐cadherin and dUTPase.3, 4 Interestingly, β‐strand swaps have also been observed for two other dimeric UbVs that we reported recently, namely UbV.XR and UbV.15.D. These UbVs, bind to the E3 ligase XIAP and the DUSP domain of USP15, respectively.5, 6 Similarities between UbV.v27.1, UbV.XR, and UbV.15.D include similar strand exchange topologies involving strand β1 that creates an extended β1–β2 strand fusion [Fig. 4(B,C)—note that in the case of UbV.15.D, the extended β1–β2 strand fusion is partially interrupted by a helical kink]. Moreover, dimerization of UbV.v27.1 and UbV.XR is also dependent on substitutions at position 10. However, whereas dimerization of UbV.v27.1 is dependent on a Val at position 10, UbV.XR is dependent on an Ala [Fig. 1(A)] at that position.5 UbV.15.D also differs from Ub.wt at position 10 [Fig. 1(A)] but whether its Gly10Phe substitution is required for UbV.15.D dimerization remains to be determined. We posit that systematic sampling of the residues at position 10 might prove a useful and general means for generating dimeric UbVs with enhanced affinities and specificities for specific ligands.
Materials and Methods
Protein expression and preparative scale purification
We engineered UbV.v27.1 by a phage display process.1, 7, 8, 9 Genes encoding UbV.v27.1 and Ub.wt were cloned into pHH0239 or pProEX‐HTA (Life Technologies, Carlsbad, CA), respectively, for expression as TEV cleavable N‐terminally 6xHis‐tagged fusion proteins. Protein was expressed in Escherichia coli BL21(DE3) as previously described.1 Cells were resuspended in Lysis buffer (50 mM HEPES pH 7.5, 5 mM imidazole, 500 mM NaCl, 5 mM β‐mercaptoethanol, 5% glycerol) and lysed by sonication. Cell lysate was loaded onto a 1‐ml HiTrap Chelating HP column (GE Healthcare, Chicago, IL) and eluted by an imidazole buffer gradient from 5 to 300 mM. Fractions containing UbV.v27.1 or Ub.wt were pooled, dialyzed in Dialysis buffer (50 mM HEPES pH 7.5, 500 mM NaCl, 5 mM β‐mercaptoethanol) to remove imidazole, and incubated overnight with TEV protease to cleave the 6xHis tag. After overnight incubation, TEV protease and uncleaved UbV.v27.1 or Ub.wt was removed by applying the reaction mixtures to a 1‐ml HiTrap Chelating HP column (GE Healthcare, Chicago, IL). UbV.v27.1 or Ub.wt in the flow through fractions was concentrated with a 3‐kDa cutoff Amicon Ultra‐4 concentrator (EMD Millipore). Proteins were then injected (1 mL of ~1–9 mg/mL) on a preparative 120 mL bed volume Superdex 75 16/600 column (GE Healthcare, Chicago, IL) previously equilibrated with size exclusion buffer (25 mM HEPES pH 7.5, 100 mM NaCl, 1 mM DTT). Other UbVs were purified in the same manner. Absorbance at 280 nm was used to monitor eluted proteins. Eluted fractions were analyzed by SDS‐PAGE and pure proteins were pooled, concentrated, flash frozen in liquid nitrogen, and stored at −80°C.
Peptide synthesis for analytical scale SEC
The yUIM‐1 peptide used for SEC analysis was obtained from Bio Basic Inc. The sequence, YPEDEEELIRKAIELSLKESRNSAK, corresponds to residues 256–278 of Vps27 with the addition of a nonnative N‐terminal tyrosine to aid concentration determination and a nonnative C‐terminal lysine to enable covalent labeling with 5/6‐carboxyfluorescein succinimidyl ester (NHS‐FITC). To prevent charge effects from the termini affecting peptide binding, the C‐terminus was amidated and the N‐terminus was acetylated. The peptide was resuspended in a buffered solution as previously described.1
Analytical scale SEC
For the analyses of UbV.v.27.1 and Ub.wt, proteins were thawed and resuspended in size exclusion buffer to a volume of 500 μL and a concentration of 2 mg/mL. For the analysis of yUIM‐1, peptide was thawed and resuspended in size exclusion buffer to a volume of 500 μL and a concentration of 0.4 mg/mL. For the analysis of the UbV.v27.1/yUIM‐1 complex, UbV.v27.1 was resuspended with yUIM‐1 in size exclusion buffer to a volume of 500 μL and a final concentration of 2 and 1 mg/mL of UbV.v27.1 and yUIM‐1 (representing a 1:1.5 molar ratio), respectively. The UbV.v27.1/yUIM‐1 complex was equilibrated on ice for 40 minutes. Proteins were then injected onto an Enrich SEC 70 10 × 300 mm column (Bio‐Rad Laboratories, Hercules, CA) column previously equilibrated with size exclusion buffer. Molecular weight standards (Bio‐rad, product #1511901) including γ‐globulin, ovalbumin, myoglobin, and vitamin B12 were analyzed according to the manufacturers instructions. A standard curve was generated by plotting log(molecular weight) versus Kav (Kav = (V e − V o)/(V t − V o), where V e is the observed elution volume, V o is the void volume (approximated as 1/3 of the total column volume), and V t is the total column volume.
Refinement and structural analysis
To generate the monomeric β1–β2 loop model, we took the previously reported model with Thr9 deleted (PDB:5UCL) and modeled Thr9 into a β1–β2 loop configuration using Coot.10 To generate the β‐swap model we took the previously reported model with Thr9 deleted (PDB:5UCL) and replaced residues 1–8 with residues 1–8 from the corresponding symmetry related UbV.v27.1 partner. We then modeled in Thr9 using Coot. Both the original model and the two new models were then refined against the previously collected, integrated and scaled dataset1 using the same resolution ranges, refinement strategy (refining XYZ [reciprocal‐space], XYZ [real‐space], occupancies, individual B‐factors, and TLS parameters11 [UbV.v27.1 had 10 TLS groups consisting of residues 1–5, 6–10, 11–15, 16–22, 23–34, 35–44, 45–56, 57–67, and 68–76; yUIM‐1 had 1 TLS group consisting of residues 0–16]) and optimizing the same weights (X‐ray/stereochemistry and X‐ray/ADP) in Phenix.12 Interactions between UbV.v27.1 and yUIM‐1 were analyzed using the PyMOL Molecular Graphics System (Schrodinger, LLC, New York, NY) and the protein interfaces, surfaces and assemblies (PISA) tool.13 Figures were generated using the PyMOL Molecular Graphics System.
yUIM‐1 peptide synthesis for mutational analysis
yUIM‐1 peptide (sequence corresponds to residues 256–278 of Vps27) and yUIM‐1 peptides harboring substitutions were produced by solid‐phase peptide synthesis in‐house using 9‐fluorenylmethoxycarbonyl chemistry on Rink amide MBHA resin (Novabiochem) on a Prelude peptide synthesizer (Protein Technologies, Inc.). Peptides were deprotected in the cleavage cocktail trifluoroacetic acid, phenol, water, thioanisole, 1,2‐ethanedithiol (82.5%:5%:5%:5%:2.5% v/v) for 90 minutes at room temperature and then precipitated in t‐butyl methyl ether. Crude peptides were purified using C‐18 reverse phase HPLC (Waters) and authenticity was confirmed by mass spectrometry on an Orbitrap Elite (Thermo Fisher Scientific, Waltham, MA). C‐terminal labeling was achieved through Cysteine derivation using 5‐(Iodoacetamido) fluorescein (Sigma Aldrich). The peptide contained a non‐native N‐terminal tyrosine to aid concentration determination and a non‐native C‐terminal cysteine to allow covalent labeling with 5/6‐carboxyfluorescein succinimidyl ester (NHS‐FITC). Peptides were resuspended in 20 mM HEPES pH 7.0 and concentrations were determined from absorbance measurements at 495 nm using the extinction coefficient of FITC (75,000 L mol−1 cm−1).
Fluorescence polarization binding assay
Binding measurements were performed in FP buffer (25 mM HEPES pH 7.5, 100 mM NaCl, 1 mM DTT, 0.1 mg/mL BSA, 0.03% BRIJ‐35) by mixing, in a 384‐well plate, 25 nM FITC‐labeled peptides with the sequence of yUIM‐1, or yUIM‐1 with the indicated substitutions, with serial dilutions of UbV.v.27.1 ranging from 0.28 to 50 μM. Samples were equilibrated at room temperature for 30 minutes before reading plates on a HTS Multi‐Mode Microplate Reader (Synergy Neo) using an excitation filter of 485 nm and an emission filter of 530 nm. Dissociation constants were determined using Prism (GraphPad Software Inc, San Diego, CA) using a log(inhibitor) versus response model.
Accession number
The updated coordinate file (PDB:6NJG) for PDB:5UCL has been deposited to the PDB for release upon publication.
Supporting information
Supplementary Figure 1 Comparison of Thr9 delete, β1‐β2 loop and β1‐swap models of UbV.v27.1. The panels depict the respective Fo‐Fc electron density maps contoured at 2.4 σ for the indicated models shown in stereo view. Models were refined in the same manner using identical data cutoffs.
Supplementary Figure 2 Size exclusion analysis of molecular weight markers. Elution profile of molecular weight marker proteins are highlighted.
Supplementary Table 1 Data collection and refinement statistics for the Thr9 delete, β1‐ β2 loop, and β1‐swap models of the UbV.v27.1‐yUIM‐1 crystal structure.
Acknowledgments
This work was supported by Canadian Institutes of Health Research (CIHR) grants FDN 143277 to F.S. and MOP‐136956 to S.S. The authors declare no competing interests.
Contributor Information
Sachdev S. Sidhu, Email: sachdev.sidhu@utoronto.ca
Frank Sicheri, Email: sicheri@lunenfeld.ca.
References
- 1. Manczyk N, Yates BP, Veggiani G, Ernst A, Sicheri F, Sidhu SS (2017) Structural and functional characterization of a ubiquitin variant engineered for tight and specific binding to an alpha‐helical ubiquitin interacting motif. Protein Sci 26:1060–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Swanson KA, Kang RS, Stamenova SD, Hicke L, Radhakrishnan I (2003) Solution structure of vps27 uim‐ubiquitin complex important for endosomal sorting and receptor downregulation. EMBO J 22:4597–4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Boggon TJ, Murray J, Chappuis‐Flament S, Wong E, Gumbiner BM, Shapiro L (2002) C‐cadherin ectodomain structure and implications for cell adhesion mechanisms. Science 296:1308–1313. [DOI] [PubMed] [Google Scholar]
- 4. Mol CD, Harris JM, McIntosh EM, Tainer JA (1996) Human dUTP pyrophosphatase: uracil recognition by a beta hairpin and active sites formed by three separate subunits. Structure 4:1077–1092. [DOI] [PubMed] [Google Scholar]
- 5. Gabrielsen M, Buetow L, Nakasone MA, Ahmed SF, Sibbet GJ, Smith BO, Zhang W, Sidhu SS, Huang DT (2017) A general strategy for discovery of inhibitors and activators of ring and u‐box e3 ligases with ubiquitin variants. Mol Cell 68:456–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Teyra JSA, Schmitges FW, Jaynes P, Kit Leng Lui S, Polyak MJ, Fodil N, Krieger JR, Tong J, Moffat J, Sicheri F, Moran MF, Gros P, Eichhorn PJA, Lenter M, Boehmelt G, Sidhu SS (2019) Structural and functional characterization of ubiquitin variant inhibitors of usp15. Structure . 10.1016/j.str.2019.01.002. [DOI] [PubMed] [Google Scholar]
- 7. Lowman HB, Bass SH, Simpson N, Wells JA (1991) Selecting high‐affinity binding proteins by monovalent phage display. Biochemistry 30:10832–10838. [DOI] [PubMed] [Google Scholar]
- 8. Levin AM, Weiss GA (2006) Optimizing the affinity and specificity of proteins with molecular display. Mol Biosyst 2:49–57. [DOI] [PubMed] [Google Scholar]
- 9. Ernst A, Avvakumov G, Tong J, Fan Y, Zhao Y, Alberts P, Persaud A, Walker JR, Neculai AM, Neculai D, Vorobyov A, Garg P, Beatty L, Chan P‐K, Juang Y‐C, Landry M‐C, Yeh C, Zeqiraj E, Karamboulas K, Allali‐Hassani A, Vedadi M, Tyers M, Moffat J, Sicheri F, Pelletier L, Durocher D, Raught B, Rotin D, Yang J, Moran MF, Dhe‐Paganon S, Sidhu SS (2013) A strategy for modulation of enzymes in the ubiquitin system. Science 339:590–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Emsley P, Cowtan K (2004) Coot: model‐building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 11. Painter J, Merritt EA (2006) Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D Biol Crystallogr 62:439–450. [DOI] [PubMed] [Google Scholar]
- 12. Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse‐Kunstleve RW, McCoy AJ, Moriarity NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH (2010) PHENIX: a comprehensive python‐based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Krissinel E, Henrick K (2007) Inference of macromolecular assemblies from crystalline state. J Mol Biol 372:774–797. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1 Comparison of Thr9 delete, β1‐β2 loop and β1‐swap models of UbV.v27.1. The panels depict the respective Fo‐Fc electron density maps contoured at 2.4 σ for the indicated models shown in stereo view. Models were refined in the same manner using identical data cutoffs.
Supplementary Figure 2 Size exclusion analysis of molecular weight markers. Elution profile of molecular weight marker proteins are highlighted.
Supplementary Table 1 Data collection and refinement statistics for the Thr9 delete, β1‐ β2 loop, and β1‐swap models of the UbV.v27.1‐yUIM‐1 crystal structure.


