Abstract
Many peptide chemistry scientists have been reporting extremely interesting work on the basis of chemical peptides for which the only characterization was their purity, mass, and biological activity. It seems slightly overenthusiastic, as many of these structures should be thoroughly characterized first to demonstrate the uniqueness of the structure, as opposed to the uniqueness of the sequence. Among the peptides of identical sequences in the final chemical preparation, what amount of well‐folded peptide supports the measured activity? The activity of a peptide preparation cannot prove the purity of the desired peptide. Therefore, greater care should be taken in characterizing peptides, particularly those coming from chemical synthesis. At a time when the pharmaceutical industry is changing its paradigm by moving substantially from small molecules to biologics to better serve patients' needs, it is important to understand the limitations of the descriptions of these products and to start to apply the same “good laboratory practices” to our peptide research. Here, we attempt to delineate how synthetic peptides are described and characterized and what will be needed to describe them in regards to how they are well‐folded and homogeneous in their tertiary structure. Older studies were done when the tools were not yet discovered, but more recent publications are still lacking proper descriptions of these peptides. Modern tools of analysis are capable of segregating folded and unfolded peptides, even if the preparation is biologically active.
Keywords: peptides, solid‐phase synthesis, structure, characterization
Introduction: Peptides? Which Peptides?
In the newest modern approach to pharmacology and therapeutics, a growing space is reserved for biologics. Sharfstein1 stated that one can separate biologics into several categories: proteins, cells, peptides, nucleic acids, carbohydrates, and viruses. This is a change of paradigm compared to small molecules, which used to form ~95% of the pharmacopeia for the last few decades. Without stating whether this trend is temporary or progress, these new molecular entities have changed the landscape of some areas of the pharmacology world, including structural biology, industrial chemistry, governmental agency recommendations, and molecular research. Peptides also form a family of tools used to better understand physiopathological processes, even before they are turned into potential drugs. Such examples are numerous and frequent in the literature. Peptides have also been seen as a family of chemicals that can fill the gap between small molecules and antibodies, with special mention of their theoretical capacities to drug the undruggable regions, such as the protein–protein interfaces, a surface that is often too large for small molecules, with scarce hot spots, too distant from one another to be covered by a single chemical molecule. Biochemists also put forward a very simple idea: proteins are made of bricks (amino acids) more or less modified in living organisms (by post‐translational modifications). Most biological research, including therapeutic research, attempts to understand how these proteins function. For such a task, we use compounds that fall into two categories: peptides and nonpeptides. The former are formed of the same bricks as proteins, the latter are not. Which category is best suited to complement the architecture of proteins—the ones made of the same bricks or those made mostly of flat aromatic moieties? Our main concern, here, is proteins and peptides, particularly those obtained chemically. Not only can they be made of the 20 natural amino acids, but they can also benefit from the integration of several hundreds of exotic (i.e., nonproteinogenic) amino acids with the availability of protected amino acids of increasing purity and increasing diversity (see examples of such diversity in Xiao and Schultz2 or in Gates et al.3). The current status of peptides as drugs in therapeutics has been covered by interesting reviews, such as those from Henninot et al.,4 Erak et al.,5 and Kaspar and Reichert.6 Peptides containing ~50 amino acid residues form an important family of biologics, with 70 peptide therapeutics on the market in 2016. Over the past few years, they have been accepted by the Food and Drug Administration (FDA) with a frequency of 2–3 per year. The number of new preclinical studies on peptides has grown to more than 500. Interestingly, the FDA made a cutoff of 100 amino acids for peptide/protein.
As the technologies have progressed over the years, the lengths of the peptides entering clinical development have grown, though moderately for those beyond 50 amino acids (Fig. 1)7 up to a point where it is probable that in the next decade or so, chemically obtained proteins will be generated at both the research and therapeutic levels.
Figure 1.
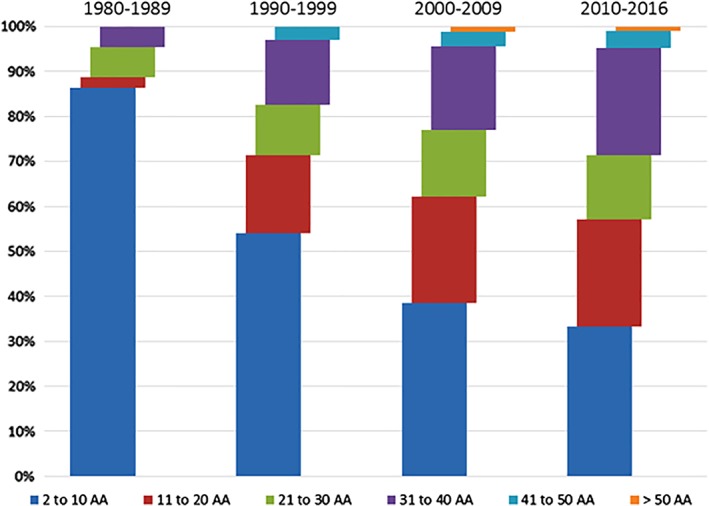
Length of peptides entering clinical development. Adapted from Lau and Dunn.7
In the present review, we chose to survey the nature of the characterization of the synthetic peptides as opposed to recombinant ones. We rather limited ourselves to synthetic peptides, essentially because (i) most of the peptides in the Pharmacopeia are from chemical origins; (ii) research peptides are often isolated from living sources (insect skin, various poisons, organ homogenates), purified, sequenced, and then synthesized; and (iii) their analogs are mostly obtained through chemical synthesis. Nevertheless, the existence of many problems linked to the nature and characterization of peptides used in research, whether from chemical or recombinant origins, must be addressed because the often limited quality of these peptides contributes to the limited reproducibility of biological results.
Bad Reputation
Less than 10 years after the revolution of solid‐phase synthesis by Merrifield,8 it was often thought that protein chemistry was in its decline.9 Peptides have a bad reputation of being expensive to produce, not following Lipinski rules, and not easily crossing membranes, and that even if they do, they are fairly unstable in the human body. This reputation has blocked progress, including in research areas in which the peptide chemistry has often been considered the Cinderella of medicinal chemistry, if not in academia, then at least in industry. However, 64 therapeutic peptides are available in the pharmacopeia in the USA and/or Europe and more than 150 are in active development, even when excluding insulin from this catalog. They have generated more than 10 billion dollars in market value. By being “easier” to produce, and with longer structures becoming available to research laboratories, we were interested in outlining some of the limitations associated with the descriptions of such “long” peptides. In contrast to common beliefs, the in‐depth characterization of compounds remains a high standard for good science and research. Peptides that are synthesized, purified by reversed phase high pressure liquid chromatography (RP‐HPLC), and characterized by mass spectrometry (MS) are seen frequently. On this basis, the peptide can be determined to be 95% pure. Furthermore, a simple biological test in which the peptide expectedly exhibits a given activity leads the reader to assume that once purified, peptides presenting the right mass in a mass spectrum are pure and homogeneous, despite indirect evidence that some S‐S bridges are not formed spontaneously and/or wrongly assembled.
Thus, peptides have a poor reputation among most medicinal chemists. Among the liabilities attributed to this modality, most can be addressed by the skills of peptide chemists, as demonstrated by the steady flow of approved peptide therapeutics.7 Here are some counterexamples:
“Peptides will ultimately be replaced by small molecules discovered by high throughput screening”: Following the discovery of the first nonpeptide antagonist of cholecystokinin receptors derived from a naturally occurring benzodiazepine in 1985,10 random screening of small molecule libraries has been predicted to provide direct access to both antagonists and agonists for any peptide receptor. Although it has been shown to be true mainly for antagonists of Class A receptors with great success, such as angiotensin 2 receptor antagonists, peptides still dominate the field of gonadotropin‐releasing hormone (GnRH) modulators. In addition, peptide analogs dominate the Class B family of G protein‐coupled receptor therapeutic agonists. Notoriously, the incretin family with GLP‐1 and GLP‐2 agonists has still not seen a small molecule competitor on the market. Furthermore, several attempts to find small molecule MCHR1 antagonists have also failed because most of these small molecules that were extremely powerful at the receptor were hErg‐positive, and this association characteristic was very difficult to read (see discussion in Johansson and Löfberg11).
“Peptides are not orally available”: desmopressin, a synthetic analog of the naturally occurring vasopressin, has been developed as an orally disintegrating tablet, although oral availability is very low. Since its initial approval in Finland in 2005, desmopressin has been approved in more than 80 countries. More recently, Novo‐Nordisk has reported several positive clinical trials of its oral Semaglutide®, a once‐daily lipopeptide GLP‐1 analog.12, 13
“Peptides are not metabolically stable and are too short lived”: Semaglutide® is currently approved by both the EMA and FDA as a once‐weekly subcutaneous injection for the treatment of patients with type 2 diabetes.14 This astonishing half‐life extension is attributable to protection against proteolysis and binding of its lipid moiety to serum albumin. Another example is Degarelix®, a selective GnRH receptor antagonist. After subcutaneous injection, it forms a gel from which the drug is released over a period of 1 month, achieving testosterone levels corresponding to medical castration in 97–98% of patients.15 Another example is Lanreotide, a somatostatin analog initially formulated as a PLGA nanoparticle formulation (Somatuline LA) that provided 10 days of coverage. The observation that at high concentration of peptide became an amyloid semisolid led to the approval of Somatuline autogel, which provides 42–56 days of coverage following subcutaneous injection.16
“Peptides require multistep synthesis and cannot be produced cost‐effectively on a large scale.” This led to the assumption that only highly active peptides, active at microgram doses, can be cost‐effective. For example, teriparatide, a 34‐amino‐acid peptide approved in 2002 for the treatment of osteoporosis, was produced by recombinant technology. Dosed at 20 μg once a day, it required only 7.3 mg per patient per year. The next year, the 34‐amino‐acid HIV fusion inhibitor enfuvurtide was approved. It requires 90‐mg injections twice a day, corresponding to 66 g of peptide per patient per year. Major improvements in both solid‐phase synthesis and RP‐HPLC purification allow large batches to be made and make the active pharmaceutical ingredient (API) available at an affordable cost.17
Thus, more and more studies are being published to describe longer and longer peptides. Because FDA approvals go together with up‐to‐date technical characterizations of a drug candidate, particularly biologics, more and more techniques have been developed to characterize these chemical entities. More and more evidence strongly suggests that Malcolm's statement9 was far from what really happened over the last few years and will happen in the coming decades.
Thoughts of the Agencies
Interestingly, a chemically synthesized polypeptide is not defined as a “biological product” and will be regulated as a drug, regardless of its length. Due to continuous improvements in synthetic methods and analytical tools, chemical synthesis appears to be a viable approach for manufacturing generic peptide drugs. This led the FDA to clarify its definition of biological products in the Biologics Price Competition and Innovation Act of 2009 (https://www.fda.gov/downloads/drugs/guidances/ucm444661.pdf). In particular, the definition of a “biological product” was amended to include “a protein (except any chemically synthesized polypeptide).” In the absence of scientific consensus, the FDA decided to base the statuary distinction on size only. According to these regulatory definitions, a “protein” means any alpha amino acid polymer with a specifically defined sequence >40 amino acids in size (the total number of amino acids is not limited to the number of amino acids in a contiguous sequence); thus, peptides <40 amino acids in size are excluded independent of their mode of production (synthetic or recombinant). This definition excludes insulin but includes peptides such as glucagon, liraglutide, nesiritide, teriparatide, and teduglutide (of rDNA origin). Current peptide synthesis technologies are a viable alternative to producing generic copies of these drugs that, due to the availability of orthogonal techniques, can be characterized extensively (https://www.fda.gov/Drugs/ScienceResearch/ucm578111.htm). The FDA now considers it possible to demonstrate that the active ingredient in a proposed generic synthetic peptide is the same as the reference active ingredient of rDNA origin, allowing the submission of an abbreviated new drug application. A “chemically synthesized polypeptide,” according to the FDA definition, is not a “biological product” and will be regulated as a drug under the Federal Food, Drug, and Cosmetic (FD&C) Act. Although several definitions exist, the FDA interprets the statutory exclusion for “chemically synthesized polypeptide” to mean any molecule that is made entirely by chemical synthesis and composed of up to 99 amino acids. Such molecules will be regulated as drugs under the FD&C Act. A “chemically synthesized polypeptide” composed of more than 99 amino acids, according to these definitions, will be considered a biological product. Thus, the FDA is prepared to examine polypeptides produced by chemical synthesis that contain between 41 and 99 amino acids as drugs and not biologics. This acknowledges the current and future achievements of polypeptide chemical synthesis and characterization.
Peptides? Which Structure, Which Refolding?
Roughly half a decade ago, we decided to enter a series of programs aimed at synthesizing a series of proteins of growing length from ubiquitin18 and calstabin19 to quinone reductase (which contains 226 amino acids and with which we failed). The difficulties encountered in developing a universal methodology to obtain such proteins, most of which were enzymes, led us to limit ourselves to ~200 amino acids. The most advanced program concerned calstabin, a 120 amino‐acid proline isomerase. We successfully obtained it by solid‐phase synthesis after struggling with multiple steps of native chemical ligation solutions. In brief, this native chemical ligation technique20 involves the chemoselective conjugation between a pair of unprotected peptide fragments, one functionalized as a C‐terminal thioester and the other with an N‐terminal cysteine (Cys) residue leading to a conjugate dipeptide in a single step after several steps, including a rearrangement, leading to the recovery of the initial cysteine side chain. Although the original description was limited to cystein‐containing peptides, it permitted the development of a series of techniques that are less limited to this point of view (see Conibear et al. for a complete review21). The process that we used for these chemical approaches was the standard process used for shorter peptides over the last decade.22 We treated the large, unfolded peptide like a regular peptide: precipitation, lyophilization, purification by RP‐HPLC, and MS analysis. We obtained a unique liquid chromatography (LC) peak, strictly symmetrical, strongly suggesting that the peptide was pure and ready to enter the refolding process. We realized that nothing can be farther from the notion of purity than such a “simple” observation of the RP‐HPLC profile and/or mass spectrogram. The refolding process, used on the basis of several years of refolding recombinant proteins expressed in insoluble fractions of bacteria, led to a mere 50% of the total protein being correctly refolded. Separation onto a gel filtration column led to two pools: one active and the other not.19 These two fractions could not be distinguished from one another using these analytical approaches. A closer analysis with circular dichroism (CD) spectrometry showed a slight difference between both, whereas MS, as expected, gave a single mass corresponding to the desired product. Furthermore, modification of the CD spectrum of the mixture due to the presence of the unfolded portion was not significantly altered compared to the pure, recombinant sample.19
After many years in the peptide area, these results were unexpected, as most reports indicate minimal analytical characterization of the samples. For long peptides, generally >40 amino acids, the main characterization steps performed these last few years are MS and CD. Table 1 provides some of the reports on “long” peptides randomly picked over the last three decades as examples of the way such synthetic peptides were characterized over the years. Furthermore, in no way this table could be considered as exhaustive. From the research standpoint, though, the situation is less constrained than from the therapeutic one. A close examination of Table 1 shows that peptides followed the historical changes in technical skill, with more and more characterizations in recently published papers. Nevertheless, due to the lack of obligations and rules, it is not rare to find papers describing peptides with minimal characterization of the peptide (obtained chemically) when it is RP‐HPLC/MS pure and the expected biological activity is at the rendezvous because the peptide is pure and forms a homogeneous entity. For example, the folding of the synthetic Anaphylatoxin C3a was demonstrated because the peptide has the same activities than the commercial (recombinant) one on two biological tests.52 None of those methods deliver any information on the peptide conformation (3D structure). However, in contrast to recombinant expression, in peptide synthesis, no chaperone is present to assist in the refolding of the nascent peptide chain. Thus, refolding is either spontaneous (e.g., ubiquitin18) or a long and partial process in which the end product must be thoroughly characterized. As a particularly interesting example, the recent paper by Kuroha et al.69 on a new lasso peptide essentially aimed at describing the structure of the material. Such papers are not easy to publish, essentially because they do not “tell a story,” but they are important to notice, as they pave the road to a standard procedure leading to accurately described pure peptides. Of course, other complete characterizations of synthesized peptides have been reported (e.g., Wei et al.44 for the HNP1 peptide analogs or Henriques et al.43 for LEAP2). In these two instances, the peptides were even crystallized.
Table 1.
Examples of chemically obtained peptides and their characterization(s)
| Year | Peptide/protein | Reference | Number of amino acids | Biophysical characterizationa | Biological assay/activity |
|---|---|---|---|---|---|
| 1983 | hPTH | 23 | 84 | LC | Functional binding |
| 1988 | TGF alpha | 24 | ~15 fragments | LC/tlc | Cell growth |
| 1992 | Elafin | 25 | 57 | LC | Enzyme inhibition |
| 1992 | NPY | 26 | 36 | LC | Binding |
| 1992 | Calciseptine | 27 | 60 | LC | Channel blocking |
| 1994 | IL8 | 20 | 72 | LC/MS | NRb |
| 1994 | Ubiquitin | 28 | 76 | Crystallization | NR |
| 1994 | Ubiquitin | 29 | 76 | MS | NR |
| 1996 | Midkine | 30 | 121 | LC | Neurite extension |
| 1997 | Secretoneurin | 31 | 33 | NMR, CD | NR |
| 1998 | TSR and EGF1 modules | 32 | 29 and 41 | LC/MS, CD | NR |
| 1998 | GFP precursor | 33 | 238 | Fluorescencec | |
| 1999 | Peptide E | 34 | 25 | CD | Anesthesia |
| 2001 | Octadecaneuropeptide | 35 | 18 | LC/MS, NMR | Calcium |
| 2002 | Transthyretin | 36 | 127 | LC/MS, NMR | NR |
| 2005 | 26Rfa | 37 | 26 | CD, NMR | NR |
| 2006 | Seleno‐glutaredoxin 3 | 38 | 82 | LC | Activation of reduction rate |
| 2008 | CGRP | 39 | 37 | LC/MS | Binding |
| 2009 | Kisspeptin‐52 | 40 | 52 | LC/MS, NMR | Calcium |
| 2010 | Polytheonamide B | 41 | 48 | LC, NMR | Cellular toxicity |
| 2010 | Insulin | 42 | 51 | LC/MS, NMR | Binding |
| 2010 | LEAP‐2 | 43 | 40 | NMR | Antimicrobial among others |
| 2010 | HNP1 | 44 | 30 | LC/MS, NMR, crystallization | Antimicrobial |
| 2011 | Tetraubiquitin | 45 | 304 | LC/MS | Enzymatic degradation |
| 2012 | EPO | 46 | 166 | LC/MS, CD | Colony formation |
| 2012 | D‐VEGF | 47 | 101 | LC/MS, Crystallization | Binding |
| 2012 | Cyclotide 2v | 48 | 42 | LC/MS | NR |
| 2013 | EPO | 49 | 166 | CD | Colony formation |
| 2013 | α‐Scorpio toxin OD1 | 50 | 65 | Crystallization | Na+ Current |
| 2013 | PYP | 51 | 134 | LC/MS, absorbance | Fluorescence |
| 2013 | Anaphylatoxin C3a | 52 | 77 | LC/MS | Binding |
| 2013 | HGF | 53 | 127 | LC/MS, CD | Binding |
| 2013 | Dengue capsid Prot C | 54 | 80 | LC/MS, CD | Dimerization |
| 2013 | INSL3 | 55 | 46 | LC/MS, CD | Binding |
| 2014 | Hepcidin | 56 | 25 | NMR (Supp. Inf.) | Ferroportin degradation |
| 2014 | GM‐CSF | 57 | 127 | LC/MS, CD | Cell proliferation |
| 2014 | Lucifensin | 58 | 40 | LC/MS, CD | Antimicrobial |
| 2014 | Influenza virus M2d | 59 | 97 | CD | Single channel current |
| 2014 | SUMO | 60 | 91 | LC/MS | Used as substrate of SUMO E1 |
| 2014 | SUMO | 61 | 96 | LC/MSe | NR |
| 2015 | G‐CSF | 62 | 174 | SDS‐PAGE, MS | Cell proliferation |
| 2015 | Caenopore‐5 | 63 | 82 | LC/MS, 1D 1H NMR, CD | Permeabilization |
| 2015 | Histone 2B | 64 | 125 | LC/MS, CD | NR |
| 2016 | HGF | 65 | 85 | LC/MS | NR |
| 2016 | d‐ASFV pol Xf | 66 | 174 | LC/MS, CD | d‐Polymerase activity |
| 2016 | NK1 | 65 | 180 | LC/MS | NR |
| 2016 | AS48 | 67 | 70 | LC/MS, CD | Antimicrobial |
| 2016 | K27 Ubiquitin | 68 | 151 | LC/MS, CD, crystallography | Biochemistry |
| 2017 | Subterisin | 69 | 16 | NMR, MS/MS | NR |
| 2018 | CIGB‐330 | 70 | 25 | LC/MS | Cell proliferation |
| 2018 | GsMTx4 | 71 | 34 | LC/MS, CD, crystallography | NR |
It should be reminded that LC and LC/MS are not structural techniques. The information issued from such experiments is only used to purify the material. NMR, CD, and crystallization could be considered as structural indicating techniques.
NR means not reported in this particular reference.
In this context, fluorescence is both a biophysical characterization and a biological activity (fluorescent protein).
This is an integral membrane protein.
Only the chemical conjugate between this protein and a peptide (RASIKAEGR) was analyzed for its structure.
d‐ASFV pol X stands for all D‐aminoacid African swine fever virus polymerase X.
Peptide Characterizations: What Is at Our Disposal?
The main point of peptide synthesis, especially since the use of robots became common, as well as the use of capping at every cycle of the step‐by‐step synthesis of such compounds, is that the sequence is fine, corresponding to a list of iterative steps entered into the robot by the operator and easily independently checked. In other words, the sequence entered should correspond to the desired sequence, with a minimal risk of mistake. Interestingly, the solid‐phase synthesis of peptides has covered a number of coupling sequences between almost any natural amino acids, which is still rare to find in short sequences (e.g., ~20 amino acid length), with massive failure in the succession of these coupling sequences, though we showed that the rates of coupling may be extremely different from one amino acid to another.72 Nevertheless, these observations concerned few, if any, exotic amino acids. We observed that coupling rates between natural and exotic or between exotic amino acids could be extremely slow. The possible incorporation of nonencoded amino acids into a pseudo‐peptide sequence remains one of the vastest possibilities of peptides, with no limit in terms of the nature of exotic amino acids that can be included. In contrast, using recombinant techniques, the incorporation of amino acids of various structures is possible, but limited to two or three different amino acids per sequence due to the limitations of genetic code manipulations.73, 74 Thus, for the analysis of the newly formed peptides, the routine initial step is purification via chromatography, up to a point in which the main peak in RP‐HPLC is purified and usually corresponds to the desired product. MS added the qualitative notion that the overall mass of the purified product corresponds to the theoretical mass, as calculated by the succession of amino acids in the sequence. The next two steps are verification of the peptide sequence and assessing its conformational homogeneity. The landscape for full amino acid sequence peptides dramatically evolved during the last decade with two well‐established techniques, both based on MS: fully automated amino acid sequence verification with LC–MS/MS of digest developed in the context of proteomic analyses, and conformational/folding information by ion mobility MS (IM‐MS) developed in the field of native spectrometry.75
The input of proteomic analysis methods
The development of proteomic analysis provided a very powerful tool that allows the rapid and nonambiguous full sequence verification of a given peptide or protein. The LC–MS/MS analysis performed on a series of digestion products using different enzymes allows determination of not only the amino acid sequence, but also the presence of exotic amino acids. The power of this method is well illustrated by its routine use in sequence verification of monoclonal antibodies containing ~1500 amino acids and establishing the heterogeneity of N‐glycosylations for the evaluation of the consistency of batch‐to‐batch production. A good example is the quantitative mass spectrometry multi‐attribute method (MAM)76 widely used by many manufacturers for the full characterization of monoclonal antibodies. A single amino acid substitution can be detected.77 Thus, the tools assessed for the recombinant approach, particularly medicinal antibodies, can be applied to the much simpler situation of synthetic peptides, even if their sizes are in the 100 amino acid range. Amino acid sequence determination based on proteomic analysis78 can easily be adapted to synthetic peptides <100 amino acids and provides in depth characterization when used in conjunction with already well‐established techniques, such as chromatography with absorbance or mass spectroscopy detection. In addition, they yield information on both the purity of the desired peptide and the possible impurities present in the preparation. This is well illustrated in the case of identifying and quantifying hundreds of contaminating host cell proteins present at ppm in recombinant proteins.79 However, the most basic tool is gel filtration because it can be used in a preparative mode. Because of the potential unfolded nature of some of the molecules underneath the purified product, the behavior of these species will be different in a simple gel filtration analysis, as we demonstrated previously with our synthetic version of the protein calstabine.19
After sequence verification, detection, quantification, and identification of possible impurities, one cannot claim that the peptide is active in whatever biological assay, because one does not know unequivocally the quantity of peptide that is folded appropriately. Several conformations are possibly formed for a given sequence, together with the completely, nonfolded linear peptide, and the differences between these cannot be appreciated by either RP‐HPLC or basic MS. Various tools exist for such structural characterization. The most common ones involve NMR, with the major limitation that these experiments require a large amount of material. Nevertheless, observation of the shapes of the peaks in the spectrum of a given peptide will tell if the compound is unique, regardless of whether it is well‐folded, or if the preparation is composed of several entities more or less folded. Despite providing a unique method for assessing the identity of a peptide, it is questionable whether it will be able to detect small amounts of micro‐heterogenic impurities.
Another possibility is the use of CD. Again, the main obstacle for the general use of this technique will be the need of a control peptide, such as a known, well‐folded peptide of the same sequence. Studying the CD spectra will reveal the various forms the peptide has adopted (e.g., barrels, coils, etc.). This technique also requires quite a large amount of product. In addition to these well‐established methods, IM‐MS can provide crucial information.
Conformation information by IM‐MS
This method provides a measurement of the collision cross section (CCS) of a molecule in the gas phase. This CCS is obviously linked to the size and shape of the molecular ions in the gas phase. If experimental parameters are selected carefully, the gas phase CCS can provide reliable information on the conformation in the liquid phase. For example, IM‐MS allowed the characterization of peptide−synthetic polymer conjugates,80 the study of conformer preferences for hydrophobic antimicrobial peptides,81 the identification of lasso peptide topologies,82, 83 and the separation of d‐amino‐acid‐containing peptides.84 IM‐MS can even be coupled with 1D or 2D LC as shown for the characterization of antibody–drug conjugates,85 in which initial separation by size is achieved by hydrophobic interaction chromatography.
Crystallization
There is a common belief that peptides are not easy to crystalize, though crystallization is the ultimate way to acquire information on their 3D structures, as opposed to the more cumbersome ways of gaining information by NMR. We provide a series of examples of such reports mostly extracted from the Protein Data Base (www.rcsb.org) in Table 2, ranging in length from 20 to 106 amino acids. This approach does not solve our problem (segregation between unfolded and folded peptides) because only the final product (in the crystal) will be present, by definition, and will show one or the other of possible forms that “accepted” to crystalize. Furthermore, crystallization is itself a method of purification.
Table 2.
Examples of crystallographic data on peptides (extracted from the Protein Data Bank)
| PDB ID | Name of the peptide (if any) | Date | MW | Residue count | Primary citation author | Reference |
|---|---|---|---|---|---|---|
| 1COI | COIL‐VALD | 1996 | 3302.8 | 31 | Ogihara et al. | 86 |
| 1ALG | P11 | 1997 | 2442.84 | 24 | Nordhoff et al. | 87 |
| 1ZDC | Stable miniprotein A domain, Z34C | 1997 | 4188.71 | 35 | Starovasnik et al. | 88 |
| 2A3D | Protein (de novo 3‐helix bundle) | 1999 | 8120.21 | 73 | Walsh et al. | 89 |
| 1E0M | WW‐prototype | 2000 | 4358.77 | 37 | Macias et al. | 90 |
| 2JWU | Gb88 | 2007 | 6457.42 | 56 | He et al. | 91 |
| 2KIR | Designer toxin | 2009 | 3890.8 | 34 | Takacs et al. | 92 |
| 2FD7 | [V15]crambin | 2009 | 4707.0 | 46 | Bang et al. | 93 |
| 3OVJ | KLVFFA hexapeptide segment from amyloid beta | 2010 | 3716.48 | 24 | Landau et al. | 94 |
| 2L96 | LAK160‐P7 | 2011 | 2666.54 | 24 | Vermeer et al. | 95 |
| 4H8M | CC‐Hex‐H24‐A5/7C | 2012 | 6648.03 | 64 | Zaccai et al. | 96 |
| 2LR2 | Immunoglobulin G‐binding protein A | 2012 | 9875.64 | 88 | Barb et al. | 97 |
| 2MT8 | MTAbl13 of grafted MCoTI‐II | 2014 | 4148.76 | 39 | Huang et al. | 98 |
| 2MSQ | Conotoxin cBru9a | 2014 | 2809.19 | 27 | Akcan et al. | 99 |
| 4P6K | Computationally designed transporter of Zn(II) and proton | 2014 | 3016.33 | 26 | Joh et al. | 100 |
| 4D5M | Triptorelin | 2014 | 5534.81 | 44 | Valéry et al. | 101 |
| 4OWI | p53LZ2 | 2014 | 7861.36 | 66 | Lee et al. | 102 |
| 5ET3 | Fullerene organizing protein (C60Sol‐COP‐3) | 2015 | 7097.77 | 60 | Kim et al. | 103 |
| – | [V15 A]crambin | 2015 | 4708.5 | 46 | Tang et al. | 104 |
| 5KWX | Designed peptide NC_EEH_D1 | 2016 | 58989.76 | 25 | Bhardwaj et al. | 105 |
| 2NAU | Entity | 2016 | 3283.89 | 28 | Datta et al. | 106 |
Conclusions
A trend has emerged in government agencies recommending that the regulation of market‐oriented peptides (and proteins, including antibodies) be reinforced, with more validation of sequences and minute details of structures. Although these recommendations are peripheral to research, one must realize that many peptides synthesized/discovered these last few years have been poorly described from the point of view of their structure.
We stress that, at the research level, the characterization of long peptides should be enforced imperatively in order not to try/test preparations in which only an unknown portion of the peptide is properly folded and thus active. It is clear for us that the introduction of proteomic methodologies and IM‐MS offers a rather easy way to produce batches of peptides with much better characterization than in the last decade.
Statement: Synthetic peptides of 20–100 amino acids are too often uncharacterized in regard to their folding and three‐dimensional structure, whereas their purity and ad hoc sequence are reported. This may lead to underestimating the peptide's biological activity. New tools, particularly from mass spectrometry, should be used to validate the structures of these peptides.
References
- 1. Sharfstein ST (2018) Non‐protein biologic therapeutics. Curr Opin Biotechnol 53:65–75. [DOI] [PubMed] [Google Scholar]
- 2. Xiao H, Schultz PG (2016) At the interface of chemical and biological synthesis: an expanded genetic code. Cold Spring Harb Perspect Biol 8:a023945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gates ZP, Vinogradov AA, Quartararo AJ, Bandyopadhyay A, Choo Z‐N, Evans ED, Halloran KH, Mijalis AJ, Mong SK, Simon MD, Standley EA, Styduhar ED, Tasker SZ, Touti F, Weber JM, Wilson JL, Jamison TF, Pentelute BL (2018) Xenoprotein engineering via synthetic libraries. Proc Natl Acad Sci U S A 115:E5298–E5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Henninot A, Collins JC, Nuss JM (2018) The current state of peptide drug discovery: back to the future? J Med Chem 61:1382–1414. [DOI] [PubMed] [Google Scholar]
- 5. Erak M, Bellmann‐Sickert K, Els‐Heindl S, Beck‐Sickinger AG (2018) Peptide chemistry toolbox ‐ transforming natural peptides into peptide therapeutics. Bioorg Med Chem 26:2759–2765. [DOI] [PubMed] [Google Scholar]
- 6. Kaspar AA, Reichert JM (2013) Future directions for peptide therapeutics development. Drug Discov Today 18:807–817. [DOI] [PubMed] [Google Scholar]
- 7. Lau JL, Dunn MK (2018) Therapeutic peptides: historical perspectives, current development trends, and future directions. Bioorg Med Chem 26:2700–2707. [DOI] [PubMed] [Google Scholar]
- 8. Merrifield RB (1963) Solid phase peptide synthesis. I. The synthesis of a tetrapeptide. J Am Chem Soc 85:2149–2154. [Google Scholar]
- 9. Malcolm AD (1978) The decline and fall of protein chemistry? Nature 275:90–91. [DOI] [PubMed] [Google Scholar]
- 10. Chang RS, Lotti VJ, Monaghan RL, Birnbaum J, Stapley EO, Goetz MA, Albers‐Schönberg G, Patchett AA, Liesch JM, Hensens OD (1985) A potent nonpeptide cholecystokinin antagonist selective for peripheral tissues isolated from Aspergillus alliaceus . Science 230:177–179. [DOI] [PubMed] [Google Scholar]
- 11. Johansson A, Löfberg C (2015) Novel MCH1 receptor antagonists: a patent review. Expert Opin Ther Pat 25:193–207. [DOI] [PubMed] [Google Scholar]
- 12. Bain SC, Mosenzon O, Arechavaleta R, Bogdański P, Comlekci A, Consoli A, Deerochanawong C, Dungan K, Faingold MC, Farkouh ME, Franco DR, Gram J, Guja C, Joshi P, Malek R, Merino‐Torres JF, Nauck MA, Pedersen SD, Sheu WH‐H, Silver RJ, Tack CJ, Tandon N, Jeppesen OK, Strange M, Thomsen M, Husain M (2019) Cardiovascular safety of oral semaglutide in patients with type 2 diabetes: rationale, design and patient baseline characteristics for the PIONEER 6 trial. Diabetes Obes Metab 21:499–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Davies M, Pieber TR, Hartoft‐Nielsen M‐L, Hansen OKH, Jabbour S, Rosenstock J (2017) Effect of oral semaglutide compared with placebo and subcutaneous semaglutide on glycemic control in patients with type 2 diabetes: a randomized clinical trial. JAMA 318:1460–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dhillon S (2018) Semaglutide: first global approval. Drugs 78:275–284. [DOI] [PubMed] [Google Scholar]
- 15. Klotz L, Boccon‐Gibod L, Shore ND, Andreou C, Persson B‐E, Cantor P, Jensen J‐K, Olesen TK, Schröder FH (2008) The efficacy and safety of degarelix: a 12‐month, comparative, randomized, open‐label, parallel‐group phase III study in patients with prostate cancer. BJU Int 102:1531–1538. [DOI] [PubMed] [Google Scholar]
- 16. Cendros JM, Peraire C, Trocóniz IF, Obach R (2005) Pharmacokinetics and population pharmacodynamic analysis of lanreotide autogel. Metab Clin Exp 54:1276–1281. [DOI] [PubMed] [Google Scholar]
- 17. Bray BL (2003) Large‐scale manufacture of peptide therapeutics by chemical synthesis. Nat Rev Drug Discov 2:587–593. [DOI] [PubMed] [Google Scholar]
- 18. Bacchi M, Fould B, Jullian M, Kreiter A, Maurras A, Nosjean O, Coursindel T, Puget K, Ferry G, Boutin JA (2017) Screening ubiquitin specific protease activities using chemically synthesized ubiquitin and ubiquitinated peptides. Anal Biochem 519:57–70. [DOI] [PubMed] [Google Scholar]
- 19. Bacchi M, Jullian M, Sirigu S, Fould B, Huet T, Bruyand L, Antoine M, Vuillard L, Ronga L, Chavas LMG, Nosjean O, Ferry G, Puget K, Boutin JA (2016) Total chemical synthesis, refolding, and crystallographic structure of fully active immunophilin calstabin 2 (FKBP12.6). Protein Sci 25:2225–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dawson PE, Muir TW, Clark‐Lewis I, Kent SB (1994) Synthesis of proteins by native chemical ligation. Science 266:776–779. [DOI] [PubMed] [Google Scholar]
- 21. Conibear AC, Watson EE, Payne RJ, Becker CFW (2018) Native chemical ligation in protein synthesis and semi‐synthesis. Chem Soc Rev 47:9046–9068. [DOI] [PubMed] [Google Scholar]
- 22. Audinot V, Beauverger P, Lahaye C, Suply T, Rodriguez M, Ouvry C, Lamamy V, Imbert J, Rique H, Nahon JL, Galizzi JP, Canet E, Levens N, Fauchere JL, Boutin JA (2001) Structure‐activity relationship studies of melanin‐concentrating hormone (MCH)‐related peptide ligands at SLC‐1, the human MCH receptor. J Biol Chem 276:13554–13562. [DOI] [PubMed] [Google Scholar]
- 23. Kimura T, Takai M, Yoshizawa K, Sakakibara S (1983) Solution synthesis of [ASN76]‐human parathyroid hormone (1‐84). Biochem Biophys Res Commun 114:493–499. [DOI] [PubMed] [Google Scholar]
- 24. Darlak K, Franklin G, Woost P, Sonnenfeld E, Twardzik D, Spatola A, Schultz G (1988) Assessment of biological activity of synthetic fragments of transforming growth factor‐alpha. J Cell Biochem 36:341–352. [DOI] [PubMed] [Google Scholar]
- 25. Tsunemi M, Kato H, Nishiuchi Y, Kumagaye S, Sakakibara S (1992) Synthesis and structure‐activity relationships of elafin, an elastase‐specific inhibitor. Biochem Biophys Res Commun 185:967–973. [DOI] [PubMed] [Google Scholar]
- 26. Beck‐Sickinger AG, Durr H, Hoffmann E, Gaida W, Jung G (1992) Characterization of the binding site of neuropeptide Y to the rabbit kidney receptor using multiple peptide synthesis. Biochem Soc Trans 20:847–850. [DOI] [PubMed] [Google Scholar]
- 27. Kuroda H, Chen YN, Watanabe TX, Kimura T, Sakakibara S (1992) Solution synthesis of calciseptine, an L‐type specific calcium channel blocker. Pept Res 5:265–268. [PubMed] [Google Scholar]
- 28. Alexeev D, Bury SM, Turner MA, Ogunjobi OM, Muir TW, Ramage R, Sawyer L (1994) Synthetic, structural and biological studies of the ubiquitin system: chemically synthesized and native ubiquitin fold into identical three‐dimensional structures. Biochem J 299:159–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ramage R, Green J, Muir TW, Ogunjobi OM, Love S, Shaw K (1994) Synthetic, structural and biological studies of the ubiquitin system: the total chemical synthesis of ubiquitin. Biochem J 299:151–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Inui T, Bodi J, Kubo S, Nishio H, Kimura T, Kojima S, Maruta H, Muramatsu T, Sakakibara S (1996) Solution synthesis of human midkine, a novel heparin‐binding neurotrophic factor consisting of 121 amino acid residues with five disulphide bonds. J Pept Sci 2:28–39. [DOI] [PubMed] [Google Scholar]
- 31. Oulyadi H, Davoust D, Vaudry H (1997) A determination of the solution conformation of secretoneurin, a neuropeptide originating from the processing of secretogranin II, by 1H‐NMR and restrained molecular dynamics. Eur J Biochem 246:665–673. [DOI] [PubMed] [Google Scholar]
- 32. Hackeng TM, Dawson PE, Kent SB, Griffin JH (1998) Chemical synthesis of human protein S thrombin‐sensitive module and first epidermal growth factor module. Biopolymers 46:53–63. [DOI] [PubMed] [Google Scholar]
- 33. Nishiuchi Y, Inui T, Nishio H, Bodi J, Kimura T, Tsuji FI, Sakakibara S (1998) Chemical synthesis of the precursor molecule of the Aequorea green fluorescent protein, subsequent folding, and development of fluorescence. Proc Natl Acad Sci U S A 95:13549–13554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Condamine E, Leprince J, Suaudeau C, Mayer C, Davoust D, Costentin J, Vaudry H (1999) The proenkephalin A‐processing product peptide E, which encompasses two enkephalin sequences, has a much lower opioid activity than beta‐endorphin. Peptides 20:865–871. [DOI] [PubMed] [Google Scholar]
- 35. Leprince J, Oulyadi H, Vaudry D, Masmoudi O, Gandolfo P, Patte C, Costentin J, Fauchère JL, Davoust D, Vaudry H, Tonon MC (2001) Synthesis, conformational analysis and biological activity of cyclic analogs of the octadecaneuropeptide ODN. Design of a potent endozepine antagonist. Eur J Biochem 268:6045–6057. [DOI] [PubMed] [Google Scholar]
- 36. Wilce JA, Daly NL, Craik DJ (2002) Synthesis and structural analysis of the N‐terminal domain of the thyroid hormone‐binding protein transthyretin. Clin Chem Lab Med 40:1221–1228. [DOI] [PubMed] [Google Scholar]
- 37. Thuau R, Guilhaudis L, Ségalas‐Milazzo I, Chartrel N, Oulyadi H, Boivin S, Fournier A, Leprince J, Davoust D, Vaudry H (2005) Structural studies on 26RFa, a novel human RFamide‐related peptide with orexigenic activity. Peptides 26:779–789. [DOI] [PubMed] [Google Scholar]
- 38. Metanis N, Keinan E, Dawson PE (2006) Synthetic seleno‐glutaredoxin 3 analogues are highly reducing oxidoreductases with enhanced catalytic efficiency. J Am Chem Soc 128:16684–16691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Miranda LP, Holder JR, Shi L, Bennett B, Aral J, Gegg CV, Wright M, Walker K, Doellgast G, Rogers R, Li H, Valladares V, Salyers K, Johnson E, Wild K (2008) Identification of potent, selective, and metabolically stable peptide antagonists to the calcitonin gene‐related peptide (CGRP) receptor. J Med Chem 51:7889–7897. [DOI] [PubMed] [Google Scholar]
- 40. Gutiérrez‐Pascual E, Leprince J, Martínez‐Fuentes AJ, Ségalas‐Milazzo I, Pineda R, Roa J, Duran‐Prado M, Guilhaudis L, Desperrois E, Lebreton A, Pinilla L, Tonon M‐C, Malagón MM, Vaudry H, Tena‐Sempere M, Castaño JP (2009) In vivo and in vitro structure‐activity relationships and structural conformation of Kisspeptin‐10‐related peptides. Mol Pharmacol 76:58–67. [DOI] [PubMed] [Google Scholar]
- 41. Inoue M, Shinohara N, Tanabe S, Takahashi T, Okura K, Itoh H, Mizoguchi Y, Iida M, Lee N, Matsuoka S (2010) Total synthesis of the large non‐ribosomal peptide polytheonamide B. Nat Chem 2:280–285. [DOI] [PubMed] [Google Scholar]
- 42. Sohma Y, Hua Q‐X, Whittaker J, Weiss MA, Kent SBH (2010) Design and folding of GluA4(ObetaThrB30)insulin ("ester insulin"): a minimal proinsulin surrogate that can be chemically converted into human insulin. Angew Chem Int Ed Engl 49:5489–5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Henriques ST, Tan CC, Craik DJ, Clark RJ (2010) Structural and functional analysis of human liver‐expressed antimicrobial peptide 2. Chembiochem 11:2148–2157. [DOI] [PubMed] [Google Scholar]
- 44. Wei G, Pazgier M, de Leeuw E, Rajabi M, Li J, Zou G, Jung G, Yuan W, Lu W‐Y, Lehrer RI, Lu W (2010) Trp‐26 imparts functional versatility to human alpha‐defensin HNP1. J Biol Chem 285:16275–16285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kumar KS, Bavikar SN, Spasser L, Moyal T, Ohayon S, Brik A (2011) Total chemical synthesis of a 304 amino acid K48‐linked tetraubiquitin protein. Angew Chem Int Ed Engl 50:6137–6141. [DOI] [PubMed] [Google Scholar]
- 46. Wang P, Dong S, Brailsford JA, Iyer K, Townsend SD, Zhang Q, Hendrickson RC, Shieh J, Moore MA, Danishefsky SJ (2012) At last: erythropoietin as a single glycoform. Angew Chem Int Ed Engl 51:11576–11584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mandal K, Uppalapati M, Ault‐Riché D, Kenney J, Lowitz J, Sidhu SS, Kent SBH (2012) Chemical synthesis and X‐ray structure of a heterochiral {D‐protein antagonist plus vascular endothelial growth factor} protein complex by racemic crystallography. Proc Natl Acad Sci U S A 109:14779–14784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zheng J‐S, Tang S, Guo Y, Chang H‐N, Liu L (2012) Synthesis of cyclic peptides and cyclic proteins via ligation of peptide hydrazides. Chembiochem 13:542–546. [DOI] [PubMed] [Google Scholar]
- 49. Wang P, Dong S, Shieh J‐H, Peguero E, Hendrickson R, Moore MAS, Danishefsky SJ (2013) Erythropoietin derived by chemical synthesis. Science 342:1357–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Durek T, Vetter I, Wang C‐IA, Motin L, Knapp O, Adams DJ, Lewis RJ, Alewood PF (2013) Chemical engineering and structural and pharmacological characterization of the α‐scorpion toxin OD1. ACS Chem Biol 8:1215–1222. [DOI] [PubMed] [Google Scholar]
- 51. Gordon WR, Bang D, Hoff WD, Kent SBH (2013) Total chemical synthesis of fully functional photoactive yellow protein. Bioorg Med Chem 21:3436–3442. [DOI] [PubMed] [Google Scholar]
- 52. Ghassemian A, Wang C‐IA, Yau M‐K, Reid RC, Lewis RJ, Fairlie DP, Alewood PF, Durek T (2013) Efficient chemical synthesis of human complement protein C3a. Chem Commun 49:2356–2358. [DOI] [PubMed] [Google Scholar]
- 53. Raibaut L, Vicogne J, Leclercq B, Drobecq H, Desmet R, Melnyk O (2013) Total synthesis of biotinylated N domain of human hepatocyte growth factor. Bioorg Med Chem 21:3486–3494. [DOI] [PubMed] [Google Scholar]
- 54. Zhan C, Le Z, Chen X, Lu W‐Y, Lu W (2013) Total chemical synthesis of dengue 2 virus capsid protein via native chemical ligation: role of the conserved salt‐bridge. Bioorg Med Chem 21:3443–3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Belgi A, Bathgate RAD, Kocan M, Patil N, Zhang S, Tregear GW, Wade JD, Hossain MA (2013) Minimum active structure of insulin‐like peptide 5. J Med Chem 56:9509–9516. [DOI] [PubMed] [Google Scholar]
- 56. Dekan Z, Mobli M, Pennington MW, Fung E, Nemeth E, Alewood PF (2014) Total synthesis of human hepcidin through regioselective disulfide‐bond formation by using the safety‐catch cysteine protecting group 4,4′‐dimethylsulfinylbenzhydryl. Angew Chem Int Ed Engl 53:2931–2934. [DOI] [PubMed] [Google Scholar]
- 57. Zhang Q, Johnston EV, Shieh JH, Moore MA, Danishefsky SJ (2014) Synthesis of granulocyte‐macrophage colony‐stimulating factor as homogeneous glycoforms and early comparisons with yeast cell‐derived material. Proc Natl Acad Sci U S A 111:2885–2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Stanchev S, Zawada Z, Monincová L, Bednárová L, Slaninová J, Fučík V, Čeřovský V (2014) Synthesis of lucifensin by native chemical ligation and characteristics of its isomer having different disulfide bridge pattern. J Pept Sci 20:725–735. [DOI] [PubMed] [Google Scholar]
- 59. Zheng J‐S, Yu M, Qi Y‐K, Tang S, Shen F, Wang Z‐P, Xiao L, Zhang L, Tian C‐L, Liu L (2014) Expedient total synthesis of small to medium‐sized membrane proteins via Fmoc chemistry. J Am Chem Soc 136:3695–3704. [DOI] [PubMed] [Google Scholar]
- 60. Wucherpfennig TG, Pattabiraman VR, Limberg FRP, Ruiz‐Rodríguez J, Bode JW (2014) Traceless preparation of C‐terminal α‐ketoacids for chemical protein synthesis by α‐ketoacid‐hydroxylamine ligation: synthesis of SUMO2/3. Angew Chem Int Ed Engl 53:12248–12252. [DOI] [PubMed] [Google Scholar]
- 61. Boll E, Drobecq H, Ollivier N, Raibaut L, Desmet R, Vicogne J, Melnyk O (2014) A novel PEG‐based solid support enables the synthesis of >50 amino‐acid peptide thioesters and the total synthesis of a functional SUMO‐1 peptide conjugate. Chem Sci 5:2017–2022. [Google Scholar]
- 62. Roberts AG, Johnston EV, Shieh JH, Sondey JP, Hendrickson RC, Moore MA, Danishefsky SJ (2015) Fully synthetic granulocyte colony‐stimulating factor enabled by isonitrile‐mediated coupling of large, side‐chain‐unprotected peptides. J Am Chem Soc 137:13167–13175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Medini K, Harris PWR, Hards K, Dingley AJ, Cook GM, Brimble MA (2015) Chemical synthesis of a pore‐forming antimicrobial protein, caenopore‐5, by using native chemical ligation at a glu‐cys site. Chembiochem 16:328–336. [DOI] [PubMed] [Google Scholar]
- 64. Seenaiah M, Jbara M, Mali SM, Brik A (2015) Convergent versus sequential protein synthesis: the case of ubiquitinated and glycosylated H2B. Angew Chem Int Ed Engl 54:12374–12378. [DOI] [PubMed] [Google Scholar]
- 65. Raibaut L, Cargoët M, Ollivier N, Chang YM, Drobecq H, Boll E, Desmet R, Monbaliu J‐CM, Melnyk O (2016) Accelerating chemoselective peptide bond formation using bis(2‐selenylethyl)amido peptide selenoester surrogates. Chem Sci 7:2657–2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wang Z, Xu W, Liu L, Zhu TF (2016) A synthetic molecular system capable of mirror‐image genetic replication and transcription. Nat Chem 8:698–704. [DOI] [PubMed] [Google Scholar]
- 67. Hemu X, Qiu Y, Nguyen GK, Tam JP (2016) Total synthesis of circular bacteriocins by butelase 1. J Am Chem Soc 138:6968–6971. [DOI] [PubMed] [Google Scholar]
- 68. Pan M, Gao S, Zheng Y, Tan X, Lan H, Tan X, Sun D, Lu L, Wang T, Zheng Q, Huang Y, Wang J, Liu L (2016) Quasi‐racemic X‐ray structures of K27‐linked ubiquitin chains prepared by total chemical synthesis. J Am Chem Soc 138:7429–7435. [DOI] [PubMed] [Google Scholar]
- 69. Kuroha M, Hemmi H, Ohnishi‐Kameyama M, Kodani S (2017) Isolation and structure determination of a new lasso peptide subterisin from Sphingomonas subterranea. Tetrahedron Lett 58:3429–3432. [Google Scholar]
- 70. Garay H, Espinosa LA, Perera Y, Sánchez A, Diago D, Perea SE, Besada V, Reyes O, González LJ (2018) Characterization of low‐abundance species in the active pharmaceutical ingredient of CIGB‐300: a clinical‐grade anticancer synthetic peptide. J Pept Sci 24:e3081. [DOI] [PubMed] [Google Scholar]
- 71. Qu Q, Gao S, Li Y‐M (2018) Racemic crystal structures of peptide toxins, GsMTx4 prepared by protein total synthesis. J Pept Sci 24:e3112. [DOI] [PubMed] [Google Scholar]
- 72. Boutin JA, Gesson I, Henlin JM, Bertin S, Lambert PH, Volland JP, Fauchère JL (1997) Limitations of the coupling of amino acid mixtures for the preparation of equimolar peptide libraries. Mol Divers 3:43–60. [DOI] [PubMed] [Google Scholar]
- 73. Kim CH, Axup JY, Schultz PG (2013) Protein conjugation with genetically encoded unnatural amino acids. Curr Opin Chem Biol 17:412–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Sun SB, Schultz PG, Kim CH (2014) Therapeutic applications of an expanded genetic code. Chembiochem 15:1721–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Leney AC, Heck AJR (2017) Native mass spectrometry: what is in the name? J Am Soc Mass Spectrom 28:5–13. [DOI] [PubMed] [Google Scholar]
- 76. Rogers RS, Nightlinger NS, Livingston B, Campbell P, Bailey R, Balland A (2015) Development of a quantitative mass spectrometry multi‐attribute method for characterization, quality control testing and disposition of biologics. MAbs 7:881–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Beck A, Diemer H, Ayoub D, Debaene F, Wagner‐Rousset E, Carapito C, van Dorsselaer A, Sanglier‐Cianférani S (2013) Analytical characterization of biosimilar antibodies and Fc‐fusion proteins. Trends Analyt Chem 48:81–95. [Google Scholar]
- 78. Vandermarliere E, Stes E, Gevaert K, Martens L (2016) Resolution of protein structure by mass spectrometry. Mass Spectrom Rev 35:653–665. [DOI] [PubMed] [Google Scholar]
- 79. Husson G, Delangle A, O'Hara J, Cianferani S, Gervais A, van Dorsselaer A, Bracewell D, Carapito C (2018) Dual data‐independent acquisition approach combining global HCP profiling and absolute quantification of key impurities during bioprocess development. Anal Chem 90:1241–1247. [DOI] [PubMed] [Google Scholar]
- 80. Alalwiat A, Tang W, Gerişlioğlu S, Becker ML, Wesdemiotis C (2017) Mass spectrometry and ion mobility characterization of bioactive peptide‐synthetic polymer conjugates. Anal Chem 89:1170–1177. [DOI] [PubMed] [Google Scholar]
- 81. Patrick JW, Gamez RC, Russell DH (2015) Elucidation of conformer preferences for a hydrophobic antimicrobial peptide by vesicle capture‐freeze‐drying: a preparatory method coupled to ion mobility‐mass spectrometry. Anal Chem 87:578–583. [DOI] [PubMed] [Google Scholar]
- 82. Jeanne Dit Fouque K, Afonso C, Zirah S, Hegemann JD, Zimmermann M, Marahiel MA, Rebuffat S, Lavanant H (2015) Ion mobility‐mass spectrometry of lasso peptides: signature of a rotaxane topology. Anal Chem 87:1166–1172. [DOI] [PubMed] [Google Scholar]
- 83. Jeanne Dit Fouque K, Moreno J, Hegemann JD, Zirah S, Rebuffat S, Fernandez‐Lima F (2018) Identification of lasso peptide topologies using native nanoelectrospray ionization‐trapped ion mobility spectrometry‐mass spectrometry. Anal Chem 90:5139–5146. [DOI] [PubMed] [Google Scholar]
- 84. Jeanne Dit Fouque K, Garabedian A, Porter J, Baird M, Pang X, Williams TD, Li L, Shvartsburg A, Fernandez‐Lima F (2017) Fast and effective ion mobility‐mass spectrometry separation of d‐amino‐acid‐containing peptides. Anal Chem 89:11787–11794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. D'atri V, Ehkirch A, Rouviere F, Hernandez‐Alba O, Goyon A, Colas O, Sarrut M, Beck A, Heinisch S, Cianferani S, Guillarme D (2018) Characterization of antibody drug conjugates by an innovative on‐line four‐dimensional HICxSEC‐IM‐MS methodology. N Biotechnol 44:S35. [DOI] [PubMed] [Google Scholar]
- 86. Ogihara NL, Weiss MS, Degrado WF, Eisenberg D (1997) The crystal structure of the designed trimeric coiled coil coil‐VaLd: implications for engineering crystals and supramolecular assemblies. Protein Sci 6:80–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Nordhoff A, Tziatzios C, van den Broek JA, Schott MK, Kalbitzer HR, Becker K, Schubert D, Schirmer RH (1997) Denaturation and reactivation of dimeric human glutathione reductase‐‐an assay for folding inhibitors. Eur J Biochem 245:273–282. [DOI] [PubMed] [Google Scholar]
- 88. Starovasnik MA, Braisted AC, Wells JA (1997) Structural mimicry of a native protein by a minimized binding domain. Proc Natl Acad Sci U S A 94:10080–10085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Walsh ST, Cheng H, Bryson JW, Roder H, Degrado WF (1999) Solution structure and dynamics of a de novo designed three‐helix bundle protein. Proc Natl Acad Sci U S A 96:5486–5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Macias MJ, Gervais V, Civera C, Oschkinat H (2000) Structural analysis of WW domains and design of a WW prototype. Nat Struct Biol 7:375–379. [DOI] [PubMed] [Google Scholar]
- 91. He Y, Chen Y, Alexander P, Bryan PN, Orban J (2008) NMR structures of two designed proteins with high sequence identity but different fold and function. Proc Natl Acad Sci U S A 105:14412–14417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Takacs Z, Toups M, Kollewe A, Johnson E, Cuello LG, Driessens G, Biancalana M, Koide A, Ponte CG, Perozo E, Gajewski TF, Suarez‐Kurtz G, Koide S, Goldstein SAN (2009) A designer ligand specific for Kv1.3 channels from a scorpion neurotoxin‐based library. Proc Natl Acad Sci U S A 106:22211–22216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Bang D, Tereshko V, Kossiakoff AA, Kent SBH (2009) Role of a salt bridge in the model protein crambin explored by chemical protein synthesis: X‐ray structure of a unique protein analogue, V15Acrambin‐alpha‐carboxamide. Mol Biosyst 5:750–756. [DOI] [PubMed] [Google Scholar]
- 94. Landau M, Sawaya MR, Faull KF, Laganowsky A, Jiang L, Sievers SA, Liu J, Barrio JR, Eisenberg D (2011) Towards a pharmacophore for amyloid. PLoS Biol 9:e1001080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Vermeer LS, Lan Y, Abbate V, Ruh E, Bui TT, Wilkinson LJ, Kanno T, Jumagulova E, Kozlowska J, Patel J, McIntyre CA, Yam WC, Siu G, Atkinson RA, Lam JKW, Bansal SS, Drake AF, Mitchell GH, Mason AJ (2012) Conformational flexibility determines selectivity and antibacterial, antiplasmodial, and anticancer potency of cationic α‐helical peptides. J Biol Chem 287:34120–34133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Zaccai NR, Chi B, Thomson AR, Boyle AL, Bartlett GJ, Bruning M, Linden N, Sessions RB, Booth PJ, Brady RL, Woolfson DN (2011) A de novo peptide hexamer with a mutable channel. Nat Chem Biol 7:935–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Barb AW, Ho TG, Flanagan‐Steet H, Prestegard JH (2012) Lanthanide binding and IgG affinity construct: potential applications in solution NMR, MRI, and luminescence microscopy. Protein Sci 21:1456–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Huang Y‐H, Henriques ST, Wang CK, Thorstholm L, Daly NL, Kaas Q, Craik DJ (2015) Design of substrate‐based BCR‐ABL kinase inhibitors using the cyclotide scaffold. Sci Rep 5:12974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Akcan M, Clark RJ, Daly NL, Conibear AC, de Faoite A, Heghinian MD, Sahil T, Adams DJ, Marí F, Craik DJ (2015) Transforming conotoxins into cyclotides: backbone cyclization of P‐superfamily conotoxins. Biopolymers 104:682–692. [DOI] [PubMed] [Google Scholar]
- 100. Joh NH, Wang T, Bhate MP, Acharya R, Wu Y, Grabe M, Hong M, Grigoryan G, DeGrado WF (2014) De novo design of a transmembrane Zn2+‐transporting four‐helix bundle. Science 346:1520–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Valéry C, Deville‐Foillard S, Lefebvre C, Taberner N, Legrand P, Meneau F, Meriadec C, Delvaux C, Bizien T, Kasotakis E, Lopez‐Iglesias C, Gall A, Bressanelli S, Le Du M‐H, Paternostre M, Artzner F (2015) Atomic view of the histidine environment stabilizing higher‐pH conformations of pH‐dependent proteins. Nat Commun 6:7771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Lee J‐H, Kang E, Lee J, Kim J, Lee KH, Han J, Kang HY, Ahn S, Oh Y, Shin D, Hur K, Chae SY, Song PH, Kim Y‐I, Park JC, Lee JI (2014) Protein grafting of p53TAD onto a leucine zipper scaffold generates a potent HDM dual inhibitor. Nat Commun 5:3814. [DOI] [PubMed] [Google Scholar]
- 103. Kim K‐H, Ko D‐K, Kim Y‐T, Kim NH, Paul J, Zhang S‐Q, Murray CB, Acharya R, DeGrado WF, Kim YH, Grigoryan G (2016) Protein‐directed self‐assembly of a fullerene crystal. Nat Commun 7:11429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Tang S, Si Y‐Y, Wang Z‐P, Mei K‐R, Chen X, Cheng J‐Y, Zheng J‐S, Liu L (2015) An efficient one‐pot four‐segment condensation method for protein chemical synthesis. Angew Chem Int Ed Engl 54:5713–5717. [DOI] [PubMed] [Google Scholar]
- 105. Bhardwaj G, Mulligan VK, Bahl CD, Gilmore JM, Harvey PJ, Cheneval O, Buchko GW, Pulavarti SVSRK, Kaas Q, Eletsky A, Huang P‐S, Johnsen WA, Greisen P Jr, Rocklin GJ, Song Y, Linsky TW, Watkins A, Rettie SA, Xu X, Carter LP, Bonneau R, Olson JM, Coutsias E, Correnti CE, Szyperski T, Craik DJ, Baker D (2016) Accurate de novo design of hyperstable constrained peptides. Nature 538:329–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Datta A, Bhattacharyya D, Singh S, Ghosh A, Schmidtchen A, Malmsten M, Bhunia A (2016) Role of aromatic amino acids in lipopolysaccharide and membrane interactions of antimicrobial peptides for use in plant disease control. J Biol Chem 291:13301–13317. [DOI] [PMC free article] [PubMed] [Google Scholar]


