Abstract
There is great interest in the role epigenetic variation induced by non-genetic exposures may play in the context of health and disease. In particular, DNA methylation has previously been shown to be highly dynamic during the earliest stages of development and is influenced by in utero exposures such as maternal smoking and medication. In this study we sought to identify the specific DNA methylation differences in blood associated with prenatal and birth factors, including birth weight, gestational age and maternal smoking. We quantified neonatal methylomic variation in 1263 infants using DNA isolated from a unique collection of archived blood spots taken shortly after birth (mean = 6.08 days; s.d. = 3.24 days). An epigenome-wide association study (EWAS) of gestational age and birth weight identified 4299 and 18 differentially methylated positions (DMPs) respectively, at an experiment-wide significance threshold of p < 1 × 10−7. Our EWAS of maternal smoking during pregnancy identified 110 DMPs in neonatal blood, replicating previously reported genomic loci, including AHRR. Finally, we tested the hypothesis that DNA methylation mediates the relationship between maternal smoking and lower birth weight, finding evidence that methylomic variation at three DMPs may link exposure to outcome. These findings complement an expanding literature on the epigenomic consequences of prenatal exposures and obstetric factors, confirming a link between the maternal environment and gene regulation in neonates.
This article is part of the theme issue ‘Developing differences: early-life effects and evolutionary medicine’.
Keywords: DNA methylation, birth weight, gestational age, maternal smoking, epigenome-wide association study, mediation analysis
1. Introduction
Epigenetic mechanisms developmentally regulate gene expression via modifications to DNA, histone proteins and chromatin. Because epigenetic processes can be influenced by exposure to a range of external environmental factors [1–4] and also by genetic variation [5,6], there is great interest in the role that epigenetic variation may play in the context of health and disease [7]. As epigenetic marks are inherited mitotically in somatic cell lineages, they provide a mechanism by which disruption early in life can be propagated through development, producing long-term phenotypic variation. DNA methylation is the best-characterized epigenetic modification, stably influencing gene expression via the disruption of transcription factor binding and recruitment of methyl-binding proteins that initiate chromatin compaction and gene silencing. Despite being often regarded as a mechanism of transcriptional repression, DNA methylation is associated with both increased and decreased gene expression [8], and also influences other genomic functions, including alternative splicing and promoter usage [9].
The availability of high-throughput profiling methods for quantifying DNA methylation across the genome at single base resolution in large numbers of samples has enabled researchers to perform epigenome-wide association studies (EWAS) aimed at identifying methylomic variation associated with environmental exposures and disease [7]. Although these studies are inherently more complex to design and interpret than genetic association studies [10–12], recent analyses have documented differences in DNA methylation in neonates and children following exposure to a wide range of environmental factors during gestation, including maternal smoking, maternal diet and pollution [2,3,13]. There is also interest in the role that DNA methylation may play as mediator through which environmental exposures can influence long-term health outcomes. For example, there is evidence that the causal relationship between maternal smoking during pregnancy and low birth weight is mediated through differences in DNA methylation at specific loci across the genome [14,15]. While noteworthy, these analyses have been based on moderate samples sizes, have generally not replicated the same loci, and may have overestimated mediation effects because of invalid assumptions and misclassification of the exposure [16].
Another active area of research concerns the utility of DNA methylation as a biomarker for clinical monitoring and screening. The potential of a DNA methylation based predictor has been most robustly demonstrated for age, with a number of algorithms available, referred to as ‘epigenetic clocks’ [17–19]. There is particular interest in how measures of age derived from DNA methylation data correlate with actual chronological age, and also whether ‘accelerated’ epigenetic age predicts ageing phenotypes such as mortality, cancer and dementia [18,20,21]. There is also interest in whether other exposures (or phenotypes) can be inferred from an epigenetic profile. The development of an epigenetic biomarker using neonatal blood samples might enable the evaluation of in utero exposures, which are hard to measure objectively, and could be a useful prospective predictor for future health outcomes. To this end, a biomarker of maternal smoking during pregnancy was recently developed using cord blood samples that demonstrates high specificity (97%)—but only moderate sensitivity (58%) [22]—demonstrating the potential application of such approaches. Because DNA methylation is known to be highly dynamic during the earliest stages of development [23,24], insults during this period may have important functional consequences or impact upon disease susceptibility later in life. Of note, several psychiatric disorders are hypothesized to have important neurodevelopmental origins [25–27] and have been associated with a number of prenatal and perinatal risk factors. For example, epidemiological studies have reported a higher risk of autism in those born with a low birth weight [28,29] or born pre-term [30]. Although DNA methylation predictors for age [18] and smoking [31] developed in childhood or adult samples have been shown to work reasonably well, methods developed specifically in either cord blood or neonatal samples have superior performance at these ages [19].
In this study, we first sought to identify specific patterns of DNA methylation in neonatal blood samples associated with three obstetric and neonatal influences measured in the same individuals: birth weight, gestational age and exposure to maternal smoking. We subsequently used our results to explore whether variable DNA methylation mediates the relationship between maternal smoking and low birth weight. We attempted to address this issue by quantifying methylomic variation in 1263 infants using DNA isolated from archived blood spots taken shortly after birth (mean = 6.08 days; s.d. = 3.24 days) originally profiled in a case–control study of autism [32]. Although we cannot exclude the role of DNA methylation changes occurring during the first few days after birth, our study extends previous research into the early-life epigenome that has used samples collected either later in childhood or from cord blood [13,24,33], which has the limitation that it may be contaminated by maternal blood [34,35]. Our findings complement the expanding literature on the epigenomic consequences of prenatal exposures and obstetric factors, confirming a link between the maternal environment and markers of gene regulation in neonates.
2. Methods
(a). Overview of the MINERvA cohort
A description of the MINERvA cohort was recently published alongside extensive details of the profiling of DNA methylation and data quality control steps [32]. Briefly, MINERvA contains a subset of 1316 samples from the iPSYCH autism spectrum disorder case–control sample [36]. All perinatal data used for case–control sample matching, plus additional information on birth weight and maternal smoking were obtained from the Danish Medical Birth Register or the Central Person Register. An overview of the demographic characteristics of the MINERvA cohort is given in electronic supplementary material, table S1. Of note, cases and controls were matched as closely as possible. Although rates of maternal smoking were higher in autism cases, there was no significant difference in birth weight between autism cases and controls.
(b). DNA methylation profiling in MINERvA
Neonatal dried blood spot samples collected on standard Guthrie cards and stored within the Danish Neonatal Screening Biobank [37] were retrieved as part of the iPSYCH study [36]. Neonatal DNA extractions and DNA methylation quantification were performed at the Statens Serum Institut (SSI, Copenhagen, Capital Region, Denmark). Briefly, DNA was converted with sodium bisulfite using the EZ-96 DNA Methylation Kit (Zymo Research, CA, USA) and DNA methylation was quantified across the genome using the Infinium HumanMethylation450K array (‘450K array’) (Illumina, CA, USA). After a stringent quality control process (outlined in [32]), 1263 samples (96.0%) were included for subsequent analysis. Normalization of the DNA methylation data was performed used the dasen() function in the wateRmelon package [38]. For each sample, we derived nine additional variables from the DNA methylation data using established algorithms: DNA methylation age [18], gestational age [19], smoking [31] and six blood cell composition variables [39,40]. Our previous publication on these data demonstrated that the smoking score, despite being trained in adults who smoked, correlated with reported maternal smoking status from the registry data [32]. All quality control and statistical analyses were performed using the R statistical environment v. 3.2.1 [41].
(c). Epigenome-wide association analyses (EWAS)
We performed an EWAS of three obstetric/neonatal factors that were robustly measured in our cohort. First, to identify DNA methylation sites associated with birth weight (measured in grams (g)) and gestational age (measured in weeks), a linear model was fitted for each DNA methylation site with DNA methylation as the dependent variable, both birth weight and gestational age as independent variables, and a set of possible confounders as covariates: sex, experimental array number (i.e. chip), days to sampling, maternal smoking (using the continuous variable estimated from the DNA methylation data) and six derived cell composition variables. Given the strong concordance between the findings of our maternal smoking EWAS and those of previous EWAS analyses of smoking (see Results), we used the derived maternal smoking score to make up for missing data in the registry data and maximize power for analyses. To compare results between autism spectrum disorder (ASD) cases and controls, we tested for a heterogeneous effect by including an interaction term between (i) birth weight and case–control status, and (ii) gestational age and case–control status. In these interaction models, ASD case–control status was also included as a main effect. Second, to identify differentially methylated positions (DMPs) associated with registry-reported maternal smoking exposure, a linear model was fitted for each DNA methylation site with DNA methylation as the dependent variable, and a binary indicator variable for in utero exposure to smoking in addition to a set of possible confounders as covariates: sex, birth weight, gestational age, experimental array number (i.e. chip), and six derived blood cell composition variables. Significant DMPs were identified at an experiment-wide multiple testing adjusted threshold of p < 1 × 10−7. Clustering of significant DMPs into loci was performed by taking each significant site in turn, starting with the one with the smallest p value (referred to as the index association), identifying all other significant sites within 5 kb upstream and downstream and merging these into a single locus. Any less significant (i.e. larger p value) DMPs merged with an index site were then excluded from consideration as an index association. This procedure was repeated until all significant DMPs were either merged with a more significant association or considered as an index site. Conditional analyses were performed within loci with at least two DMPs by repeating the original association analysis for the secondary signal (i.e. the less significant site), including the most significant DNA methylation site in that loci as an additional covariate.
(d). Replication dataset
The Accessible Resource for Integrated Epigenomic Studies (ARIES; http://www.ariesepigenomics.org.uk) cohort consists of a sub-sample of 1018 ALSPAC (http://www.bristol.ac.uk/alspac/) child–mother pairs with Illumina 450K array DNA methylation data generated from cord blood (n = 914), and whole blood at two time points during childhood (age 7 (n = 973) and age 15 or 17 years (n = 974)). The results used in this manuscript are taken from the gestational age and birth weight EWAS performed by Simpkin et al. [24] and presented in electronic supplementary material, tables S1 and S3 published alongside this manuscript.
(e). Mediation analyses
Mediation analyses were performed using the criteria outlined by Baron & Kenny [42] and the Sobel test [43]. We considered DMPs associated with registry-reported maternal smoking without controlling for birth weight in our dataset (n = 143 DMPs) and tested whether the following criteria were met for each site:
-
(i)
smoking significantly correlated with DNA methylation level (p < 1 × 10−7; sex, gestational age, batch and cell composition included as covariates);
-
(ii)
smoking significantly correlated with birth weight without adjusting the model for DNA methylation (sex and gestational age included as covariates);
-
(iii)
DNA methylation significantly correlated with birth weight (p < 1 × 10−7; sex, gestational age, batch and cell composition included as covariates);
-
(iv)
the association between smoking and birth weight decreased upon addition of DNA methylation to the model (i.e. p value got larger; sex, gestational age, batch and cell composition included as covariates);
-
(v)
the Sobel test gave p < 3.50 × 10−4 (corrected for 143 DMPs considered, implemented through the R bda package (https://cran.r-project.org/web/packages/bda/index.html).
DMPs meeting the criteria for mediation were taken forward for a sensitivity analysis that accounted for misclassification of the exposure following the method outlined in Valeri et al. [16] using the SIMEX procedure [44]. Our naive outcome regression model between birth weight and registry-reported maternal smoking exposure and naive mediator regression model between DNA methylation and birth weight included covariates for gestational age, sex, cellular composition and experimental chip. The naive direct effect is then the coefficient from the outcome regression model for maternal smoking and the naive indirect effect is the naive direct effect multiplied by the estimated coefficient for maternal smoking from the mediator regression. Applying SIMEX to outcome and mediator regressions, we obtained corrected estimates of the regression parameters which were then used to calculate the direct and indirect effects. We set the parameter for specificity to 1.0 as we assume that all smokers are likely to have reported correctly, whereas we assume that some non-smokers will have reported incorrectly and therefore the sensitivity parameter was set to 0.6. A bootstrap method was used to estimate the standard errors of the estimated effects and their 95% confidence intervals (CIs).
3. Results
(a). Blood cell proportions derived from DNA methylation data correlate with birth weight and gestational age in neonatal blood
Our first analyses aimed to explore whether measures derived from DNA methylation data at birth (i.e. gestational age and blood cell composition estimates) are associated with birth weight. We previously demonstrated the robustness of DNA methylation data derived from neonatal blood spots by implementing two DNA methylation clock algorithms to derive estimates for (i) age in years [18] and (ii) gestational age in weeks [19] for each sample. As expected, we observed a strong positive correlation between estimated and actual gestational age (r = 0.602, 95% CI = (0.566, 0.636), p = 3.80 × 10−125) and a weaker positive correlation between estimated chronological age and actual gestational age (r = 0.139, 95% CI = (0.0849, 0.193), p = 6.52 × 10−7) [32]. We next extended these analyses to investigate how birth weight correlates with these predicted ages and whether variable birth weight explains the difference between predicted age (derived from DNA methylation data) and actual age. Birth weight was significantly correlated with predicted measures of both age (r = 0.119, 95% CI = (0.0638, 0.173), p = 2.40 × 10−5; electronic supplementary material, figure S1A) and gestational age (r = 0.333, 95% CI = (0.0849, 0.193), p = 5.15 × 10−34; electronic supplementary material, figure S1B). However, the age acceleration residuals—which are adjusted for reported gestational age—are not significantly associated with birth weight (p > 0.05, electronic supplementary material, figure S1C,D), indicating that variation in birth weight does not explain the difference between reported gestational age and predicted age and that other pregnancy and/or obstetric factors may be influencing derived age estimates. Given the difficulties in collecting large volumes of blood from neonates, little is known about blood cell-type variation at this stage of life. We therefore explored how predicted cellular composition variables derived from the DNA methylation data (see Methods) correlate with birth weight and reported gestational age. Gestational age was positively correlated with the estimated proportions of CD8 T-cells (r = 0.140; 95% CI = (0.0857, 0.194), p = 5.63 × 10−7) and natural killer cells (r = 0.0722, 95% CI = (0.0171, 0.127), p = 0.0103), and negatively correlated with the estimated proportion of B-cells (r = −0.231, 95% CI = (−0.282, −0.178), p = 1.06 × 10−16) (electronic supplementary material, figure S2). Birth weight was significantly negatively correlated with the estimated proportion of monocytes (r = −0.0604, 95% CI = (−0.115, −0.00529), p = 0.0317) and positively correlated with the estimated proportion of granulocytes (r = 0.0624, 95% CI = (0.00727, 0.117), p = 0.0266) (electronic supplementary material, figure S3). Given the potential confounding influence of cellular heterogeneity in EWAS analyses using blood, these derived variables were included in all subsequent analyses.
(b). Birth weight and gestational age are associated with variable DNA methylation in neonatal blood
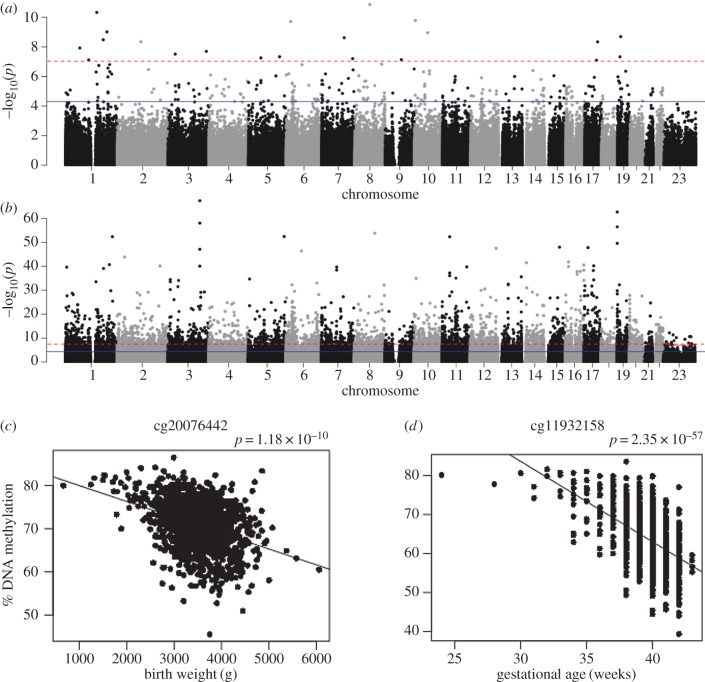
To identify DNA methylation sites associated with reported gestational age and birth weight we next performed an EWAS across all Illumina 450K array sites on the autosomes and X-chromosome (n = 430 676 sites), undertaking the analyses simultaneously to minimize confounding resulting from the strong correlation between these two obstetric variables (r = 0.491, 95% CI = (0.448, 0.532), p = 1.14 × 10−77; electronic supplementary material, figure S4). In total, we identified 18 differentially methylated positions (DMPs) associated with birth weight (figure 1; electronic supplementary material, figures S5 and S6) and 4299 DMPs associated with gestational age (figure 1; electronic supplementary material, figures S7 and S8) at an experiment-wide significance threshold (p < 1 × 10−7) (electronic supplementary material, tables S2 and S3). The associated DMPs were characterized by a median shift in DNA methylation of 1.40% (s.d. = 0.368%) per kg and 0.406% (s.d. = 0.275) per gestational week. Seven sites were significantly associated with both birth weight and gestational age (table 1). Sensitivity analyses repeating the EWAS excluding samples from ‘outlier’ individuals born (i) before 35 weeks (N = 23) or (ii) before 32 weeks (N = 5) revealed high concordance with our primary analysis (electronic supplementary material, figure S9), suggesting that the results are robust to the presence of premature individuals. Although the majority of DMPs associated with increased birth weight were associated with reduced DNA methylation (66.7%) there was not a significant bias (sign test p = 0.238) due to the small total number of DMPs identified. In contrast, there was a highly-significant bias towards increased DNA methylation at sites associated with older gestational age (73.2%, sign test p = 2.29 × 10−135). Although this contradicts results from a previous study using cord blood derived DNA from the ARIES cohort (n = 914), which identified a smaller number of DMPs that were enriched for sites showing a decrease in DNA methylation with older gestational age [24], our most significant DMPs are characterized by reduced DNA methylation with age (electronic supplementary material, table S3) and associations at sites showing this pattern of change are significantly stronger (Mann–Whitney p = 2.58 × 10−228). Furthermore, we find a significant excess of consistent effects between studies indicating that age-associated changes are similar in cord and neonatal whole blood (electronic supplementary material, figure S10). Of 148 DMPs associated with gestational age in the ARIES cohort, 146 (98.6%) had the same direction of effect (sign test p = 6.18 × 10−41), with 110 being significantly associated (p < 1 × 10−7) in both cohorts. We found similar consistency for our EWAS of birth weight; all 21 DMPs associated with birth weight in the ARIES cohort had the same direction of effect in our data (sign test p < 2 × 10−323), with two DMPs being significantly associated (p < 1 × 10−7) in both cohorts (electronic supplementary material, figure S10). While the 18 DMPs associated with birth weight are annotated to distinct genomic loci, the 4299 DMPs associated with gestational age are clustered into 3550 distinct locations with up to 25 additional DMPs located within 5 kb of the index DMP characterized by the most significant association. Conditional analyses within each genomic locus containing at least two DMPs (n = 483; 13.6%) revealed evidence for independent secondary signals for 240 DMPs in 193 of these loci (conditional p < 5 × 10−5), while 100 DMPs within 66 loci were only associated as a result of their correlation with the most significant DMP in the same loci (conditional p > 0.05; electronic supplementary material, table S4). Finally, given the association between both low birth weight and pre-term birth and autism we tested whether the DNA methylation differences we identified were consistent between individuals who later went on to develop a childhood diagnosis of autism (n = 629) and matched controls (n = 634). There were no significant differences between autism cases and controls for DMPs associated with birth weight (min. p = 0.0446) or for DMPs associated with gestation age (min. p = 3.51 × 10−5) after correcting for the number of DMPs significant in each analysis (birth weight: p < 0.00278 corrected for 18 DMPs; gestational age: p < 1.16 × 10−5 corrected for 4299 DMPs) (electronic supplementary material, tables S2 and S3).
Figure 1.
Methylomic variation associated with birth weight and gestational age in neonates. Manhattan plots of p-values from an epigenome-wide association study (EWAS) of (a) birth weight (g) and (b) gestational age (weeks). The red (dashed) horizontal line indicates experiment-wide significance (p < 1 × 10−7); the blue (solid) horizontal line indicates a ‘discovery’ significance threshold (p < 5 × 10−5). Shown are scatterplots of the top-ranked differentially methylated positions (DMPs) associated with (c) birth weight (cg20076442) and (d) gestational age (cg11932158). (Online version in colour.)
Table 1.
DNA methylation in neonatal blood is associated with birth weight and gestational age. In total, seven differentially methylated positions (DMPs) were associated with both birth weight and gestational age at an experiment-wide threshold of p < 1 × 10−7. Genomic locations are based on hg19.
| EWAS of birth weight (g) |
EWAS of gestational age (weeks) |
gene annotation |
||||||
|---|---|---|---|---|---|---|---|---|
| probe ID | p-value | regression coefficient | p-value | regression coefficient | chr | position | UCSC gene name | UCSC genic region |
| cg04411893 | 9.88×10−8 | −8.41×10−6 | 2.28×10−12 | −0.003758699 | chr3 | 185 300 709 | ||
| cg05937055 | 4.66×10−9 | −1.20×10−5 | 5.20×10−10 | −0.004302303 | chr1 | 181 128 764 | ||
| cg06870470 | 2.97×10−9 | −1.68×10−5 | 6.93×10−28 | −0.01068899 | chr19 | 11 315 767 | DOCK6 | body |
| cg13066703 | 2.50×10−8 | −1.11×10−5 | 3.68×10−36 | −0.008742825 | chr1 | 211 526 705 | TRAF5 | body |
| cg19744173 | 1.87×10−10 | −1.41×10−5 | 8.41×10−20 | −0.006889143 | chr2 | 112 913 178 | FBLN7 | body |
| cg20068209 | 5.19×10−8 | −1.37×10−5 | 2.00×10−41 | −0.011959147 | chr6 | 75 988 568 | TMEM30A | body |
| cg20076442 | 1.18×10−10 | −1.96×10−5 | 3.23×10−25 | −0.010843225 | chr8 | 72 745 197 | ||
(c). Maternal smoking influences DNA methylation in neonates at multiple loci
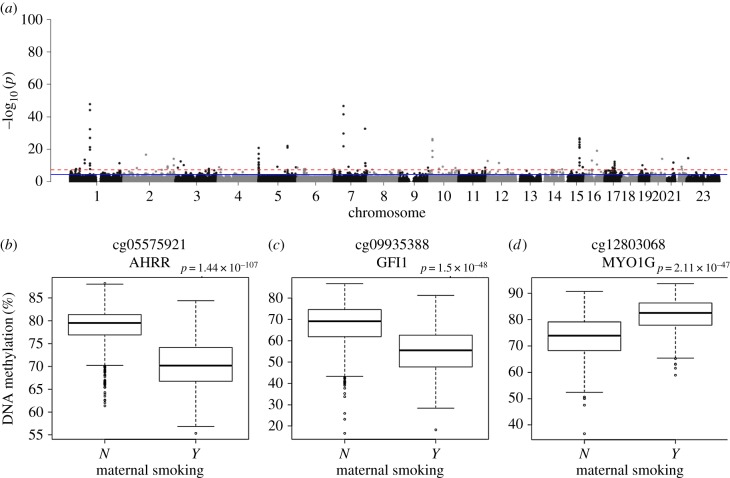
Exposure to tobacco smoke is known to be associated with widespread alterations in DNA methylation in whole blood [1], and previous analyses have demonstrated that these effects can be detected in cord blood from neonates exposed to prenatal smoking [13]. One limitation of using cord blood is that DNA methylation estimates can be influenced by contamination with maternal blood, and we therefore sought to test these whether these associations were detectable in neonatal whole blood samples. Mothers were asked about their smoking status at their first prenatal visit, early in the second trimester of pregnancy. Among the mothers of neonates in our cohort, 294 (25.1%) reported smoking during pregnancy and 879 (74.9%) reported not smoking during pregnancy; we excluded 36 mothers who reported giving up smoking at some time during the pregnancy and 54 mothers for whom smoking data were not available. First, we assessed whether maternal smoking influenced derived measures of age generated from DNA methylation data in these samples. Neither age, gestational age nor age acceleration was associated with in utero exposure to smoking (electronic supplementary material, figure S11). Next, we performed an EWAS of maternal smoking exposure (n = 1173, controlling for sex, birth weight, gestational age, experimental batch, and derived cell composition variables), identifying 110 neonatal blood DMPs associated with maternal smoking (p < 1 × 10−7) representing 70 discrete genomic loci (figure 2; electronic supplementary material, figure S12 and table S5). Conditional analyses within each genomic locus with at least two DMPs (n = 13) identified seven loci where a single DMP was associated with maternal smoking (conditional p for other sites greater than 0.05) and four loci characterized by secondary semi-independent effects (conditional p for other sites less than 5 × 10−5; electronic supplementary material, table S6). There was no significant bias towards a particular direction of effect (50.9% hypomethylated; 49.1% hypermethylated, sign test p = 0.924) and the median effect was a difference of 2.28% DNA methylation (s.d. = 2.19%) (electronic supplementary material, figure S13). There was considerable overlap with DMPs reported in a large EWAS of maternal smoking performed in cord blood [13] (n = 6685; electronic supplementary material, figure S14). Of note, 4847 (84.0%) of the 5768 DMPs reported in that study were characterized by the same direction of effect (sign test p < 2 × 10−323) in our analysis of neonatal blood, with 102 meeting criteria for experiment-wide significance (p < 1 × 10−7) in both studies. This included previously reported DMPs associated with tobacco smoking in adults [1,31], such as AHRR, where five additional DMPs were clustered with the lead signal at cg05575921, none of which remained significant in the conditional analysis, GFI1, which had nine DMPs including multiple independent associations, and MYO1G, which had four DMPs with evidence of multiple independent associations (electronic supplementary material, table S6). These findings confirm that smoking behaviour by mothers during pregnancy has a profound influence on DNA methylation in their offspring at birth and in the first few days of life.
Figure 2.
DNA methylation at birth is associated with maternal smoking during pregnancy. The red (dashed) horizontal line indicates experiment-wide significance (p < 1 × 10−7), the blue (solid) horizontal line indicates a ‘discovery’ significance threshold (p < 5 × 10−5). (a) Manhattan plots from an EWAS of maternal smoking. Boxplots of the three top-ranked DNA methylation sites associated with maternal smoking: (b) cg05575921, (c) cg09935388 and (d) cg12803068. Shown are unadjusted DNA methylation values.
(d). DNA methylation mediates the relationship between maternal smoking and low birth weight
Having established that the DNA methylation signatures associated with prenatal smoking exposure are robustly detectable in neonatal blood, and that variable DNA methylation is associated with an established outcome of maternal smoking [45,46] we next asked whether methylomic variation might mediate the relationship between maternal smoking in pregnancy and lower birth weight. Previous attempts to explore this using data from cord blood have been relatively inconsistent [14,15], supporting a mediation role for DNA methylation at non-overlapping DNA methylation sites. We repeated our EWAS of maternal smoking, excluding birth weight as a covariate, and identified an extended set of 143 DMPs (electronic supplementary material, table S7) which contained 105 (95.5%) of the 110 DMPs we identified in the analysis adjusting for birth weight. Mediation analyses, performed using the Sobel test (see Methods), showed that DNA methylation at three sites, annotated to three different genomic loci, met the five criteria set out by Baron & Kenny [42] (see Methods) as providing evidence for mediating the association between smoking and birth weight (electronic supplementary material, figure S15 and table S7). This included one site (cg09935388) annotated to GFI1 that has been reported previously [15] and two novel mediation sites (cg05575921 annotated to AHRR, and cg26889659 annotated to EXOC2) (table 2). Because smoking behaviour is prone to misclassification—not only owing to smokers claiming to be non-smokers, especially during pregnancy [47], but also because a complex behaviour is simplified into a dichotomous variable—we repeated the analysis for the three significant DNA methylation sites estimating the natural direct effect between maternal smoking and low birth weight (i.e. not via DNA methylation) and the natural indirect effect (i.e. via DNA methylation) using the SIMEX (simulation and extrapolation) procedure which incorporates misclassification [44]. Stringently accounting for misclassification suggests that the estimated mediation effect via DNA methylation identified using the Sobel test is potentially overestimated; although the results robustly support a significant mediation effect for DNA methylation at cg26889659, under scenarios of more extreme misclassification (see Methods) the effects of mediation via DNA methylation at cg05575921 and cg09935388 are no longer significantly different from 0 (electronic supplementary material, table S8).
Table 2.
Three DNA methylation sites meet the criteria for significant mediation between birth weight and maternal smoking.
| birth weight ∼ maternal smoking |
EWAS of maternal smoking |
EWAS of birth weight |
EWAS of birth weight adjusted for maternal smoking |
Sobel test p-value | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| See table 1 | chr. | position | gene annotation | p-value | regression coefficient | p-value | regression coefficient | p-value | regression coefficient | p-value | regression coefficient | ||
| cg09935388 | chr1 | 92 947 588 | GFI1 | 3.21 × 10−14 | −282.418 | 1.39 × 10−55 | −0.1208 | 2.58 × 10−8 | 4.18 × 10−5 | 2.72 × 10−8 | −229.556 | 3.58 × 10−9 | |
| cg05575921 | chr5 | 373 378 | AHRR | 3.21 × 10−14 | −282.418 | 8.29 × 10−119 | −0.08196 | 1.17 × 10−10 | 2.37 × 10−5 | 1.51 × 10−4 | −178.772 | 8.52 × 10−14 | |
| cg26889659 | chr6 | 684 090 | EXOC2 | 3.21 × 10−14 | −282.418 | 7.42 × 10−13 | −0.03981 | 3.84 × 10−8 | 2.89 × 10−5 | 2.41 × 10−10 | −238.068 | 1.12 × 10−6 | |
4. Discussion
In this study we quantified neonatal variation in DNA methylation in 1263 infants using samples isolated from archived blood spots taken shortly after birth. Our study finds that gestational age, birth weight and maternal smoking are all associated with significant DNA methylation differences in neonatal blood, with gestational age having the most effects widespread across the genome. These data add to a growing literature demonstrating that prenatal and obstetric exposures can influence epigenetic variation in early life [3,13,24,48–50], providing a potential mechanism linking them to altered gene function and long-term health and disease outcomes.
Our use of neonatal DNA samples means that we are uniquely positioned to identify epigenetic variation at birth, avoiding the confounding exposures that could influence the results from samples collected later in childhood (for example, health and disease, nutrition, medication, and stress). Although there have been larger studies of prenatal exposures, a strength of our study is that we profiled whole blood from neonatal infants rather than cord blood, minimizing the contamination of our samples with maternal blood DNA and meaning our data can be more easily compared with the extensive blood DNA methylation datasets derived from samples later in life. A limitation of our sampling strategy, however, is that no blood cell reference DNA methylation datasets specifically for use on neonatal blood are yet available, likely reflecting the difficulties of obtaining sufficient volumes of neonatal blood for cell sorting and methylomic profiling. Although there has been much written on the importance of selecting an appropriate tissue to profile for epigenetic studies [10], the goal of this study was to identify biomarkers of exposure, and therefore our use of a peripheral tissue is justified. Furthermore, although blood spots were collected only a few days after birth (mean = 6.08 days; s.d. = 3.24 days), it is possible that some postnatal exposure to passive smoking during the first few days of life could also have influenced our results, although the amount of exposure in approximately 6 days is likely to be negligible.
As well as identifying specific loci at which DNA methylation is associated with early-life factors such as smoking exposure and gestational age, we tested the hypothesis that DNA methylation mediates established epidemiological relationships between exposures and outcomes. We explored the association between maternal smoking and lower birth weight, finding evidence that methylomic variation at several DMPs may be mechanistically involved in linking exposure (maternal smoking) to outcome (birth weight). While our results are consistent with previous reports, such analyses can be influenced by misclassification bias [16]. Smoking, in particular, is prone to misclassification not only owing to participants claiming to be non-smokers when they are in fact smokers, a circumstance known to be worse when reflecting smoking during pregnancy[47], but also as a result of simplifying a complex behaviour into a single dichotomous variable representing the entire period of pregnancy. Given our robust prenatal smoking exposure associations, it is plausible that the mediator in our analyses—DNA methylation—is in fact a better measure of smoking exposure than self-reported status [16]. Of note, applying a methodology that accounts for misclassification of an exposure reduced the magnitude of the mediation effect at all three significant loci, suggesting that these results need validation using an alternative approach such as Mendelian randomization [51].
While we explored whether DNA methylation lies on the causal pathway between maternal smoking and low birth weight, results from EWAS analyses do not distinguish cause from effect. In fact, it is likely that the DNA methylation differences we report for birth weight and gestational age reflect other in utero exposures or processes. For example birth weight is known to be associated with maternal body mass index, blood pressure and fasting glucose levels [52–54] and the epigenetic changes we report may reflect the downstream influences of these pathways. It is also possible that the birth-weight-associated DMPs identified in our study reflect exposures or influences occurring in the immediate neonatal period before the blood samples were collected. Similarly, the DNA methylation differences observed in neonates exposed to prenatal smoking might also be influenced by exposure to passive smoking from the mother or father immediately after birth, although the amount of exposure in approximately 6 days is likely to be negligible. Another potential limitation of our design is that the analyses were performed within the context of an autism case–control study [32]. Of note, however, although autism cases had a higher exposure to maternal smoking than non-autism controls, there was not significant difference in birth weight between infants who went on to develop autism compared with those who did not (electronic supplementary material, table S1). Finally, the nature of the samples we profiled in this study (i.e. small amounts of neonatal blood) meant that additional DNA was not available for technical validation experiments. However, the Illumina 450K array has been shown to yield highly reproducible measures of DNA methylation, and the observed consistency with previous studies of maternal smoking, gestational age, and birth weight suggests our findings are robust.
5. Conclusion
Our data demonstrate that in utero exposures are associated with detectable patterns of DNA methylation in neonatal blood samples, highlighting the role that the prenatal environment plays in influencing gene regulation in neonates. While previous studies have shown that maternal smoking effects persist into later childhood [13], these have found that the effects are attenuated, suggesting that obtaining a biomarker as close to birth as possible will have maximal sensitivity regarding exposures during gestation.
Supplementary Material
Supplementary Material
Acknowledgements
We acknowledge iPSYCH and The Lundbeck Foundation for providing samples and funding.
Ethics
The MINERvA study was approved by the Regional Scientific Ethics Committee in Denmark and the Danish Data Protection Agency.
Data accessibility
Given the nature of the MINERvA cohort, access to data can only be provided through secured systems which comply with the current Danish and EU data standards. To comply with the study's ethical approval, access to the raw data is only available to qualified researchers upon request. All summary statistics and analysis scripts are available directly from the authors (please contact Jonas Bybjerg-Grauholm at JOGR@ssi.dk). R scripts used to perform the quality control of these data are available on GitHub: https://github.com/ejh243/MinervaASDEWAS.git and have been archived in Zenado: https://zenodo.org/badge/latestdoi/116149862 and scripts for the analyses reported in this manuscript are available on GitHub: https://github.com/ejh243/MinervaNeonatalEWAS.git and have been archived in Zenado: https://doi.org/10.5281/zenodo.1303340.
Authors' contributions
E.H., J.M., A.R. and D.S. designed and coordinated the study. J.B.-G., D.M.H., M.V.H., M.B.-H. and C.S.H. led generation of DNA methylation data from dried neonatal bloodspots. E.H. led and A.R., D.S., J.M., J.B.-G., J.G., C.L.-A. and M.D.F. oversaw implementation of the data analyses. D.M.H., O.M., P.B.M., A.D.B., T.W. and M.N. are principal investigators of the iPSYCH study. E.H. and J.M. drafted the manuscript, with input from A.R., D.S., C.L.-A., J.G. and J.B.-G. All co-authors read and approved the final manuscript.
Competing interests
T.W. has acted as advisor and lecturer to H. Lundbeck A/S. None of the other authors report any potential conflict of interest.
Funding
This study was supported by grant no. HD073978 from the Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institute of Environmental Health Sciences, and National Institute of Neurological Disorders and Stroke; and by the Beatrice and Samuel A. Seaver Foundation. The iPSYCH (The Lundbeck Foundation Initiative for Integrative Psychiatric Research) team acknowledges funding from The Lundbeck Foundation (grant no. R102-A9118 and R155-2014-1724), the Stanley Medical Research Institute, the European Research Council (project no: 294838), the Novo Nordisk Foundation for supporting the Danish National Biobank resource, and grants from Aarhus and Copenhagen Universities and University Hospitals, including support to the iSEQ Center, the GenomeDK HPC facility, and the CIRRAU Center. This research has been conducted using the Danish National Biobank resource, supported by the Novo Nordisk Foundation. J.M. and E.H. are supported by funding from the UK Medical Research Council (K013807).
References
- 1.Joehanes R, et al. 2016. Epigenetic signatures of cigarette smoking. Circ. Cardiovasc. Genet. 9, 436–447. ( 10.1161/CIRCGENETICS.116.001506) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tobi EW, et al. 2014. DNA methylation signatures link prenatal famine exposure to growth and metabolism. Nat. Commun. 5, 5592 ( 10.1038/ncomms6592) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gruzieva O, et al. 2017. Epigenome-wide meta-analysis of methylation in children related to prenatal NO2 air pollution exposure. Environ. Health Perspect. 125, 104–110. ( 10.1289/EHP36) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Panni T, et al. 2016. A genome-wide analysis of DNA methylation and fine particulate matter air pollution in three study populations: KORA F3, KORA F4, and the Normative Aging Study. Environ. Health Perspect. 124, 983–990. ( 10.1289/ehp.1509966) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gaunt TR, et al. 2016. Systematic identification of genetic influences on methylation across the human life course. Genome Biol. 17, 61 ( 10.1186/s13059-016-0926-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hannon E, et al. 2015. Methylation QTLs in the developing brain and their enrichment in schizophrenia risk loci. Nat. Neurosci. 19, 48–54. ( 10.1038/nn.4182) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murphy TM, Mill J. 2014. Epigenetics in health and disease: heralding the EWAS era. Lancet 383, 1952–1954. ( 10.1016/S0140-6736(14)60269-5) [DOI] [PubMed] [Google Scholar]
- 8.Wagner JR, Busche S, Ge B, Kwan T, Pastinen T, Blanchette M. 2014. The relationship between DNA methylation, genetic and expression inter-individual variation in untransformed human fibroblasts. Genome Biol. 15, R37 ( 10.1186/gb-2014-15-2-r37) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maunakea AK, et al. 2010. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature 466, 253–257. ( 10.1038/nature09165) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mill J, Heijmans BT. 2013. From promises to practical strategies in epigenetic epidemiology. Nat. Rev. Genet. 14, 585–594. ( 10.1038/nrg3405) [DOI] [PubMed] [Google Scholar]
- 11.Relton CL, Davey Smith G. 2010. Epigenetic epidemiology of common complex disease: prospects for prediction, prevention, and treatment. PLoS Med. 7, e1000356 ( 10.1371/journal.pmed.1000356) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rakyan VK, Down TA, Balding DJ, Beck S. 2011. Epigenome-wide association studies for common human diseases. Nat. Rev. Genet. 12, 529–541. ( 10.1038/nrg3000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joubert BR, et al. 2016. DNA methylation in newborns and maternal smoking in pregnancy: genome-wide consortium meta-analysis. Am. J. Hum. Genet. 98, 680–696. ( 10.1016/j.ajhg.2016.02.019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Witt SH, et al. 2018. Impact on birth weight of maternal smoking throughout pregnancy mediated by DNA methylation. BMC Genomics 19, 290 ( 10.1186/s12864-018-4652-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Küpers LK, et al. 2015. DNA methylation mediates the effect of maternal smoking during pregnancy on birthweight of the offspring. Int. J. Epidemiol. 44, 1224–1237. ( 10.1093/ije/dyv048) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Valeri L, et al. 2017. Misclassified exposure in epigenetic mediation analyses. Does DNA methylation mediate effects of smoking on birthweight? Epigenomics 9, 253–265. ( 10.2217/epi-2016-0145) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hannum G, et al. 2013. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol. Cell 49, 359–367. ( 10.1016/j.molcel.2012.10.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horvath S. 2013. DNA methylation age of human tissues and cell types. Genome Biol. 14, R115 ( 10.1186/gb-2013-14-10-r115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knight AK, et al. 2016. An epigenetic clock for gestational age at birth based on blood methylation data. Genome Biol. 17, 206 ( 10.1186/s13059-016-1068-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen BH, et al. 2016. DNA methylation-based measures of biological age: meta-analysis predicting time to death. Aging (Albany, NY) 8, 1844–1865. ( 10.18632/aging.101020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levine ME, Lu AT, Bennett DA, Horvath S. 2015. Epigenetic age of the pre-frontal cortex is associated with neuritic plaques, amyloid load, and Alzheimer's disease related cognitive functioning. Aging (Albany, NY) 7, 1198–1211. ( 10.18632/aging.100864) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reese SE, et al. 2017. DNA methylation score as a biomarker in newborns for sustained maternal smoking during pregnancy. Environ. Health Perspect. 125, 760–766. ( 10.1289/EHP333) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spiers H, Hannon E, Schalkwyk LC, Smith R, Wong CC, O'Donovan MC, Bray NJ, Mill J. 2015. Methylomic trajectories across human fetal brain development. Genome Res. 25, 338–352. ( 10.1101/gr.180273.114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simpkin AJ, et al. 2015. Longitudinal analysis of DNA methylation associated with birth weight and gestational age. Hum. Mol. Genet. 24, 3752–3763. ( 10.1093/hmg/ddv119) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murray RM, Lewis SW. 1987. Is schizophrenia a neurodevelopmental disorder? Br. Med. J. (Clin. Res. Edn) 295, 681–682. ( 10.1136/bmj.295.6600.681) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weinberger DR. 1987. Implications of normal brain development for the pathogenesis of schizophrenia. Arch. Gen. Psychiatry 44, 660–669. ( 10.1001/archpsyc.1987.01800190080012) [DOI] [PubMed] [Google Scholar]
- 27.Ecker C, Bookheimer SY, Murphy DG. 2015. Neuroimaging in autism spectrum disorder: brain structure and function across the lifespan. Lancet Neurol. 14, 1121–1134. ( 10.1016/S1474-4422(15)00050-2) [DOI] [PubMed] [Google Scholar]
- 28.Maimburg RD, Vaeth M. 2006. Perinatal risk factors and infantile autism. Acta Psychiatr. Scand. 114, 257–264. ( 10.1111/j.1600-0447.2006.00805.x) [DOI] [PubMed] [Google Scholar]
- 29.Gardener H, Spiegelman D, Buka SL. 2011. Perinatal and neonatal risk factors for autism: a comprehensive meta-analysis. Pediatrics 128, 344–355. ( 10.1542/peds.2010-1036) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Larsson HJ, et al. 2005. Risk factors for autism: perinatal factors, parental psychiatric history, and socioeconomic status. Am. J. Epidemiol. 161, 916–925; discussion 926–928 ( 10.1093/aje/kwi123) [DOI] [PubMed] [Google Scholar]
- 31.Elliott HR, et al. 2014. Differences in smoking associated DNA methylation patterns in South Asians and Europeans. Clin. Epigenetics 6, 4 ( 10.1186/1868-7083-6-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hannon E, et al. 2018. Elevated polygenic burden for autism is associated with differential DNA methylation at birth. Genome Med. 10, 19 ( 10.1186/s13073-018-0527-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ladd-Acosta C, et al. 2016. Presence of an epigenetic signature of prenatal cigarette smoke exposure in childhood. Environ. Res. 144, 139–148. ( 10.1016/j.envres.2015.11.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Masuzaki H, et al. 2004. Labor increases maternal DNA contamination in cord blood. Clin. Chem. 50, 1709–1711. ( 10.1373/clinchem.2004.036517) [DOI] [PubMed] [Google Scholar]
- 35.Bauer M, Orescovic I, Schoell WM, Bianchi DW, Pertl B. 2002. Detection of maternal deoxyribonucleic acid in umbilical cord plasma by using fluorescent polymerase chain reaction amplification of short tandem repeat sequences. Am. J. Obstet. Gynecol. 186, 117–120. ( 10.1067/mob.2002.118306) [DOI] [PubMed] [Google Scholar]
- 36.Pedersen CB, et al. 2017. The iPSYCH2012 case-cohort sample: new directions for unravelling genetic and environmental architectures of severe mental disorders. Mol. Psychiatry 23, 6–14. ( 10.1038/mp.2017.196) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nørgaard-Pedersen B, Hougaard DM. 2007. Storage policies and use of the Danish Newborn Screening Biobank. J. Inherit. Metab. Dis. 30, 530–536. ( 10.1007/s10545-007-0631-x) [DOI] [PubMed] [Google Scholar]
- 38.Pidsley R, Wong CCY, Volta M, Lunnon K, Mill J, Schalkwyk LC. 2013. A data-driven approach to preprocessing Illumina 450K methylation array data. BMC Genomics 14, 293 ( 10.1186/1471-2164-14-293) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Houseman EA, et al. 2012. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics 13, 86 ( 10.1186/1471-2105-13-86) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koestler DC, et al. 2013. Blood-based profiles of DNA methylation predict the underlying distribution of cell types: a validation analysis. Epigenetics 8, 816–826. ( 10.4161/epi.25430) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.R Development Core Team. 2008. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; See http://www.R-project.org/. [Google Scholar]
- 42.Baron RM, Kenny DA. 1986. The moderator-mediator variable distinction in social psychological research: conceptual, strategic, and statistical considerations. J. Pers. Soc. Psychol. 51, 1173–1182. ( 10.1037/0022-3514.51.6.1173) [DOI] [PubMed] [Google Scholar]
- 43.Sobel ME. 1982. Asymptotic confidence intervals for indirect effects in structural equation models. Sociol. Methodol. 13, 290–312. ( 10.2307/270723) [DOI] [Google Scholar]
- 44.Küchenhoff H, Mwalili SM, Lesaffre E. 2006. A general method for dealing with misclassification in regression: the misclassification SIMEX. Biometrics 62, 85–96. ( 10.1111/j.1541-0420.2005.00396.x) [DOI] [PubMed] [Google Scholar]
- 45.Durmus B, et al. 2011. Parental smoking during pregnancy, early growth, and risk of obesity in preschool children: the Generation R Study. Am. J. Clin. Nutr. 94, 164–171. ( 10.3945/ajcn.110.009225) [DOI] [PubMed] [Google Scholar]
- 46.Windham GC, Hopkins B, Fenster L, Swan SH. 2000. Prenatal active or passive tobacco smoke exposure and the risk of preterm delivery or low birth weight. Epidemiology 11, 427–433. ( 10.1097/00001648-200007000-00011) [DOI] [PubMed] [Google Scholar]
- 47.Dietz PM, Homa D, England LJ, Burley K, Tong VT, Dube SR, Dube SR, Bernert JT. 2011. Estimates of nondisclosure of cigarette smoking among pregnant and nonpregnant women of reproductive age in the United States. Am. J. Epidemiol. 173, 355–359. ( 10.1093/aje/kwq381) [DOI] [PubMed] [Google Scholar]
- 48.Agha G, et al. 2016. Birth weight-for-gestational age is associated with DNA methylation at birth and in childhood. Clin. Epigenetics 8, 118 ( 10.1186/s13148-016-0285-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Non AL, Binder AM, Kubzansky LD, Michels KB. 2014. Genome-wide DNA methylation in neonates exposed to maternal depression, anxiety, or SSRI medication during pregnancy. Epigenetics 9, 964–972. ( 10.4161/epi.28853) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schroeder JW, et al. 2011. Neonatal DNA methylation patterns associate with gestational age. Epigenetics 6, 1498–1504. ( 10.4161/epi.6.12.18296) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Relton CL, Davey Smith G. 2012. Two-step epigenetic Mendelian randomization: a strategy for establishing the causal role of epigenetic processes in pathways to disease. Int. J. Epidemiol. 41, 161–176. ( 10.1093/ije/dyr233) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tyrrell J, et al. 2016. Genetic evidence for causal relationships between maternal obesity-related traits and birth weight. JAMA 315, 1129–1140. ( 10.1001/jama.2016.1975) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lawlor DA, Relton C, Sattar N, Nelson SM. 2012. Maternal adiposity—a determinant of perinatal and offspring outcomes? Nat. Rev. Endocrinol. 8, 679–688. ( 10.1038/nrendo.2012.176) [DOI] [PubMed] [Google Scholar]
- 54.Metzger BE, et al. 2008. Hyperglycemia and adverse pregnancy outcomes. N. Engl. J. Med. 358, 1991–2002. ( 10.1056/NEJMoa0707943) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Given the nature of the MINERvA cohort, access to data can only be provided through secured systems which comply with the current Danish and EU data standards. To comply with the study's ethical approval, access to the raw data is only available to qualified researchers upon request. All summary statistics and analysis scripts are available directly from the authors (please contact Jonas Bybjerg-Grauholm at JOGR@ssi.dk). R scripts used to perform the quality control of these data are available on GitHub: https://github.com/ejh243/MinervaASDEWAS.git and have been archived in Zenado: https://zenodo.org/badge/latestdoi/116149862 and scripts for the analyses reported in this manuscript are available on GitHub: https://github.com/ejh243/MinervaNeonatalEWAS.git and have been archived in Zenado: https://doi.org/10.5281/zenodo.1303340.