Abstract
Purpose:
The purpose of our study was to investigate the effect of different slice thicknesses and/or interslice gaps on longitudinal and transverse relaxation times (T1 and T2) measured by a multi-dynamic, multi-echo (MDME) sequence.
Materials and Methods:
This retrospective study included nine healthy subjects who underwent MDME sequence (at 3T) with four different combinations of slice thicknesses and/or interslice gaps: slice thickness of 4 mm and interslice gap of 0 mm (TH4/G0), TH4/G1, TH5/G0, and TH5/G1. T1 and T2 were measured in various brain regions by a qualified neuroradiologist with 8 years of clinical experience: the frontal white matter (WM), occipital WM, genu, splenium, frontal cortex, thalamus, putamen, caudate head, and cerebrospinal fluid (CSF). The paired samples t-test was used to investigate the effect of different slice thicknesses and interslice gaps (TH4/G0 versus TH4/G1 and TH5/G0 versus TH5/G1). P < 0.013 was considered statistically significant.
Results:
T2 in all brain regions and T1 in the frontal WM, putamen, and CSF did not significantly change for different slice thicknesses and/or gaps (Ps > 0.013). In addition, T1 in all brain regions of interest did not significantly change between TH4/G0, TH4/G1, TH5/G0 and TH5/G1. However, T1 in some of the brain regions was higher with TH4/G0 than with TH5/G0 (occipital WM, frontal cortex, and caudate head) and with TH4/G1 than with TH5/G1 (occipital WM, genu, splenium and thalamus, all Ps < 0.013).
Conclusion:
T2 estimated using the MDME sequence was stable regardless of slice thickness or gap. Although the sequence seems to provide stable relaxation values, identical slice thicknesses need to be used for follow-up to prevent potential T1 changes.
Keywords: interslice gap, multi-dynamic multi-echo sequence, slice thickness, T1-relaxation time, T2-relaxation time
Introduction
A novel quantitative MRI sequence, the 2D fast spin echo (FSE) multi-dynamic multi-echo (MDME) sequence, enable rapid and simultaneous sampling of physical properties to constitute an MR image.1 Using the sequence, longitudinal and transverse relaxation times (T1 and T2) and proton density (PD) are simultaneously quantified with good accuracy and reproducibility.2 This is achieved by the design of an MDME sequence that consists of two repeated data acquisition phases.1 In the first phase, a slice-selective saturation pulse saturates one slice. In the second phase, a slice-selective excitation pulse and a series of slice-selective refocusing pulses generate a train of spin echoes for another slice. This mismatch between the saturated slice and the image slice allows for different degrees of saturation recovery in a controlled manner. Echo trains with different saturation delays are used to estimate T1 and T2.1 The magnetization at thermal equilibrium (Mo) that is also obtained with T1 via curve fitting is scaled to yield the PD.1 The saturation recovery curve is also used for B1 inhomogeneity correction. The high temporal efficiency provided by the MDME sequence can greatly facilitate the clinical application of quantitative MRI. In addition, the resulting absolute quantitative values enable the formation of synthetic images with the intended contrast that is consistent regarding imperfections of the scanner and variations in the pulse sequence.3,4 Although the quality of these synthetic images is perceived to be inferior, diagnosis based on synthetic MRI and conventional MRI has shown good agreement.5,6 Using the MDME sequence, several previous studies reported promising results for multiple sclerosis, brain metastases, idiopathic normal pressure hydrocephalus, and meningitis.7–13
The current MDME sequence, quantification of relaxation times and proton density by multi-echo acquisition of a saturation recovery using turbo spin-echo readout (QRAPMASTER) is an upgraded version of quantification of relaxation times and proton density by twin-echo saturation-recovery turbo-field echo (QRAPTEST) in which the previous small flip angle excitation pulse was replaced with a 90° pulse and a series of 180° refocusing pulses, which were incorporated to generate a train of spin echoes instead of gradient echoes.1,14 The resulting improvements in signal-to-noise ratio (SNR) and the robustness against susceptibility effects are clearly beneficial. However, three different slice-selective radiofrequency (RF) pulses with large flip angles (90°, 120°, and 180°) are indispensably used in the sequence, which may, therefore, be more subject to the imperfection of the RF pulses, particularly at a higher field. Although the MDME sequence allows for simultaneous B1 inhomogeneity correction, residual inhomogeneity may persist at a higher field as previously discussed.15 In clinical practice, it is sometimes necessary to modify the slice thickness and gap due to individual differences in brain size and a limited acquisition time. Consequently, the extent of imperfections in RF pulses may also be variable. While previous studies demonstrated the robust performance of the sequence, the majority of studies have been conducted at 1.5T.1,2,5,7,16,17 In this report, we investigated the effect of varying the slice thickness and/or interslice gap on the estimated T1 and T2 by the MDME sequence in anticipation of further expanded applications of the sequence in neuroimaging. A set of data acquired from healthy subjects at 3T was retrospectively collected where the potential impact of RF pulse imperfections is expected to be more pronounced.18
Materials and Methods
Study population
Our institutional review board approved the study protocol, and informed consent was waived. Ten healthy subjects underwent brain MRI scans with the MDME sequence for health check-ups in October 2015. Data from one subject were excluded due to motion artifacts. Finally, nine subjects (all men; mean age of 24 years; age range of 23–27 years) were included in this study. No abnormal radiologic signs were revealed in the brain MRI scans.
MR examination
All subjects underwent MR examination using a 3T clinical scanner (Discovery MR750w; GE Medical Systems, WI, USA) with a 32-channel head coil. Axial images were acquired using the 2D MDME sequence 1) (flip angles = 120° [saturation], 90° [excitation], and 180° [refocusing]). All subjects were scanned using four combinations of different slice thicknesses (TH:mm) and interslice gaps (G:mm): TH4/G0, TH4/G1, TH5/G0, and TH5/G1. The in-plane resolution was maintained at 0.75 × 0.75 mm2. The scan was set to cover the same range of the brain by adjusting the number of slices. Consequently, TRs were automatically adjusted accordingly. The detailed sequence parameters are shown in Table 1.
Table 1.
Multi-dynamic, Multi-echo sequence parameters
| TH4/G0 | TH4/G1 | TH5/G0 | TH5/G1 | |
|---|---|---|---|---|
| TR (ms) | 6073 | 4510 | 4553 | 4000 |
| TE (ms) | 21.4/85.8 | 21.6/85.8 | 21.6/86.6 | 21.6/86.6 |
| Number of sections | 38 | 30 | 30 | 26 |
| Scan time | 7 min 42 s | 5 min 43 s | 5 min 46 s | 5 min 04 s |
| TI (ms) | Automatically calculated 4 different TI | |||
| FOV (mm2) | 240 × 192 | |||
| Scan matrix | 320 × 256 | |||
| Asset factor | 2 | |||
| Echo-train length | 12 | |||
| Bandwidth (kHz) | 22.73 | |||
| NEX | 1 | |||
G, interslice gaps (mm); NEX, the number of excitations; TI, inversion delay; TH, slice thicknesses (mm).
Data post-processing
Least squares fitting was performed on the signal intensity (S) of each pixel of the images per section to calculate T1 by assuming a single exponential decay according to the following Eq. (1):
| (1) |
where A is the overall intensity scaling factor considering the coil sensitivity, RF chain amplification and voxel volume; α is the applied excitation flip angle; and θ is the saturation pulse flip angle. T2 was also calculated by least squares fitting. Quantitative MRI maps of T1 and T2 were generated using vendor-provided software (SyMRI 7.2; Synthetic MR, Linköping, Sweden).
Image analysis
To accurately and consistently place ROIs in the intended brain regions across the T1 and T2 maps obtained from different slice thicknesses and/or gaps, first, all quantitative maps were downloaded as Digital Imaging and COmmunications in Medicine (DICOM) files after data post-processing using the SyMRI software. Second, a folder of all quantitative maps for each subject was loaded into a DICOM viewer (RadiAnt DICOM Viewer, version 3.4.2; Meixant, Poznan, Poland). The number of slices was 38 for TH4/G0, 30 for TH4/G1 and TH5/G0, and 26 for TH5/G1 to encompass the entire brain. Finally, ROIs were placed in a total of nine brain regions by a qualified neuroradiologist with 8 years of clinical experience: the frontal white matter (WM), occipital WM, genu, splenium, frontal cortex, thalamus, putamen, head of the caudate nucleus, and cerebral spinal fluid (CSF). All quantitative maps were adjusted to show the same slice and magnified focusing on the same brain region to minimize the partial volume effect. An ROI was drawn on each of the most representative regions in the T1 maps with TH4/G0 and copied and pasted onto the remaining quantitative maps for each subject (Figs. 1 and 2). The positions of the pasted ROIs were examined, and, if necessary, carefully adjusted. A mean pixel value was obtained for each ROI. As a result, four T1 and four T2 values were simultaneously obtained for each of the nine brain regions.
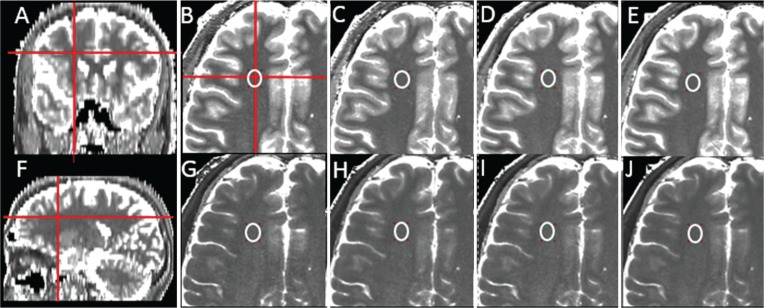
Fig. 1.
Representative T1 and T2 maps from a subject and the location of the ROI for the frontal white matter (WM). (A and F) coronal and sagittal T1 maps (generated from the axial T1 maps by the RadiAnt DICOM Viewer; Meixant, Poznan, Poland). (B–E) magnified axial T1 maps. (G–J) magnified axial T2 maps (B and G) slice thickness of 4 mm and interslice gap of 0 mm (TH4/G0), (C and H) TH4/G1, (D and I) TH5/G0, and (E and J) TH5/G1. The ROI was carefully drawn to avoid the partial volume effect on the axial T1 map with TH4/G0 (B) and copied and pasted on the rest of the T1 (C–E) and T2 (G–J) maps. These pasted ROIs were further adjusted if necessary. The mean T1 and T2 values in the frontal WM were not significantly different for the four slice groups with different thicknesses and gaps.
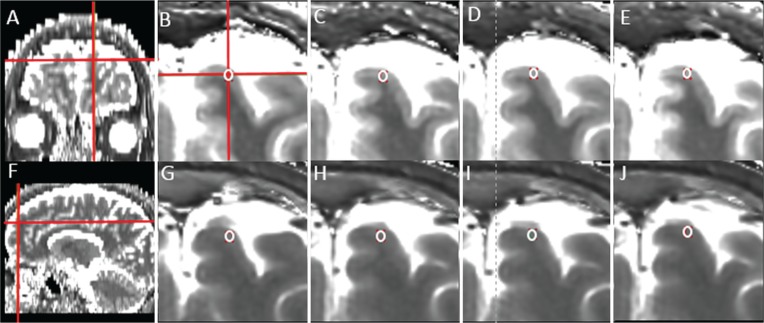
Fig. 2.
Representative T1 and T2 maps from the same subject shown in Fig. 1 and the location of the ROI for the frontal cortex. (A and F) coronal and sagittal T1 maps (generated from the axial T1 maps by the RadiAnt DICOM Viewer; Meixant, Poznan, Poland). (B–E) magnified axial T1 maps. (G–J) magnified axial T2 maps (B and G) slice thickness of 4 mm and interslice gap of 0 mm (TH4/G0), (C and H) TH4/G1, (D and I) TH5/G0, and (E and J) TH5/G1. The ROI was carefully drawn to avoid the partial volume effect on the axial T1 map with TH4/G0 (B) and copied and pasted on the other T1 (C–E) and T2 (G–J) maps. These pasted ROIs were further adjusted, if necessary. The mean T2 values in the frontal cortex were not significantly different for the four slice groups with different thicknesses and gaps, whereas the mean T1 values were significantly different between TH4/G1 and TH5/G1.
Statistical analysis
Continuous data were expressed as mean ± standard deviation (SD). The Kolmogorov–Smirnov test was applied to the quantitative values for normality (P < 0.05 indicates a non-normal distribution). If data distributions were normal (P- value > 0.05), the paired samples t-test was used to investigate the effect of different slice thicknesses (TH4/G0 versus TH5/G0 and TH4/G1 versus TH5/G1) and interslice gaps (TH4/G0 versus TH4/G1 and TH5/G0 versus TH5/G1). If the data distributions were non-normally distributed (P-value ≤ 0.05), the Wilcoxon signed rank test (paired) was used. The significance level for paired samples t-test or Wilcoxon signed rank test was adjusted to P < 0.013 for the Bonferroni correction. Statistical analyses were performed using commercially available software (IBM SPSS Statistics, version 20 [IBM Corporation, Armonk, NY, USA]).
Results
Figures 1 and 2 show representative T1 and T2 maps of the brain from a subject. The high SNR and contrast in the quantitative maps are clearly shown for all combinations of slice thicknesses and interslice gaps. Representative locations of the ROIs are also shown for the frontal WM (Fig. 1) and frontal cortex (Fig. 2), which are the largest (∼122 pixels) and the smallest (∼6 pixels) in ROI size, respectively. The potential effect of the partial volume is clearly negligible.
Table 2 summarizes the T1 and T2 values for the nine regions of the brain for the four slice groups. Data distributions of all brain regions except the CSF were normal (Ps > 0.05, Kolmogorov–Smirnov test). The P-values calculated from the paired samples t-tests or the Wilcoxon signed rank test between the slice groups are summarized in Table 3.
Table 2.
T1 and T2 for the four combinations of slice thicknesses and interslice gaps
| ROI size (mm2) | T1 relaxation time (ms) | T2 relaxation time (ms) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| TH4/G0 | TH4/G1 | TH5/G0 | TH5/G1 | TH4/G0 | TH4/G1 | TH5/G0 | TH5/G1 | ||
| Frontal WM | 68.6 ± 32.5 | 734 ± 33 | 722 ± 20 | 700 ± 33 | 690 ± 23 | 68 ± 5 | 67 ± 4 | 67 ± 5 | 66 ± 4 |
| Occipital WM | 19.9 ± 6.3 | 725 ± 32 | 721 ± 31 | 686 ± 27 | 687 ± 29 | 75 ± 5 | 75 ± 2 | 75 ± 2 | 75 ± 4 |
| Genu | 25.0 ± 10.5 | 641 ± 43 | 640 ± 41 | 609 ± 29 | 608 ± 44 | 62 ± 2 | 61 ± 3 | 61 ± 2 | 60 ± 2 |
| Splenium | 31.6 ± 5.6 | 717 ± 42 | 705 ± 19 | 681 ± 34 | 656 ± 34 | 70 ± 3 | 69 ± 2 | 69 ± 3 | 68 ± 2 |
| Frontal cortex | 3.4 ± 2.4 | 1206 ± 108 | 1254 ± 158 | 1135 ± 99 | 1138 ± 97 | 82 ± 5 | 81 ± 5 | 82 ± 6 | 79 ± 5 |
| Thalamus | 64.3 ± 10.9 | 913 ± 33 | 897 ± 32 | 872 ± 39 | 870 ± 38 | 66 ± 3 | 66 ± 3 | 66 ± 3 | 66 ± 2 |
| Putamen | 38.3 ± 11.9 | 1142 ± 53 | 1119 ± 49 | 1097 ± 43 | 1094 ± 58 | 65 ± 2 | 65 ± 2 | 65 ± 2 | 64 ± 3 |
| Head of caudate nucleus | 18.5 ± 5.5 | 1153 ± 64 | 1142 ± 71 | 1101 ± 57 | 1097 ± 46 | 70 ± 2 | 69 ± 2 | 70 ± 2 | 69 ± 2 |
| CSF | 26.1 ± 14.3 | 4088 ± 158 | 4154 ± 43 | 4152 ± 38 | 4141 ± 18 | 1359 ± 401 | 1024 ± 392 | 1125 ± 483 | 914 ± 336 |
Data are means ± standard deviation. CSF, cerebral spinal fluid; G, interslice gaps (mm); TH, slice thicknesses (mm); WM, white matter.
Table 3.
Results of the paired samples t-tests
| T1 relaxation time | T2 relaxation time | |||||||
|---|---|---|---|---|---|---|---|---|
| TH4/G0 versus TH4/G1 | TH5/G0 versus TH5/G1 | TH4/G0 versus TH5/G0 | TH4/G1 versus TH5/G1 | TH4/G0 versus TH4/G1 | TH5/G0 versus TH5/G1 | TH4/G0 versus TH5/G0 | TH4/G1 versus TH5/G1 | |
| Frontal WM | 0.370 | 0.366 | 0.043 | 0.026 | 0.237 | 0.174 | 0.426 | 0.174 |
| Occipital WM | 0.609 | 0.794 | 0.003a | 0.002a | 0.860 | 0.642 | 0.638 | 0.642 |
| Genu | 0.905 | 0.984 | 0.044 | 0.009a | 0.332 | 0.244 | 0.081 | 0.244 |
| Splenium | 0.527 | 0.028 | 0.055 | 0.004a | 0.362 | 0.408 | 0.023 | 0.408 |
| Frontal cortex | 0.136 | 0.912 | 0.006a | 0.021 | 0.383 | 0.104 | 0.953 | 0.104 |
| Thalamus | 0.268 | 0.807 | 0.015 | 0.004a | 0.247 | 0.195 | 0.886 | 0.195 |
| Putamen | 0.238 | 0.868 | 0.033 | 0.247 | 0.645 | 0.179 | 1.000 | 0.179 |
| Head of caudate nucleus | 0.562 | 0.809 | 0.007a | 0.076 | 0.499 | 0.360 | 0.777 | 0.360 |
| CSF | 0.373 | 0.859 | 0.373 | 1.000 | 0.038 | 0.110 | 0.110 | 0.441 |
Adjusted P < 0.013 values were considered statistically significant. The P values were calculated from the paired samples t-tests (all brain regions except the cerebral spinal fluid [CSF]) or the Wilcoxon signed rank test (CSF). G, interslice gaps (mm); TH, slice thicknesses (mm); WM, white matter.
For all brain regions, T2 did not differ between any of the slice groups (Ps > 0.013). For all brain regions, T1 did not differ significantly between TH5/G0 and TH5/G1 and between TH4/G0 and TH4/G1 (Ps > 0.013), despite the large difference in TR for the latter. However, the use of different slice thicknesses influenced the T1 values for various brain regions. The T1 values of the occipital WM, frontal cortex and head of caudate nucleus were significantly higher with TH4/G0 than with TH5/G0 (P = 0.003 for the occipital WM, P = 0.006 for the frontal cortex and P = 0.007 for the head of caudate nucleus). In addition, the T1 values of the occipital WM, genu, splenium, and thalamus were found to be higher with TH4/G1 than with TH5/G1 (P = 0.002 for the occipital WM, P = 0.009 for the genu, P = 0.004 for the splenium and P = 0.004 for the thalamus). Box-and-whisker representations in Fig. 3 illustrated the T1 and T2 values in the frontal WM and occipital WM with different slice thicknesses and/or interslice gaps.
Fig. 3.
Box-and-whisker plots show the mean and range of the T1 and T2 relaxation times in the frontal white matter (WM). (A and B) and occipital WM (C and D) with different slice thicknesses and/or interslice gaps. Although the T2 values were robust regardless of the slice thicknesses and presence of interslice gaps in both regions, significantly different T1 values were obtained in the occipital WM upon increasing the slice thickness. G, interslice gaps (mm); TH, slice thicknesses (mm).
Discussion
The MDME sequence enabled simultaneous acquisition of the tissue relaxation parameters T1 and T2, whereas most of the past quantitative MRI methods allowed only one parameter per scan. One advantage of this method is that the quantitative MRI results are in absolute values for tissue properties, which are independent of scanners or variations in the pulse sequence.
The MDME sequence used in our study employs three different RF pulses for saturation, excitation, and refocusing with relatively large flip angles of 120°, 90° and 180°, respectively. In a prior article,1 Warntjes et al. thoroughly investigated the impact of the resulting imperfections of the pulse profiles on the estimation of T1, T2, and PD with different uses of an agarose phantom and simulations at 1.5T. The fitting algorithms for quantification were also modified accordingly by fine-tuning the intensity of the echoes for the T2 correction and the relationship between the effective flip angles of the saturation and excitation pulses for PD estimation.1 The promising applicability of the sequence in neuroimaging has been previously reported.7–13 However, to our knowledge, the majority of previous studies investigated the robustness of the sequence at 1.5T.1,2,5,7,16,17 Given that the potential impact of the imperfection of RF pulses on quantitative MRI can be more pronounced at a higher field,15,18 we investigated the effect of differences in slice thickness (4 versus 5 mm) and interslice gap (with and without a 1-mm gap) at 3T. For further expansion of the clinical application of the MDME sequence, the effect of such fundamental sequence variations, such as the slice thickness and interslice gap, needs to be investigated at a higher field. This was the motivation for our study. A previous study reported comparable performances for sequences at 1.5 and 3T; however, these were performed only qualitatively based on the quality of synthesized images (or semi-quantitatively based on 5-level image quality scores) not quantitatively in terms of the actually measured T1 and T2 values.19 To exclude the partial volume effect in our analysis, all ROIs were carefully selected.
Our T1 and T2 results at 3T are comparable with previous findings (T1 of gray matter [GM]: 1200–1820 ms, T1 of WM: 650–847 ms, T2 of GM: 71–99 ms and T2 of WM: 56–69 ms).18–22 In response to the presence/absence of interslice gaps, we did not find significant difference in T1 and T2 values for all brain regions. Therefore, although further investigation is required with a larger number of subjects, the effect of cross-talk may not result in substantial quantitative errors in the T1 and T2 estimation for slice thicknesses of less than 5 mm. Our results support the advantage of the linear slice order employed by the MDME sequence instead of the standard interleaved slice order.1 Our comparison of the T1 and T2 values for TH4/G0 and TH4/G1 also demonstrates that the influence of the large difference in TR is minimal.
By increasing the slice thickness from TH4/G0 to TH5/G0, significantly different T1 values were obtained in the occipital WM, frontal cortex, and head of caudate nucleus. Given that the ROI sizes of these brain regions are the three smallest, the different SNRs of the images for the two slice groups could be responsible for the different T1 values. Upon the negligible effects of partial volume and cross-talk, these spatially dependent changes in T1 in response to the increased slice thickness might also be due to spatially dependent residual inhomogeneity of B1 at 3T as previously discussed.15 By increasing the slice thickness from TH4/G1 to TH5/G1 in the presence of interslice gaps, significantly different T1 values were obtained in the occipital WM, genu, splenium, and thalamus. These brain regions do not coincide with those for TH4/G0 versus TH5/G0, and they include those with a relatively large ROI size, such as the thalamus. These spatially dependent changes in T1 might also be due to spatially dependent residual B1 inhomogeneity. Given the proximity of these brain regions to the ventricle, flow artifacts may also be responsible for the different T1 values. While the brain coverages are all identical for the four different slice groups, the coverages of the individual slices are all different for the different slice groups due to different slice thicknesses and the presence/absence of interslice gaps. Therefore, T1 changes in response to the increased slice thickness with and without interslice gaps can occur in different brain regions due to the presence of factors that render the performance of the sequence spatially dependent, such as residual B1 inhomogeneity and flow artifacts.
In our study, although the T1 values for several brain regions were measured differently in response to increased slice thicknesses, the T2 values were robust regardless of the slice thickness or presence of interslice gaps. Thus, our study corroborates the advantage of the MDME sequence, which is that there is no propagation of errors between T1 and T2, even if the quantification is simultaneously performed.1 Several previous studies reported T2 values for various brain pathologies, such as multiple sclerosis,23,24 stroke,25 epilepsy,26 Parkinson’s disease,27 and in a developing preterm brain.28 In terms of the clinical follow-up, the robust performance of the MDME sequence for T2 estimation in our study is quite promising. Finally, the substantially large T1 and T2 values of the CSF did not differ between any of the slice groups. Therefore, our results demonstrate the excellent dynamic range of the sequence using the current setting.
There are several limitations of our study. First, as was noted in the “Methods” section, we covered the same range of the brain for the subjects by adjusting the number of slices. In the current version of the sequence, TR is automatically adjusted according to the number of slices for minimum scan time. As a result, different TRs were used for all four groups. However, the shortest TR used for the TH5/G1 slice group is already as long as 4000 ms at 3T. Therefore, potential quantitative errors resulting from the different TRs can be negligible in most brain tissues. Further shortening of the TR due to a fewer number of slices would increase the weight of the short relaxation components of the total T1 relaxation behavior, and hence, the effective mono-exponential approximation would be somewhat shorter. However, changes in the number of slices and TR result in only minor changes in the echo times of the acquisition (Table 1) and have a negligible influence on the T2 estimation. Second, because our study was a retrospective study, the number of subjects was limited. This may also explain that the previously reported extent of the cross-talk effect of the sequence1 was not observed in our study. Third, we did not compare our results with thinner slices, which might be useful for various CNS diseases. Although a scan with a 3-mm slice thickness was tested in a preliminary study, it was not included in this study due to the poor SNR of the images. Finally, a separate B1 mapping using a gold standard method would have clarified the extent of the potential residual B1 inhomogeneity in T1 estimation, which may not completely be excluded in our study at 3T as discussed above and previously.15 Nonetheless, the simultaneous B1 inhomogeneity correction provided by the MDME sequence is clearly beneficial and of great importance in quantitative MRI.
Conclusion
In conclusion, while the slightly variable T1 values obtained using the MDME sequence for the different slice thicknesses require further investigation with a larger sample size, the robust performance of the sequence for T2 estimation at 3T supports the expanded applicability of the sequence in neuroimaging.
Acknowledgments
This study was supported by a grant from the Korea Healthcare Technology R&D Projects, Ministry for Health, Welfare & Family Affairs (HI16C1111), by the Brain Research Program through the National Research Foundation of Korea funded by the Ministry of Science, ICT & Future Planning (2016M3C7A1914002), by Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Science, ICT & Future Planning (2017R1A2B2006526 and 2017R1A2B2008412), by Creative-Pioneering Researchers Program through Seoul National University, and by Project Code (IBS-R006-D1).
Footnotes
Conflicts of Interest
None. Technical support was provided by a GE Healthcare employee (Moonjung Hwang) regarding the sequence modifications and implementation with our MRI. Authors not associated with GE Healthcare maintained full control of the data at all times.
References
- 1.Warntjes JB, Leinhard OD, West J, Lundberg P. Rapid magnetic resonance quantification on the brain: optimization for clinical usage. Magn Reson Med 2008; 60:320–329. [DOI] [PubMed] [Google Scholar]
- 2.Krauss W, Gunnarsson M, Andersson T, Thunberg P. Accuracy and reproducibility of a quantitative magnetic resonance imaging method for concurrent measurements of tissue relaxation times and proton density. Magn Reson Imaging 2015; 33:584–591. [DOI] [PubMed] [Google Scholar]
- 3.Bobman SA, Riederer SJ, Lee JN, et al. Cerebral magnetic resonance image synthesis. AJNR Am J Neuroradiol 1985; 6:265–269. [PMC free article] [PubMed] [Google Scholar]
- 4.Riederer SJ, Lee JN, Farzaneh F, Wang HZ, Wright RC. Magnetic resonance image synthesis. Clinical implementation. Acta Radiol Suppl 1986; 369:466–468. [PubMed] [Google Scholar]
- 5.Blystad I, Warntjes JB, Smedby O, Landtblom AM, Lundberg P, Larsson EM. Synthetic MRI of the brain in a clinical setting. Acta Radiol 2012; 53:1158–1163. [DOI] [PubMed] [Google Scholar]
- 6.West H, Leach JL, Jones BV, et al. Clinical validation of synthetic brain MRI in children: initial experience. Neuroradiology 2017; 59:43–50. [DOI] [PubMed] [Google Scholar]
- 7.West J, Aalto A, Tisell A, et al. Normal appearing and diffusely abnormal white matter in patients with multiple sclerosis assessed with quantitative MR. PLoS One 2014; 9:e95161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Granberg T, Uppman M, Hashim F, et al. Clinical feasibility of synthetic MRI in multiple sclerosis: a diagnostic and volumetric validation study. AJNR Am J Neuroradiol 2016; 37:1023–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hagiwara A, Hori M, Yokoyama K, et al. Synthetic MRI in the detection of multiple sclerosis plaques. AJNR Am J Neuroradiol 2017; 38:257–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hagiwara A, Hori M, Yokoyama K, et al. Utility of a multiparametric quantitative MRI model that assesses myelin and edema for evaluating plaques, periplaque white matter, and normal-appearing white matter in patients with multiple sclerosis: a feasibility study. AJNR Am J Neuroradiol 2017; 38:237–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hagiwara A, Hori M, Suzuki M, et al. Contrast-enhanced synthetic MRI for the detection of brain metastases. Acta Radiol Open 2016; 5:2058460115626757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Virhammar J, Warntjes M, Laurell K, Larsson EM. Quantitative MRI for rapid and user-independent monitoring of intracranial CSF volume in hydrocephalus. AJNR Am J Neuroradiol 2016; 37:797–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andica C, Hagiwara A, Nakazawa M, et al. Synthetic MR imaging in the diagnosis of bacterial meningitis. Magn Reson Med Sci 2017; 16:91–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Warntjes JB, Dahlqvist O, Lundberg P. Novel method for rapid, simultaneous T1, , and proton density quantification. Magn Reson Med 2007; 57:528–537. [DOI] [PubMed] [Google Scholar]
- 15.West J, Blystad I, Engström M, Warntjes JB, Lundberg P. Application of quantitative MRI for brain tissue segmentation at 1.5 T and 3.0 T field strengths. PLoS One 2013; 8:e74795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ambarki K, Lindqvist T, Wåhlin A, et al. Evaluation of automatic measurement of the intracranial volume based on quantitative MR imaging. AJNR Am J Neuroradiol 2012; 33:1951–1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Warntjes JB, Engström M, Tisell A, Lundberg P. Brain characterization using normalized quantitative magnetic resonance imaging. PLoS One 2013; 8:e70864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang J, Mao W, Qiu M, Smith MB, Constable RT. Factors influencing flip angle mapping in MRI: RF pulse shape, slice-select gradients, off-resonance excitation, and B0 inhomogeneities. Magn Reson Med 2006; 56:463–468. [DOI] [PubMed] [Google Scholar]
- 19.Tanenbaum LN, Tsiouris AJ, Johnson AN, et al. Synthetic MRI for clinical neuroimaging: results of the magnetic resonance image compilation (MAGiC) prospective, multicenter, multireader trial. AJNR Am J Neuroradiol 2017; 38:1103–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ethofer T, Mader I, Seeger U, et al. Comparison of longitudinal metabolite relaxation times in different regions of the human brain at 1.5 and 3 Tesla. Magn Reson Med 2003; 50:1296–1301. [DOI] [PubMed] [Google Scholar]
- 21.Stanisz GJ, Odrobina EE, Pun J, et al. T1, T2 relaxation and magnetization transfer in tissue at 3T. Magn Reson Med 2005; 54:507–512. [DOI] [PubMed] [Google Scholar]
- 22.Wright PJ, Mougin OE, Totman JJ, et al. Water proton T1 measurements in brain tissue at 7, 3, and 1.5 T using IR-EPI, IR-TSE, and MPRAGE: results and optimization. MAGMA 2008; 21:121–130. [DOI] [PubMed] [Google Scholar]
- 23.Larsson HB, Frederiksen J, Kjaer L, Henriksen O, Olesen J. In vivo determination of T1 and T2 in the brain of patients with severe but stable multiple sclerosis. Magn Reson Med 1988; 7:43–55. [DOI] [PubMed] [Google Scholar]
- 24.MacKay A, Whittall K, Adler J, Li D, Paty D, Graeb D. In vivo visualization of myelin water in brain by magnetic resonance. Magn Reson Med 1994; 31:673–677. [DOI] [PubMed] [Google Scholar]
- 25.Bernarding J, Braun J, Hohmann J, et al. Histogram-based characterization of healthy and ischemic brain tissues using multiparametric MR imaging including apparent diffusion coefficient maps and relaxometry. Magn Reson Med 2000; 43:52–61. [DOI] [PubMed] [Google Scholar]
- 26.Bernasconi A, Bernasconi N, Caramanos Z, et al. T2 relaxometry can lateralize mesial temporal lobe epilepsy in patients with normal MRI. Neuroimage 2000; 12:739–746. [DOI] [PubMed] [Google Scholar]
- 27.Vymazal J, Righini A, Brooks RA, et al. T1 and T2 in the brain of healthy subjects, patients with Parkinson disease, and patients with multiple system atrophy: relation to iron content. Radiology 1999; 211:489–495. [DOI] [PubMed] [Google Scholar]
- 28.Bastin ME, Sinha S, Whittle IR, Wardlaw JM. Measurements of water diffusion and T1 values in peritumoural oedematous brain. Neuroreport 2002; 13:1335–1340. [DOI] [PubMed] [Google Scholar]