Abstract
Osteosarcoma (OS) is the most common primary bone tumour in children and adolescents. It is a highly aggressive tumor with a tendency to spread to the lungs, which are the most common site of metastasis. Advanced osteosarcoma patients with metastasis share a poor prognosis. Despite the use of chemotherapy to treat OS, the 5-year overall survival rate for patients has remained unchanged at 65–70% for the past 20 years. In addition, the 5-year survival of patients with a metastatic disease is around 20%, highlighting the need for novel therapeutic targets.
Autophagy is an intracellular degradation process which eliminates and recycles damaged proteins and organelles to improve cell lifespan. In the context of cancer, numerous studies have demonstrated that autophagy is used by tumor cells to repress initial steps of carcinogenesis and/or support the survival and growth of established tumors. In osteosarcoma, autophagy appears to be deregulated and could also act both as a pro or anti-tumoral process. In this manuscript, we aim to review these major findings regarding the role of autophagy in osteosarcoma.
Keywords: Autophagy, Osteosarcoma, Survival, Cell death
1. Introduction
Autophagy is an intracellular degradation pathway identified more than 50 years ago. The 2016 Nobel prize award to Pr Ohsumi for his work on autophagy renewed interest for the involvement of this process in various fields and autophagy is now recognized as a critical process in bone homeostasis [1].
Although autophagy's role in cancer is complex and context-dependent, pharmacological modulation of this process is emerging in clinical trials. However, understanding the role of autophagy within the tumor and its microenvironment is an essential prerequisite.
Osteosarcoma is an aggressive cancer mainly occurring in children and young adults. Osteosarcoma treatment relies on chemotherapy and surgery, leading to a 70% 5-year survival in patients with a non-metastatic disease. Nevertheless, the 5-year survival of patients with a metastatic disease is around 20%, emphasizing the importance to develop new therapeutic strategies. As autophagy modulation could be part of these new options, the aim of the present review is to summarize the major findings regarding the role of autophagy in osteosarcoma.
2. Autophagy, a major degradative pathway
Autophagy is an intracellular degradation process which eliminates and recycles damaged proteins and organelles [2], [3]. An isolation membrane, called a phagophore, is formed in the cytosol to sequester the damaged material. After elongation and closure of the phagophore, a double-membrane vesicle called an autophagosome is generated. Autophagosomes then fuse with lysosomes to generate autolysosomes, in which the content is degraded and recycled. Basal autophagy exists in all eukaryotic cells and exerts a quality control function. Autophagy can also be stimulated by various stresses such as starvation or hypoxia to sustain cell survival [2], [3]. Although autophagy is essentially a prosurvival mechanism, overactivated autophagy can lead to cell death[4]. In addition to its classical degradation role, autophagy was recently shown to participate in some secretion processes [5].
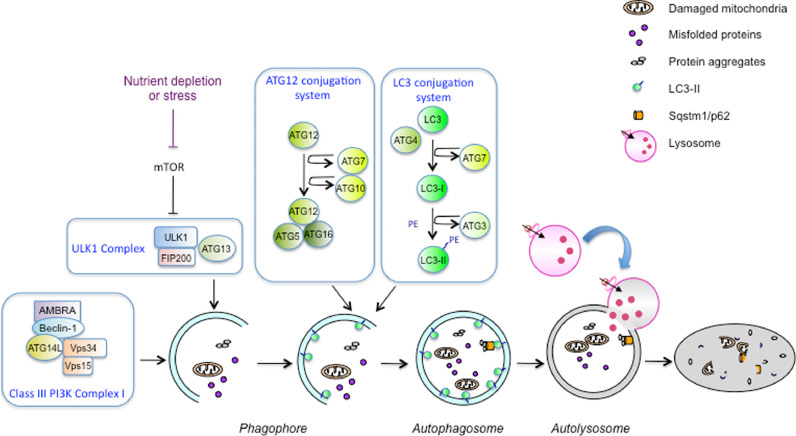
Autophagy is a complex process regulated by the coordinated action of more than 30 autophagy-related proteins (ATG) (Fig. 1). Autophagy initiation is mediated by the UNC-51-like kinase (ULK1) complex which is negatively regulated by the mammalian target of rapamycin (mTOR) in nutrient-rich conditions [6]. Upon starvation, mTOR is inactivated, leading to ULK-1 activation and autophagy induction [7]. The Class III PI3K complex I is then recruited to initiate autophagosome formation. Elongation and closure of the phagophore requires two complexes containing ubiquitin-like proteins and their respective conjugation machineries [8]. The first complex mediates the covalent conjugation of ATG12 to ATG5 due to the action of ATG7 and ATG10 enzymes. The ATG12-ATG5 conjugate then binds to ATG16 to form the ATG12-ATG5/ATG16 complex which is essential for autophagosome biogenesis. The second ubiquitin-like conjugation system allows the conjugation of phosphatidylethanolamine (PE) to the microtubule-associated light chain 3 (LC3 or ATG8). Nascent LC3 is first processed by the protease ATG4, activated by the ATG7 enzyme, transferred to the ATG3 conjugating enzyme, and then conjugated with PE [9]. LC3-PE (LC3-II) is an integral membrane protein present in autophagosomes and is used as an autophagosome marker.
Fig. 1.
Autophagy mechanism. The autophagy mechanism and the different molecular complexes involved in the process are presented. In response to different stimuli such as mTORC1 inactivation, autophagy is initiated through the action of the ULK1 complex and the class III PI3K complex. A phagophore is generated in the cytosol to isolate damaged organelles, aggregates and proteins. The ATG12 conjugation system and the LC3 conjugation system are then involved in the elongation and closure of the phagophore, leading to autophagosome formation. Finally, the autophagosome fuses with lysosomes to degrade the material which will then be recycled.
The cellular material targeted to degradation is sequestered by autophagosomes through the action of selective autophagy receptors such as SQSTM1/p62, NBR1, NDP52 or Optineurin. Due to a specific amino acid sequence binding to ATG8 protein family members (LC3-interacting region or LIR motif), these receptors mediate the cargo delivery to autophagosomes [10], [11].
3. Autophagy in bone
Bone is a complex organ in which several cell types act in a coordinated manner. Bone remodeling starts by the resorption of old mineralized bone matrix by osteoclasts (OC) followed by de novo bone formation by osteoblasts (OB). Osteocytes (OST), the multifunctional mechanosensing cells, which are embedded within the bone matrix, orchestrate this remodeling process.
Autophagy is involved in OB differentiation [12], [13], survival [14], [15] and function, i.e. bone matrix mineralization [13], [16]. In OC, autophagy is also required for differentiation [17], [18], [19] and several autophagic proteins are involved in bone resorption [20]. OST, which are long-lived key regulators of bone remodelling, are also highly dependent on autophagy for their survival and function [21], [22]. Finally, several lines of evidence suggest that an autophagy defect could be related to some bone pathologies such as osteoporosis [23], Paget's disease of bone [24], or osteopetrosis [25].
4. Autophagy in cancer
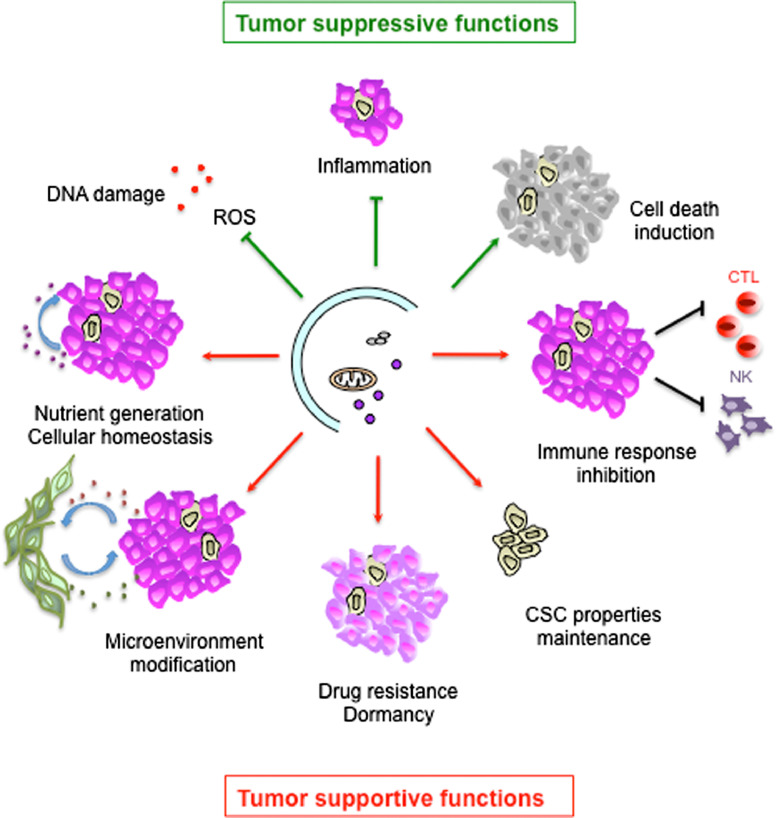
In the context of cancer, numerous studies have demonstrated that autophagy is used by tumor cells as a highly dynamic mechanism to repress initial steps in carcinogenesis and/or support the survival and growth of established tumors [26], [27] (Fig. 2). In the early stages of tumorigenesis, autophagy may be tumor suppressive by (i) limiting chromosomal instability, (ii) restricting oxidative stress which can act as an oncogenic stimulus, (iii) preventing intratumoral necrosis and local inflammation. In advanced cancers, tumor-promoting functions of autophagy rely on survival in response to stresses due to cancer progression, metastasis and treatments [27]. Tumor cells use autophagy to maintain mitochondrial function and homeostasis required by their high energy demand of unrestrained proliferation. In addition, autophagy-mediated secretion can play a role in tumor microenvironment modification [28]. Autophagy has also been involved in the generation of a dormancy state which protects cells from chemotherapeutic stress [29]. Moreover, autophagy was shown to be required in the maintenance of stem cell properties, particularly in cancer stem cells (CSC) [30]. Autophagy is also required for the motility and invasion of metastatic tumor cells by the promotion of focal adhesion disassembly in an autophagic dependent manner via paxilline degradation [31]. Lastly, autophagy can also be used by cancer cells to modify host antitumor immunosurveillance and to inhibit adaptative and innate immune responses [32].
Fig. 2.
Autophagy role in cancer. In early stages of tumor development, autophagy can exert tumor suppressive functions by DNA damage limitation through reactive oxygen species (ROS) elimination, inflammation limitation and cell death induction. In established tumors, autophagy can support tumor growth through various mechanisms such as nutrient generation, microenvironment modification, drug resistance, CSC maintenance promotion and immune response inhibition.
Hence, autophagy exerts context-dependent roles in cancer and most clinical trials, which target established tumors, were performed to inhibit autophagy [33]. These trials, associating autophagy inhibitors with conventional treatments, led to encouraging results but further research is needed to define the safety and utility of this approach in each cancer type [33]. In osteosarcoma, a clinical trial combining the autophagy inhibitor hydroxychloroquine with gemcitabine and docetaxel is presently in progress [34].
5. Autophagy in osteosarcoma
5.1. Autophagy is deregulated in osteosarcoma
Numerous genomic and epigenetic studies have revealed a striking genomic complexity and heterogeneity in osteosarcoma tumors. Several heritable genetic predisposition syndromes with germline mutations in the tumor suppressor genes such as TP53 and RB1 or in RecQL4 gene are associated with osteosarcoma [35], [36]. In addition, altered candidate driver genes with biological evidence for a role in osteosarcoma development have been identified in osteosarcoma somatic genomes [35].
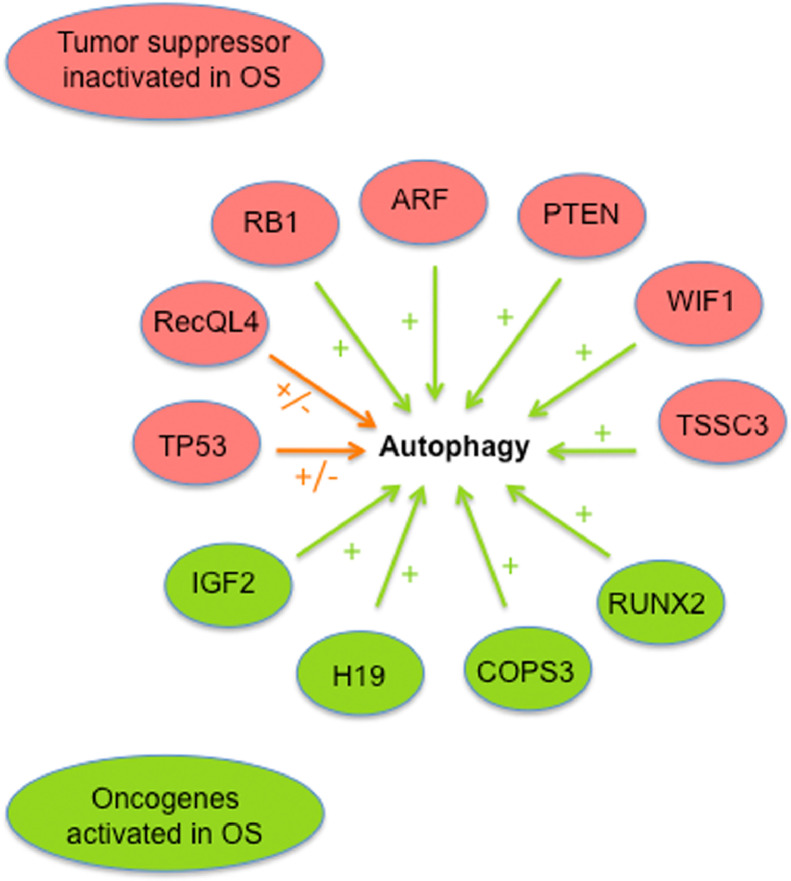
Among the tumor suppressors frequently inactivated in osteosarcoma, several ones have been demonstrated to be involved in autophagy regulation [37], [38] (Fig. 3). Indeed, RB1, ARF, WIF1, PTEN and TSSC3 were shown to trigger autophagy through various mechanisms [39], [40], [41], [42], [43]. TP53 exhibit a dual role in autophagy regulation, nuclear TP53 inducing autophagy through transcriptional regulation and cytoplasmic TP53 acting as a master autophagy repressor [38]. RECQL4 was also recently shown to exert a dual regulation of autophagy [44]. In addition, several oncogenes activated in osteosarcoma such as IGF2, H19, COPS3 and RUNX2 also regulate autophagy [29], [45], [46], [47]. Finally, different studies evidenced that the mTOR pathway was overactivated in osteosarcoma, suggesting autophagy inhibition [48], [49]. Taken together, these data indicate that numerous autophagy regulators are affected in osteosarcoma, suggesting that this critical process is deregulated. Nevertheless, autophagy can be induced in OS and the molecular mechanisms involved in autophagy triggering have been reviewed elsewhere [50].
Fig. 3.
Potential autophagy deregulation in osteosarcoma. Effect on autophagy of tumor suppressors frequently inactivated and oncogenes frequently activated in osteosarcoma. Green arrow: autophagy stimulation; Orange arrow: dual effect on autophagy. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The present review aims at updating our current knowledge regarding the complex pro and antitumoral role of autophagy in OS. Indeed, autophagy can either promote cell survival [29], [46], [51], [52], [53] or contributes to cell death [54]. Moreover, autophagy can be involved in chemosensitivity or chemoresistance during osteosarcoma therapy [54]. In the next paragraphs, we will review several studies illustrating both aspects of autophagy role in osteosarcoma.
5.2. Autophagy as a protumoral process in osteosarcoma
In normal conditions, autophagy is a prosurvival pathway that preserves organelle function, prevents cellular waste product toxicity, and produces energy and substrates for survival. In this context, tumors can use autophagy to survive in a hostile microenvironment and to increase growth and aggressiveness.
5.2.1. Autophagy role in OS tumor cells
The protumoral role of autophagy has been demonstrated in several osteosarcoma cell lines by the knockdown of key autophagy genes. It has been shown that ATG4B, the cysteine proteinase activating LC3, is crucial for osteosarcoma development in the Saos-2 cell model [51]. Indeed, treatment with an ATG4B chemical inhibitor results in autophagy deficiency and leads to a decreased proliferation in vitro and tumor growth in vivo [51]. Similarly, using a siRNA approach in three osteosarcoma cell lines (HOS, MG-63, and U2OS), Zhang et al. have shown that BECN1 knockdown induces a decrease in cell growth and invasion in vitro and reduces metastasis development in vivo [52].
5.2.2. Autophagy role in treatment resistance
In addition to the role of autophagy in osteosarcoma survival and progression, numerous studies have demonstrated its role in treatment resistance [54], [55]. First, several studies demonstrated that chemotherapy-resistant OS cells exhibit increased autophagy [56], [57], [58]. Then, different factors known to mediate drug resistance such as HMGB1, HSP90AA1 or GFRA1, exert their effects through autophagy induction [59], [60], [61]. Moreover, several microRNAs (miRNAs) inhibiting autophagy are downregulated in OS, participating in treatment resistance. miRNAs are small noncoding RNAs that regulate gene expression by binding to the 3′ untranslated region of their target mRNAs, leading to their translational suppression or degradation. MiRNAs are now recognized as major contributors in cancer development and treatment resistance, including in OS [62]. In addition, miRNAs were shown to regulate autophagy [63], and several miRNAs target autophagy genes such as Atg5 [64] or Atg16L1 [65].
MiR-410, which directly decreases the autophagy gene ATG16L1 expression, is markedly downregulated in human osteosarcoma tissues [65]. MiR-30a, which targets BECN1, is significantly reduced in doxorubicin-resistant OS cells [66]. Chemoresistance of osteosarcoma tumor cells to doxorubicin is also associated with the downregulation of miR-143 expression, leading to autophagy activation with upregulation of ATG2B, Bcl-2 and LC3-II protein levels [67]. Similarly, miR-140-5p, which suppresses autophagy through HMGN5, is downregulated in OS and associated with patient's chemoresistance [68]. More recently, it has been shown that some long non-coding RNAs are involved in autophagy activation in OS through miRNA targeting [64], [69].
Finally, autophagy inhibition through BECN1 or Barkor/ATG14 knockdown results in an increased sensitivity of OS cells to chemotherapy [52], [70].
Autophagy involvement in treatment resistance can be mediated through various mechanisms. The protective effect can be mediated by ROS reduction, as it was described for radioresistance of OS cells [71]. Treatment resistance can also involve the degradation of apoptosis-promoting proteins. This is the case in cisplatin-resistant OS cell lines exhibiting autophagic degradation of FOXO3A, which leads to decreased PUMA expression and apoptosis inhibition [56]. Autophagy was also shown to participate in the maintenance of a dormancy-like state protecting OS cells from chemotherapeutic drugs [29]. Finally, Ma et al. have shown in osteosarcoma tissues that a weak level of the autophagy substrate p62, which correlates with an active autophagic flux, may be associated with higher metastatic and chemoresistant rates in osteosarcoma patients, suggesting a protumoral effect of autophagy [72].
Considering the role of autophagy in OS treatment resistance, many studies combined the use of anti-cancer agents with autophagy inhibitors such as chloroquine, 3-methyladenine or spautin-1 (specific and potent autophagy inhibitor-1). These autophagy inhibitors enhance OS cell death induced by cisplatin [73], doxorubicin [74], bone-targeted gallium compound KP46 [75], and increase in vivo tumor regression following photodynamic therapy [76] or mTOR inhibition [77].
5.2.3. Autophagy role in OS cancer stem cells
Autophagy was also shown to participate in the maintenance of cancer stem cell (CSC) properties [78]. Regarding OS, very few studies investigated the role of autophagy in CSC. Zhang et al. have used the CD271 marker to isolate CSC from two human OS cell lines. CD271+ OS cells showed a higher autophagy activity than CD271- OS cells under hypoxia and low nutrient condition. Autophagy deficiency in the CD271+ cells decreased the stemness marker expression, restored chemotherapeutics sensitivity and restricted the advantage of CD271+ OS cells in terms of tumorigenesis in vivo [79]. More recently, calpain-6 was demonstrated to control OS CSC fate by promoting autophagy and preventing senescence [80].
5.3. Autophagy as an antitumoral process in osteosarcoma
While the cytoprotective role of autophagy is well described, particularly in response to cancer treatment, its role in cell death has long been controversial. However, several evidences now demonstrate autophagy implication in cell death, including in osteosarcoma.
Indeed, autophagy can result in an autophagic cell death, defined as an autophagy-mediated cell death that can be suppressed by the inhibition of the autophagic pathway [4], [81]. Such an autophagic cell death was observed in vitro in WT and doxorubicin-resistant U2OS cells after treatment with the bisindolic alkaloid voacamine [82]. Voacamine induced an apoptosis-independent cell death associated with autophagy stimulation, and knockdown of key autophagy genes resulted in decreased cell death [82].
In addition, autophagy can also promote apoptosis or necroptosis through various mechanisms [81]. This can be achieved, for example, through the degradation of anti-apoptotic and cell survival factors [81]. In this context, treatment of U2OS or Saos-2 osteosarcoma cell lines with Riccardin (RD), a naturally occurring macrocyclic bisbibenzyl, triggers both autophagy and apoptosis and pharmacological or genetic autophagy inhibition decreased RD-mediated cell death [83]. Similarly, celastrol, a triterpene from traditional chinese medicine induces JNK activation and ROS generation, resulting in apoptosis and autophagy in MG-63 osteosarcoma cells [84]. Pharmacological autophagy inhibition diminished caspase-3 and PARP cleavage, suggesting that autophagy promoted apoptosis.
Although a large number of studies demonstrate that chemotherapy triggers a cytoprotective autophagy, some of them indicate that autophagy can sensitize OS cells to chemotherapy. Inhibition of camptothecin-induced autophagy was shown to decrease cytotoxicity in DLM8 OS cells and to increase cytotoxicity in K7M3 OS cells [85]. Similarly, autophagy inhibition led to opposite effect in LM7 and CCH—OS-D or K7M3 OS cells [50]. In this last report, the authors showed that the expression of phosphorylated heat shock protein 27 (HSP27) after chemotherapy, predicts the effect of autophagy inhibition on OS cell survival or death.
Interestingly, a recent study of Livingston et al. demonstrated that the presence of LC3B+ puncta, an autophagosome marker, is an independent prognostic biomarker of improved survival following neoadjuvant chemotherapy [86]. This work, which was performed in tumor specimens isolated from 260 osteosarcoma patients, suggests that autophagy could be beneficial in this context.
6. Conclusion
Autophagy is a prosurvival pathway used by OS tumor cells to increase their proliferation and development, resist to cancer treatments and preserve a pool of CSC within the tumor. Nevertheless, autophagy can also be antitumoral in OS and lead to cell death but several general questions remains to be answered: which signal turns prosurvival autophagy into a death process ? Is there an autophagy threshold level required for cell death induction ? We only start to address these issues which are essential to understand the role of autophagy in OS and to use this pathway as a potential weapon against this cancer.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgements
The MATOs team is supported by grants from the France Cancer association, Fondation ARC pour la recherche sur le cancer and Cancéropôle PACA. We apologize to the authors that we were unable to cite due to space limitations.
References
- 1.Pierrefite-Carle V., Santucci-Darmanin S., Breuil V., Camuzard O., Carle G.F. Autophagy in bone: self-eating to stay in balance. Ageing Res. Rev. 2015;24:206–217. doi: 10.1016/j.arr.2015.08.004. [DOI] [PubMed] [Google Scholar]
- 2.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 3.Levine B., Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fulda S., Kögel D. Cell death by autophagy: emerging molecular mechanisms and implications for cancer therapy. Oncogene. 2015;34:5105–5113. doi: 10.1038/onc.2014.458. [DOI] [PubMed] [Google Scholar]
- 5.Ponpuak M., Mandell M.A., Kimura T., Chauhan S., Cleyrat C., Deretic V. Secretory autophagy. Curr. Opin. Cell Biol. 2015;35:106–116. doi: 10.1016/j.ceb.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim J., Kundu M., Viollet B., Guan K.-L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hosokawa N., Hara T., Kaizuka T., Kishi C., Takamura A., Miura Y., Iemura S., Natsume T., Takehana K., Yamada N., Guan J.-L., Oshiro N., Mizushima N. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol. Biol. Cell. 2009;20:1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ohsumi Y., Mizushima N. Two ubiquitin-like conjugation systems essential for autophagy. Semin. Cell Dev. Biol. 2004;15:231–236. doi: 10.1016/j.semcdb.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 9.Shpilka T., Mizushima N., Elazar Z. Ubiquitin-like proteins and autophagy at a glance. J. Cell. Sci. 2012;125:2343–2348. doi: 10.1242/jcs.093757. [DOI] [PubMed] [Google Scholar]
- 10.Mancias J.D., Kimmelman A.C. Mechanisms of Selective Autophagy in Normal Physiology and Cancer. J. Mol. Biol. 2016;428:1659–1680. doi: 10.1016/j.jmb.2016.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khaminets A., Behl C., Dikic I. Ubiquitin-Dependent And Independent Signals In Selective Autophagy. Trends Cell Biol. 2016;26:6–16. doi: 10.1016/j.tcb.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 12.Liu F., Fang F., Yuan H., Yang D., Chen Y., Williams L., Goldstein S.A., Krebsbach P.H., Guan J.-L. Suppression of autophagy by FIP200 deletion leads to osteopenia in mice through the inhibition of osteoblast terminal differentiation. J. Bone Miner. Res. 2013;28:2414–2430. doi: 10.1002/jbmr.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nollet M., Santucci-Darmanin S., Breuil V., Al-Sahlanee R., Cros C., Topi M., Momier D., Samson M., Pagnotta S., Cailleteau L., Battaglia S., Farlay D., Dacquin R., Barois N., Jurdic P., Boivin G., Heymann D., Lafont F., Lu S.S., Dempster D.W., Carle G.F., Pierrefite-Carle V. Autophagy in osteoblasts is involved in mineralization and bone homeostasis. Autophagy. 2014;10:1965–1977. doi: 10.4161/auto.36182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Z., Lai Q., Li Y., Xu C., Tang X., Ci J., Sun S., Xu B., Li Y. Acidic pH environment induces autophagy in osteoblasts. Sci. Rep. 2017;7:46161. doi: 10.1038/srep46161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bartolomé A., López-Herradón A., Portal-Núñez S., García-Aguilar A., Esbrit P., Benito M., Guillén C. Autophagy impairment aggravates the inhibitory effects of high glucose on osteoblast viability and function. Biochem. J. 2013;455:329–337. doi: 10.1042/BJ20130562. [DOI] [PubMed] [Google Scholar]
- 16.Li H., Li D., Ma Z., Qian Z., Kang X., Jin X., Li F., Wang X., Chen Q., Sun H., Wu S. Defective autophagy in osteoblasts induces endoplasmic reticulum stress and causes remarkable bone loss. Autophagy. 2018;14:1726–1741. doi: 10.1080/15548627.2018.1483807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang K., Niu J., Kim H., Kolattukudy P.E. Osteoclast precursor differentiation by MCPIP via oxidative stress, endoplasmic reticulum stress, and autophagy. J. Mol. Cell Biol. 2011;3:360–368. doi: 10.1093/jmcb/mjr021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chung Y.-H., Jang Y., Choi B., Song D.-H., Lee E.-J., Kim S.-M., Song Y., Kang S.-W., Yoon S.-Y., Chang E.-J. Beclin-1 is required for RANKL-induced osteoclast differentiation. J. Cell. Physiol. 2014;229:1963–1971. doi: 10.1002/jcp.24646. [DOI] [PubMed] [Google Scholar]
- 19.Xiu Y., Xu H., Zhao C., Li J., Morita Y., Yao Z., Xing L., Boyce B.F. Chloroquine reduces osteoclastogenesis in murine osteoporosis by preventing TRAF3 degradation. J. Clin. Invest. 2014;124:297–310. doi: 10.1172/JCI66947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeSelm C.J., Miller B.C., Zou W., Beatty W.L., van Meel E., Takahata Y., Klumperman J., Tooze S.A., Teitelbaum S.L., Virgin H.W. Autophagy proteins regulate the secretory component of osteoclastic bone resorption. Dev. Cell. 2011;21:966–974. doi: 10.1016/j.devcel.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Onal M., Piemontese M., Xiong J., Wang Y., Han L., Ye S., Komatsu M., Selig M., Weinstein R.S., Zhao H., Jilka R.L., Almeida M., Manolagas S.C., O'Brien C.A. Suppression of autophagy in osteocytes mimics skeletal aging. J. Biol. Chem. 2013;288:17432–17440. doi: 10.1074/jbc.M112.444190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Piemontese M., Onal M., Xiong J., Han L., Thostenson J.D., Almeida M., O'Brien C.A. Low bone mass and changes in the osteocyte network in mice lacking autophagy in the osteoblast lineage. Sci. Rep. 2016;6:24262. doi: 10.1038/srep24262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Camuzard O., Santucci-Darmanin S., Breuil V., Cros C., Gritsaenko T., Pagnotta S., Cailleteau L., Battaglia S., Panaïa-Ferrari P., Heymann D., Carle G.F., Pierrefite-Carle V. Sex-specific autophagy modulation in osteoblastic lineage: a critical function to counteract bone loss in female. Oncotarget. 2016;7:66416–66428. doi: 10.18632/oncotarget.12013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Daroszewska A., van ’t Hof R.J., Rojas J.A., Layfield R., Landao-Basonga E., Rose L., Rose K., Ralston S.H. A point mutation in the ubiquitin-associated domain of SQSMT1 is sufficient to cause a Paget's disease-like disorder in mice. Hum. Mol. Genet. 2011;20:2734–2744. doi: 10.1093/hmg/ddr172. [DOI] [PubMed] [Google Scholar]
- 25.Ochotny N., Voronov I., Owen C., Aubin J.E., Manolson M.F. The R740S mutation in the V-ATPase a3 subunit results in osteoclast apoptosis and defective early-stage autophagy. J. Cell. Biochem. 2013;114:2823–2833. doi: 10.1002/jcb.24630. [DOI] [PubMed] [Google Scholar]
- 26.Amaravadi R., Kimmelman A.C., White E. Recent insights into the function of autophagy in cancer. Genes Dev. 2016;30:1913–1930. doi: 10.1101/gad.287524.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat. Rev. Cancer. 2012;12:401–410. doi: 10.1038/nrc3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kraya A.A., Piao S., Xu X., Zhang G., Herlyn M., Gimotty P., Levine B., Amaravadi R.K., Speicher D.W. Identification of secreted proteins that reflect autophagy dynamics within tumor cells. Autophagy. 2015;11:60–74. doi: 10.4161/15548627.2014.984273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shimizu T., Sugihara E., Yamaguchi-Iwai S., Tamaki S., Koyama Y., Kamel W., Ueki A., Ishikawa T., Chiyoda T., Osuka S., Onishi N., Ikeda H., Kamei J., Matsuo K., Fukuchi Y., Nagai T., Toguchida J., Toyama Y., Muto A., Saya H. IGF2 preserves osteosarcoma cell survival by creating an autophagic state of dormancy that protects cells against chemotherapeutic stress. Cancer Res. 2014;74:6531–6541. doi: 10.1158/0008-5472.CAN-14-0914. [DOI] [PubMed] [Google Scholar]
- 30.Pan H., Cai N., Li M., Liu G.-H., Izpisua Belmonte J.C. Autophagic control of cell “stemness,”. EMBO Mol. Med. 2013;5:327–331. doi: 10.1002/emmm.201201999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharifi M.N., Mowers E.E., Drake L.E., Collier C., Chen H., Zamora M., Mui S., Macleod K.F. Autophagy promotes focal adhesion disassembly and cell motility of metastatic tumor cells through the direct interaction of Paxillin with LC3. Cell Rep. 2016;15:1660–1672. doi: 10.1016/j.celrep.2016.04.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Viry E., Noman M.Z., Arakelian T., Lequeux A., Chouaib S., Berchem G., Moussay E., Paggetti J., Janji B. Hijacker of the antitumor immune response: autophagy is showing its worst facet. Front Oncol. 2016:6. doi: 10.3389/fonc.2016.00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Levy J.M.M., Towers C.G., Thorburn A. Targeting autophagy in cancer. Nat. Rev. Cancer. 2017;17:528–542. doi: 10.1038/nrc.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gemcitabine, docetaxel, and hydroxychloroquine in treating participants with recurrent or refractory osteosarcoma Full text view - clinicaltrials.gov, (n.d.). https://clinicaltrials.gov/ct2/show/NCT03598595 (Accessed 11 March 2019).
- 35.Kansara M., Teng M.W., Smyth M.J., Thomas D.M. Translational biology of osteosarcoma. Nat. Rev. Cancer. 2014;14:722–735. doi: 10.1038/nrc3838. [DOI] [PubMed] [Google Scholar]
- 36.Gianferante D.M., Mirabello L., Savage S.A. Germline and somatic genetics of osteosarcoma - connecting aetiology, biology and therapy. Nat. Rev. Endocrinol. 2017;13:480–491. doi: 10.1038/nrendo.2017.16. [DOI] [PubMed] [Google Scholar]
- 37.Liu E.Y., Ryan K.M. Autophagy and cancer–issues we need to digest. J. Cell. Sci. 2012;125:2349–2358. doi: 10.1242/jcs.093708. [DOI] [PubMed] [Google Scholar]
- 38.Maiuri M.C., Tasdemir E., Criollo A., Morselli E., Vicencio J.M., Carnuccio R., Kroemer G. Control of autophagy by oncogenes and tumor suppressor genes. Cell Death Differ. 2009;16:87–93. doi: 10.1038/cdd.2008.131. [DOI] [PubMed] [Google Scholar]
- 39.Jiang H., Martin V., Gomez-Manzano C., Johnson D.G., Alonso M., White E., Xu J., McDonnell T.J., Shinojima N., Fueyo J. The RB-E2F1 pathway regulates autophagy. Cancer Res. 2010;70:7882–7893. doi: 10.1158/0008-5472.CAN-10-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pimkina J., Humbey O., Zilfou J.T., Jarnik M., Murphy M.E. ARF induces autophagy by virtue of interaction with Bcl-xl. J. Biol. Chem. 2009;284:2803–2810. doi: 10.1074/jbc.M804705200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luo X., Ye S., Jiang Q., Gong Y., Yuan Y., Hu X., Su X., Zhu W. Wnt inhibitory factor-1-mediated autophagy inhibits Wnt/β-catenin signaling by downregulating dishevelled-2 expression in non-small cell lung cancer cells. Int. J. Oncol. 2018;53:904–914. doi: 10.3892/ijo.2018.4442. [DOI] [PubMed] [Google Scholar]
- 42.Chen J.-H., Zhang P., Chen W.-D., Li D.-D., Wu X.-Q., Deng R., Jiao L., Li X., Ji J., Feng G.-K., Zeng Y.-X., Jiang J.-W., Zhu X.-F. ATM-mediated PTEN phosphorylation promotes PTEN nuclear translocation and autophagy in response to DNA-damaging agents in cancer cells. Autophagy. 2015;11:239–252. doi: 10.1080/15548627.2015.1009767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao G.-S., Gao Z.-R., Zhang Q., Tang X.-F., Lv Y.-F., Zhang Z.-S., Zhang Y., Tan Q.-L., Peng D.-B., Jiang D.-M., Guo Q.-N. TSSC3 promotes autophagy via inactivating the Src-mediated PI3K/Akt/mTOR pathway to suppress tumorigenesis and metastasis in osteosarcoma, and predicts a favorable prognosis. J. Exp. Clin. Cancer Res. 2018;37:188. doi: 10.1186/s13046-018-0856-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duan Y., Fang H. RecQL4 regulates autophagy and apoptosis in U2OS cells. Biochem. Cell Biol. 2016;94:551–559. doi: 10.1139/bcb-2016-0005. [DOI] [PubMed] [Google Scholar]
- 45.Xu J., Xia Y., Zhang H., Guo H., Feng K., Zhang C. Overexpression of long non-coding RNA H19 promotes invasion and autophagy via the PI3K/AKT/mTOR pathways in trophoblast cells. Biomed. Pharmacother. 2018;101:691–697. doi: 10.1016/j.biopha.2018.02.134. [DOI] [PubMed] [Google Scholar]
- 46.Zhang X., Shi H., Yuan X., Jiang P., Qian H., Xu W. Tumor-derived exosomes induce N2 polarization of neutrophils to promote gastric cancer cell migration. Mol. Cancer. 2018;17:146. doi: 10.1186/s12943-018-0898-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tandon M., Othman A.H., Ashok V., Stein G.S., Pratap J. The role of Runx2 in facilitating autophagy in metastatic breast cancer cells. J. Cell. Physiol. 2018;233:559–571. doi: 10.1002/jcp.25916. [DOI] [PubMed] [Google Scholar]
- 48.Hu K., Dai H.-B., Qiu Z.-L. mTOR signaling in osteosarcoma: oncogenesis and therapeutic aspects (Review) Oncol. Rep. 2016;36:1219–1225. doi: 10.3892/or.2016.4922. [DOI] [PubMed] [Google Scholar]
- 49.Ding L., Congwei L., Bei Q., Tao Y., Ruiguo W., Heze Y., Bo D., Zhihong L. mTOR: an attractive therapeutic target for osteosarcoma? Oncotarget. 2016;7:50805–50813. doi: 10.18632/oncotarget.9305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O'Farrill J.S., Gordon N. Autophagy in osteosarcoma. Adv. Exp. Med. Biol. 2014;804:147–160. doi: 10.1007/978-3-319-04843-7_8. [DOI] [PubMed] [Google Scholar]
- 51.Akin D., Wang S.K., Habibzadegah-Tari P., Law B., Ostrov D., Li M., Yin X.-M., Kim J.-S., Horenstein N., Dunn W.A. A novel ATG4B antagonist inhibits autophagy and has a negative impact on osteosarcoma tumors. Autophagy. 2014;10:2021–2035. doi: 10.4161/auto.32229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang W., Li Q., Song C., Lao L. Knockdown of autophagy-related protein 6, Beclin-1, decreases cell growth, invasion, and metastasis and has a positive effect on chemotherapy-induced cytotoxicity in osteosarcoma cells. Tumour Biol. 2015;36:2531–2539. doi: 10.1007/s13277-014-2868-y. [DOI] [PubMed] [Google Scholar]
- 53.Min L., Choy E., Pollock R.E., Tu C., Hornicek F., Duan Z. Autophagy as a potential target for sarcoma treatment. Biochim. Biophys. Acta. 2017;1868:40–50. doi: 10.1016/j.bbcan.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 54.Li J., Yang Z., Li Y., Xia J., Li D., Li H., Ren M., Liao Y., Yu S., Chen Y., Yang Y., Zhang Y. Cell apoptosis, autophagy and necroptosis in osteosarcoma treatment. Oncotarget. 2016;7:44763–44778. doi: 10.18632/oncotarget.8206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.He H., Ni J., Huang J. Molecular mechanisms of chemoresistance in osteosarcoma (Review) Oncol. Lett. 2014;7:1352–1362. doi: 10.3892/ol.2014.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jiang K., Zhang C., Yu B., Chen B., Liu Z., Hou C., Wang F., Shen H., Chen Z. Autophagic degradation of FOXO3a represses the expression of PUMA to block cell apoptosis in cisplatin-resistant osteosarcoma cells. Am. J. Cancer Res. 2017;7:1407–1422. [PMC free article] [PubMed] [Google Scholar]
- 57.Mukherjee S., Dash S., Lohitesh K., Chowdhury R. The dynamic role of autophagy and MAPK signaling in determining cell fate under cisplatin stress in osteosarcoma cells. PLoS One. 2017;12 doi: 10.1371/journal.pone.0179203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guan J., Yuan Z., He J., Wu Z., Liu B., Lin X., Mo L., Mo H. Overexpression of caveolin-1 reduces Taxol resistance in human osteosarcoma cells by attenuating PI3K-Akt-JNK dependent autophagy. Exp. Ther. Med. 2016;12:2815–2822. doi: 10.3892/etm.2016.3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang J., Ni J., Liu K., Yu Y., Xie M., Kang R., Vernon P., Cao L., Tang D. HMGB1 promotes drug resistance in osteosarcoma. Cancer Res. 2012;72:230–238. doi: 10.1158/0008-5472.CAN-11-2001. [DOI] [PubMed] [Google Scholar]
- 60.Xiao X., Wang W., Li Y., Yang D., Li X., Shen C., Liu Y., Ke X., Guo S., Guo Z. HSP90AA1-mediated autophagy promotes drug resistance in osteosarcoma. J. Exp. Clin. Cancer Res. 2018;37:201. doi: 10.1186/s13046-018-0880-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim M., Jung J.-Y., Choi S., Lee H., Morales L.D., Koh J.-T., Kim S.H., Choi Y.-D., Choi C., Slaga T.J., Kim W.J., Kim D.J. GFRA1 promotes cisplatin-induced chemoresistance in osteosarcoma by inducing autophagy. Autophagy. 2017;13:149–168. doi: 10.1080/15548627.2016.1239676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen R., Wang G., Zheng Y., Hua Y., Cai Z. Drug resistance-related microRNAs in osteosarcoma: translating basic evidence into therapeutic strategies. J. Cell. Mol. Med. 2019 doi: 10.1111/jcmm.14064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen L., Zhou Y., Sun Q., Zhou J., Pan H., Sui X. Regulation of autophagy by MiRNAs and their emerging roles in tumorigenesis and cancer treatment. Int. Rev. Cell Mol. Biol. 2017;334:1–26. doi: 10.1016/bs.ircmb.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 64.Gu Z., Hou Z., Zheng L., Wang X., Wu L., Zhang C. LncRNA DICER1-AS1 promotes the proliferation, invasion and autophagy of osteosarcoma cells via miR-30b/ATG5. Biomed. Pharmacother. 2018;104:110–118. doi: 10.1016/j.biopha.2018.04.193. [DOI] [PubMed] [Google Scholar]
- 65.Chen R., Li X., He B., Hu W. MicroRNA-410 regulates autophagy-related gene ATG16L1 expression and enhances chemosensitivity via autophagy inhibition in osteosarcoma. Mol. Med. Rep. 2017;15:1326–1334. doi: 10.3892/mmr.2017.6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xu R., Liu S., Chen H., Lao L. MicroRNA-30a downregulation contributes to chemoresistance of osteosarcoma cells through activating Beclin-1-mediated autophagy. Oncol. Rep. 2016;35:1757–1763. doi: 10.3892/or.2015.4497. [DOI] [PubMed] [Google Scholar]
- 67.Zhou J., Wu S., Chen Y., Zhao J., Zhang K., Wang J., Chen S. microRNA-143 is associated with the survival of ALDH1+CD133+ osteosarcoma cells and the chemoresistance of osteosarcoma. Exp. Biol. Med. (Maywood) 2015;240:867–875. doi: 10.1177/1535370214563893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Meng Y., Gao R., Ma J., Zhao J., Xu E., Wang C., Zhou X. MicroRNA-140-5p regulates osteosarcoma chemoresistance by targeting HMGN5 and autophagy. Sci. Rep. 2017;7:416. doi: 10.1038/s41598-017-00405-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu K., Hou Y., Liu Y., Zheng J. LncRNA SNHG15 contributes to proliferation, invasion and autophagy in osteosarcoma cells by sponging miR-141. J. Biomed. Sci. 2017;24:46. doi: 10.1186/s12929-017-0353-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhao Z., Tao L., Shen C., Liu B., Yang Z., Tao H. Silencing of Barkor/ATG14 sensitizes osteosarcoma cells to cisplatin-induced apoptosis. Int. J. Mol. Med. 2014;33:271–276. doi: 10.3892/ijmm.2013.1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Feng H., Wang J., Chen W., Shan B., Guo Y., Xu J., Wang L., Guo P., Zhang Y. Hypoxia-induced autophagy as an additional mechanism in human osteosarcoma radioresistance. J. Bone Oncol. 2016;5:67–73. doi: 10.1016/j.jbo.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ma H., Li X., Wang J., Hornicek F.J., Garbutt C.C., Chang X., Duan Z. Expression and clinical implication of autophagy-associated protein p62 in osteosarcoma. Oncology. 2018;95:52–60. doi: 10.1159/000487437. [DOI] [PubMed] [Google Scholar]
- 73.Shen C., Wang W., Tao L., Liu B., Yang Z., Tao H. Chloroquine blocks the autophagic process in cisplatin-resistant osteosarcoma cells by regulating the expression of p62/SQSTM1. Int. J. Mol. Med. 2013;32:448–456. doi: 10.3892/ijmm.2013.1399. [DOI] [PubMed] [Google Scholar]
- 74.Schott C.R., Ludwig L., Mutsaers A.J., Foster R.A., Wood G.A. The autophagy inhibitor spautin-1, either alone or combined with doxorubicin, decreases cell survival and colony formation in canine appendicular osteosarcoma cells. PLoS One. 2018;13 doi: 10.1371/journal.pone.0206427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kubista B., Schoefl T., Mayr L., van Schoonhoven S., Heffeter P., Windhager R., Keppler B.K., Berger W. Distinct activity of the bone-targeted gallium compound KP46 against osteosarcoma cells - synergism with autophagy inhibition. J. Exp. Clin. Cancer Res. 2017;36:52. doi: 10.1186/s13046-017-0527-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yu W., Wang Y., Zhu J., Jin L., Liu B., Xia K., Wang J., Gao J., Liang C., Tao H. Autophagy inhibitor enhance ZnPc/BSA nanoparticle induced photodynamic therapy by suppressing PD-L1 expression in osteosarcoma immunotherapy. Biomaterials. 2019;192:128–139. doi: 10.1016/j.biomaterials.2018.11.019. [DOI] [PubMed] [Google Scholar]
- 77.Zhu Y.-R., Zhou X.-Z., Zhu L.-Q., Yao C., Fang J.-F., Zhou F., Deng X.-W., Zhang Y.-Q. The anti-cancer activity of the mTORC1/2 dual inhibitor XL388 in preclinical osteosarcoma models. Oncotarget. 2016;7:49527–49538. doi: 10.18632/oncotarget.10389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Boya P., Codogno P., Rodriguez-Muela N. Autophagy in stem cells: repair, remodelling and metabolic reprogramming. Development. 2018:145. doi: 10.1242/dev.146506. [DOI] [PubMed] [Google Scholar]
- 79.Zhang D., Zhao Q., Sun H., Yin L., Wu J., Xu J., He T., Yang C., Liang C. Defective autophagy leads to the suppression of stem-like features of CD271+ osteosarcoma cells. J. Biomed. Sci. 2016;23:82. doi: 10.1186/s12929-016-0297-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Andrique C., Morardet L., Linares L.K., Cissé M.Y., Merle C., Chibon F., Provot S., Haÿ E., Ea H.-K., Cohen-Solal M., Modrowski D. Calpain-6 controls the fate of sarcoma stem cells by promoting autophagy and preventing senescence. JCI Insight. 2018:3. doi: 10.1172/jci.insight.121225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Doherty J., Baehrecke E.H. Life, death and autophagy. Nat. Cell Biol. 2018;20:1110–1117. doi: 10.1038/s41556-018-0201-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Meschini S., Condello M., Calcabrini A., Marra M., Formisano G., Lista P., De Milito A., Federici E., Arancia G. The plant alkaloid voacamine induces apoptosis-independent autophagic cell death on both sensitive and multidrug resistant human osteosarcoma cells. Autophagy. 2008;4:1020–1033. doi: 10.4161/auto.6952. [DOI] [PubMed] [Google Scholar]
- 83.Wang Y., Ji Y., Hu Z., Jiang H., Zhu F., Yuan H., Lou H. Riccardin D induces cell death by activation of apoptosis and autophagy in osteosarcoma cells. Toxicol. In Vitro. 2013;27:1928–1936. doi: 10.1016/j.tiv.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 84.Li H.-Y., Zhang J., Sun L.-L., Li B.-H., Gao H.-L., Xie T., Zhang N., Ye Z.-M. Celastrol induces apoptosis and autophagy via the ROS/JNK signaling pathway in human osteosarcoma cells: an in vitro and in vivo study. Cell Death Dis. 2015;6:e1604. doi: 10.1038/cddis.2014.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hollomon M.G., Gordon N., Santiago-O'Farrill J.M., Kleinerman E.S. Knockdown of autophagy-related protein 5, ATG5, decreases oxidative stress and has an opposing effect on camptothecin-induced cytotoxicity in osteosarcoma cells. BMC Cancer. 2013;13:500. doi: 10.1186/1471-2407-13-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Livingston J.A., Wang W.-L., Tsai J.-W., Lazar A.J., Leung C.H., Lin H., Advani S., Daw N., Santiago-O'Farrill J., Hollomon M., Gordon N.B., Kleinerman E.S. Analysis of HSP27 and the autophagy marker LC3B+ Puncta following preoperative chemotherapy identifies high-risk osteosarcoma patients. Mol. Cancer Ther. 2018;17:1315–1323. doi: 10.1158/1535-7163.MCT-17-0901. [DOI] [PMC free article] [PubMed] [Google Scholar]