Abstract
Human induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs) are widely used for drug safety and efficacy testing with various techniques, including high content imaging (HCI). Upon drug treatment, a significant number of hiPSC-CMs grown in regular 96-well plates coated with fibronectin detached from the bottom of the plate, complicating data acquisition. Several cell culture configurations were tested to improve cell adherence, and the effects of these configurations on total cell number, separation of feature values between the negative (DMSO 0.1%) and positive (antimycin, staurosporine) controls, scale of feature value differences, and data variability were statistically calculated. hiPSC-CMs were plated on fibronectin- (in “blanket” configuration) or MaxGel- (in “sandwich” configuration) coated plates and covered with a layer of either HydroMatrix or MaxGel 2, 7, or 11d after plating. After a total of 14d in culture, cells were treated with compounds, labeled with four fluorescent dyes (Hoechst, TMRM, NucView, and RedDot), and imaged with GE INCell2000. Based on the statistical parameters calculated, the MaxGel 25% 7d “sandwich” was superior to all other tested conditions when the cells were treated with 0.3 μM antimycin for 2 h and test compounds 10 μM crizotinib and 30 μM amiodarone for 48 h. For staurosporine treatment, the best culturing condition varied between MaxGel “sandwich” systems, depending on which parameters were under consideration. Thus, cell culturing conditions can significantly affect the ability of high content imaging to detect changes in cellular features during compound treatment and should be thoroughly evaluated before committing to compound testing.
Keywords: Human induced pluripotent stem cell-derived cardiomyocytes, hiPSC-CMs, High content imaging, Cell culture
1. Introduction
Human induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs) are gaining traction in preclinical safety testing to screen for adverse cardiac effects of new drug candidates [1,2]. The spontaneously beating hiPSC-CMs are amenable to plate-based assays, making them a preferred tool for measuring functional cardiac activity, including calcium transients [3], impedance, and surface membrane potential [2,4]. Assays including high-content imaging (HCI) have the ability to monitor concomitant changes in subcellular structures, a variety of cellular functions, cell viability, and apoptosis [[5], [6], [7]], providing a comprehensive measure of cardiotoxicity undetectable by functional cardiac electrophysiological screening assays. In addition, HCI assays yield useful information for interpretation of findings and better understanding of the mechanisms of action.
Plate-based assays including HCI require well-adhered, healthy cells with a uniform distribution and minimal well-to-well variability to obtain high quality, in-focus images that allow for a clear interpretation of effects in response to compound treatment. Traditionally, hiPSC-CMs are cultured on clear glass or plastic bottom surfaces coated with commercially available extracellular matrices (ECMs) like fibronectin, laminin, and gelatin. In this study we used commercially available hiPSC-CMs (iCells®) from Cellular Dynamics International that were grown on 96-well cell culture plates coated with fibronectin. Upon treatment with antimycin, a known respiratory chain inhibitor and positive control for mitochondrial membrane potential, we observed cells lifting off in sheets from the bottom of the wells, making it difficult to obtain high quality images. To minimize cell loss during compound treatment and to promote cell adherence to the substrate, we experimented with various commercially available and physiologically relevant extracellular matrices.
The human cardiac ECM is comprised of natural structural polymers like collagen and elastin, molecules that promote adhesion like fibronectin, vitronectin, and laminin, as well as proteins known as proteoglycans [8]. ECMs play an important role in the growth and differentiation of cardiomyocytes. In addition to promoting adherence and providing structural integrity, ECMs are also known to provide important physical and chemical cues for the maturation of stem cells [[9], [10], [11]]. Furthermore, the geometry [12] and stiffness of the matrix [11] have been shown to provide cellular information to promote maturity.
In addition to fibronectin, we experimented with several other individual primary structural and adhesive components of the human ECM such as collagen, laminin, gelatin, and vitronectin. We also explored growing the cells on a variety of 3-dimensional gels, namely, HydroMatrix™, a 3D synthetic crosslinked peptide hydrogel [13], Geltrex®, a murine-derived basement membrane containing laminin, collagen, and other growth factors [14], and MaxGel™, a human-based ECM alternative produced by in vitro co-culture of human fibroblasts and epithelial cells containing collagen, laminin, fibronectin, tenascin, elastin, a number of proteoglycans, and glycosaminoglycans [15].
In addition to growing cells on surfaces coated with an ECM matrix, studies have shown that hiPSCs cultured between two layers of ECM in a “sandwich” format show better differentiation to the cardiac phenotype, demonstrating superior structural and functional performance [10]. To test if this technique would also provide better adherence of cells and limit vertical cell movement during compound treatment in imaging experiments, we studied the effects of growing the cells under a layer of either HydroMatrix™ or MaxGel™. We demonstrated that ECM composition, dilution factor, and timing of top gel layer addition after cell seeding improved not only the adherence of the cells under control conditions in HCI experiments but also the ability to distinguish between responses to negative and positive controls, the scale of changes in the observed cellular features, and well-to-well data variability.
2. Materials and methods
2.1. hiPSC-CMs
hiPSC-CMs were obtained from Cellular Dynamics International (CDI, Madison, WI) and seeded onto 96-well μClear® cell culture plates (Greiner Bio-One North America Inc., Monroe, NC) at 15,000 viable cells/well. In order to test cell adherence to different substrates, prior to cell seeding, the plates were coated with one of the following commercially available ECMs following manufacturer’s recommendations (all received from Sigma, St. Louis, Missouri unless specified otherwise):
-
•
Fibronectin: 10 μg/mL, 50 μg/mL, 100 μg/mL, and 10 μg/mL with CaCl2 (1 mM) and MgCl2 (0.5 mM)
-
•
Collagen: 100 μg/mL
-
•
Gelatin: 0.1%
-
•
Geltrex® (Gibco/ThermoFisher, Waltham, Massachusetts): 1%
-
•
Vitronectin: 0.1 μg/cm2 and 0.4 μg/cm2
-
•
Laminin: 1.5 μg/cm2 and 6 μg/cm2
-
•
HydroMatrix™: 2.5 mg/mL
-
•
MaxGel™: 1%
All of the substrate dilutions were prepared in distilled water, except for fibronectin and MaxGel™ that were prepared in PBS and DMEM, respectively. Geltrex® was used as supplied by the manufacturer without any further dilutions.
To improve cell adherence, different 3-dimensional matrices were tested. In one set of experiments, cells were plated on fibronectin- (10 μg/mL, without Ca2+/Mg2+) coated 96-well plates and covered with a “blanket” layer of either HydroMatrix™ (2.5 mg/mL) or MaxGel™ (10%) after 7 or 11d in culture. Alternatively, cells were seeded on a layer of MaxGel™ at different concentrations (10% and 25%) and then covered with a “sandwich” layer of MaxGel™ at the corresponding concentration after 7d in culture. In a second set of experiments, the cells were plated between the two “sandwich” layers of MaxGel™ (25% and 50%) after 2 or 7d in culture.
To facilitate references throughout the text, culturing conditions are referred to in the text below by bottom layer/top layer composition with hydrogel concentration and timing of top gel layer addition. “Blanket” systems consist of a 2-dimensional thin coating of the plate well bottom with a cell adhesion component, such as fibronectin, and a 3-dimensional hydrogel top layer, whereas “sandwich” systems are comprised of two layers of a 3-dimensional hydrogel.
The cells were maintained for a period of 14d prior to experiments. Maintenance Medium (CDI, referred to as “Medium” in the text below) was exchanged every 2d.
2.2. Compound treatment
Compounds (antimycin, staurosporine, crizotinib, and amiodarone) were obtained from Sigma. Test compound stocks were prepared at 1000X in DMSO or water and then further diluted to 3X with Medium immediately before incubation with the cells. Compound incubation was performed on day 14 of cell culturing. On this day, the medium on the plate was replaced with 50 μL of fresh Medium, and after a brief equilibration period to 37 °C, 3X compound stock solutions were added to the cells to achieve a final concentration.
2.3. Imaging
On the day of imaging, the cells were incubated with a mixture of four fluorescent dyes as shown in Table 1, according to the manufacturer’s recommendations.
Table 1.
Fluorescent dyes used for multiplex image acquisition.
| Dye | Ex/Em (nM) | Targeted Readout (cell region) | Endpoints Measured |
|---|---|---|---|
| Hoechst (Life Technologies) |
350/460 | Nuclei (nucleus) | Was used to count the “number of objects” for well-based analysis and for the measurement of nuclear features such as nuclear intensity, morphology and texture in cell-based analysis |
| NucView (Biotium) |
500/530 | Apoptosis (nucleus) | Changes in intensity indicate caspase 3/7 activation and were used to calculate % of apoptotic cells in well-based analysis |
| TMRM (Life Technologies) |
548/573 | Mitochondria (cytoplasm) | Mitochondrial cell-based analysis of intensity, morphology and texture features. |
| RedDot (Biotium) |
665/695 | Cell Death (nucleus) | Nuclear staining indicates cell death and was used to calculate % of dead cells in well-based analysis and exclude them from cell-based analysis. |
Multiplex images were taken with INCell Analyzer 2000 (GE Healthcare) at two resolutions: 5X for cell count and 20X for cellular feature analysis. 3 fields randomly selected by the software per well were captured, which resulted in a total of ˜1100−1400 cells per well on average (calculated based on untreated and DMSO-treated wells).
2.4. Image analysis
Images were analyzed using Columbus™ software v.2.5.2 (Perkin Elmer Inc.) using custom image analysis routines. Data were exported and further analyzed and visualized with TIBCO®Spotfire v.7.0.1.
The dead cells, identifiable by RedDot staining of the nuclei, were excluded from the cellular feature analysis. Cellular features of live cells were analyzed from the nuclear region (intensity, morphology, texture) and the mitochondrial region (intensity, texture). For a detailed description of how the cellular features were calculated, refer to the Columbus© user manual [16]. Cell-based analysis consisted of 58 features (See Appendix A for a complete list) plus well-based parameters: cell count, % apoptotic cells, and % dead cells.
2.5. Statistical analysis
2.5.1. Data preparation
To improve the robustness of the analysis, the outliers were removed using the Feature-Bagging for Outlier Detection (FBOD) method [17]. This procedure was applied independently to each experimental setup, defined as the combination of cell culturing conditions (ECM composition, hydrogel concentration, and timing of top gel layer addition), treatment compound, and compound concentration. FBOD works by assigning each data point a mean value over multiple Local Outlier Factor (LOF) scores that are computed for random subsets of the data [18]. LOF is based on the concept of local density, which is estimated by the distances of the k nearest neighbors. The LOF score calculates how many times lower a point’s density is than that of its neighbors. Points with substantially lower local densities are marked as outliers. The mean LOF was computed over 10 random subsets of the data to obtain an estimate of the outlier score. Based on empirical evaluations [18], all data points with a score of 2 or higher were removed, which amounted to removing 0.2% of the observations (cells). After the outliers were removed, the feature values were aggregated by computing the feature’s median for each well to streamline the statistical analysis.
To evaluate the assay quality for each experimental setup, two metrics were calculated: the AUC, area under the receiver operating characteristic (ROC) curve, and the robust Z-score.
2.5.2. Area under the ROC (AUC) curve
AUC analysis is a standard method for evaluating the accuracy of diagnostic tests and was adapted to measure the ability of each feature to separate between the positive and negative controls [19]. A threshold value that is subjected to the range of distributions can be used as a classifier, where values less than the threshold are classified as negative control samples. The accuracy of this measure can be described by the confusion matrix shown in Table 2.
Table 2.
The confusion matrix.
| True Classification |
|||
|---|---|---|---|
| + | – | ||
| Assay Classification | + | True Positive (TP) |
False Positive (FP) |
| − | False Negative (FN) |
True Negative (TN) |
|
The ROC curve traces the proportion of true and false positives over the entire range of thresholds between the minimum and maximum values of the control distributions [19]. Specifically, the ROC graphs the sensitivity over 1-specificity, with sensitivity and specificity calculated by Eq. 1
| (1) |
The sensitivity is defined as the probability of correctly classifying points that belong to the positive control distribution, whereas the specificity is the probability of correctly classifying points that belong to the negative control distribution.
The area under the curve (AUC) is a summary value of the ROC that serves as a measure for how well a feature can discriminate between the controls without any assumptions about the underlying distributions [19]. In the case where there is no overlap between the feature values for each class, i.e. the feature separates the controls perfectly, the AUC is 1. On the other hand, a feature that does not distinguish between the classes any better than by random chance will yield a value of 0.5.
The feature dataset with classification labels was input into the ROC and AUC functions of the AUC package v.0.3.0 in R. Since the AUC was defined for a single feature, the individual AUC values were aggregated into an arithmetic mean AUC that measures the overall ability of each experimental setup to separate the controls.
2.5.3. Robust Z-score
The magnitude of feature value differences between the positive and negative controls was measured by a modification of the standard Z-score. The adjusted score calculates the difference between the positive and negative controls normalized by a measure of data dispersion. To best characterize the magnitude, the medians of the control values were standardized by the median absolute deviation (MAD) of the negative control (DMSO):
| (2) |
A Z-score equal to 0 represents an element equal to the median. Z-scores less than or greater than 0 represent elements less than or greater than the median scaled by the MAD. For example, a Z-score equal to 1 means the element is 1 MAD greater than the median.
2.5.4. ANOVA and t-tests
The feature values obtained from the cells treated with the negative control (DMSO) for each experimental setup were compared either by a sequence of t-tests or by analysis of variance (ANOVA). The family-wise error rate (α = 0.05) was controlled by Bonferroni correction. The results are presented in the Appendices B and E.
Furthermore, to count the number of significantly (α = 0.05) different features between the controls in each culturing condition, a t-test was conducted for each feature under each condition. The resulting p-values were adjusted by Bonferroni correction to control the family-wise error rate within each condition. The adjusted p-values are listed in the Appendices C and F. The assumptions of homogeneity of variances and normality of the used parametric tests were evaluated (α = 0.05) by Bartlett and Shapiro-Wilk tests, respectively.
3. Results and discussion
3.1. Cell treatment leads to detachment from culturing substrates
To elicit a known response on mitochondrial membrane potential, the hiPSC-CMs grown on fibronectin-coated plates were treated with the positive control antimycin (1 μM, 2 h), a respiratory chain inhibitor, and compared to negative control conditions. As illustrated in Fig. 1A, the compound treatment resulted in cells detaching from the bottom of the plate, floating in sheets or clusters, and moving out of focus or out of the imaging field. These effects rendered the collection of imaging data impossible.
Fig. 1.
Spontaneously beating hiPSC-CMs grown on fibronectin are susceptible to detachment from substrate in response to compounds. A – Cells were grown on fibronectin- (10 μg/mL) coated plates, labelled with Hoechst and TMRM, treated as indicated on each image, and imaged at 20X magnification. Significant reduction in number of cells after treatment with antimycin is apparent. Detached cells shift out of focus within the imaging field or move out of the field. Representative images are shown. Scale bar – 50 μm. B – Number of cells cultured on different substrates. Cells were imaged at 20X in control conditions (Medium and DMSO 0.1%) and after treatment with antimycin at 1 μM for 2 h. Three imaging fields were collected from each well. The number of live nuclei was summed up from all three fields within each well, and n = 8 wells were averaged for each substrate. Error bars are SEM. * P < 0.05 vs Medium and DMSO controls.  Medium,
Medium,  DMSO,
DMSO,  antimycin.
antimycin.
In order to prevent cell detachment during compound treatment, several different commercially available cell adherence matrices were subsequently tested at different concentrations, as described in the Material and Methods section. Fig.1B shows the counts of live nuclei in wells with Medium only, in the presence of 0.1% DMSO (vehicle), and with antimycin (1 μM, 2 h) treatment. Nuclei count was used as a measure of cell number since object counting is reliable and straightforward in Columbus™. The actual number of cells, however, is likely somewhat lower since about 20% of hiPSC-CMs are bi-nucleated [11]. This minor discrepancy was ignored for the purpose of this experiment, and the actual number of nuclei within the three imaging fields collected per well is referred to as “number of cells/well” and used interchangeably with “number of nuclei” and “cell number” throughout the text.
Cell adherence in control conditions (Medium and DMSO 0.1%) was highly dependent on the substrate. The following is the ranking of substrates from highest to lowest yield of average cells/well:
-
1)
Fibronectin, 10 μg/mL with Ca2+ and Mg2+
-
2)
Fibronectin, 10 μg/mL without Ca2+ and Mg2+
-
3)
Gelatin, 0.1%
-
4)
Fibronectin, 50 μg/mL without Ca2+ and Mg2+
-
5)
Geltrex®, 1%
-
6)
Collagen, 100 μg/mL
-
7)
HydroMatrix, 2.5 mg/mL
-
8)
Vitronectin, 0.1 μg/cm2
-
9)
Vitronectin, 0.4 μg/cm2
-
10)
Laminin, 1.5 μg/cm2
-
11)
Laminin, 6 μg/cm2
-
12)
MaxGel, 1%
-
13)
Fibronectin, 100 μg/mL without Ca2+ and Mg2+
After treatment with antimycin, a significant number of cells were lost due to cell detachment from the bottom of the plate in all tested conditions. The most dramatic losses were with gelatin (83% loss), collagen (78% loss), and fibronectin 10 μg/mL + Ca2+/+Mg2+ (54% loss). The highest cell retention was with fibronectin 10 μg/mL -Ca2+/-Mg2+ (20% loss). Even under the better conditions, a significant number of cells were out of focus (Fig. 1A), which was unacceptable for reliably measuring compound effects on various cellular features. Thus, none of the tested cell adherence substrates provided satisfactory cell retention during treatment.
3.2. Hydrogel “blanket” and “sandwich” culturing conditions improve retention of antimycin-treated cells
To improve cell retention, cells were plated on fibronectin- (10 μg/mL, -Ca2+/-Mg2+) coated 96-well plates and covered with a “blanket” layer of either HydroMatrix (2.5 mg/mL) or MaxGel (10%) after 7 or 11d in culture. In addition, the cells were plated on MaxGel (10% and 25%) and covered with a “sandwich” layer of MaxGel at the corresponding dilution after 7d in culture.
Minimal lateral movement of cells was observed after treatment with antimycin (Fig. 2A). Significant improvement in cell retention was observed with all MaxGel “blanket” and “sandwich” systems in both control conditions and after treatment (Fig. 2B). Since the HydroMatrix “blanket” systems resulted in the lowest number of healthy cells in untreated and DMSO-treated cells, this hydrogel was excluded from further experiments and analysis.
Fig. 2.
Cells covered with a layer of hydrogel are retained in place after treatment. A - Cells grown between two layers of MaxGel (25%, top layer added 7d after cell seeding) were treated as indicated, labeled with Hoechst, and imaged at 20x magnification. Slight lateral shift is evident, but it did not affect cell number or optical focus. Representative images are shown. Scale bar is 50 μm. B - Cells were plated either on fibronectin or a layer of MaxGel at the indicated concentration, covered with a layer of either HydroMatrix or MaxGel after 7 or 11 days in culture, and imaged at 20X in control conditions (medium and DMSO 0.1%) and after treatment with antimycin at 1 μM for 2 h. Each bar represents an average number of nuclei for n = 4–8 wells. Error bars are SEM.  Medium,
Medium,  DMSO,
DMSO,  antimycin.
antimycin.
3.3. Cell culture conditions affect the number of cellular features with significant changes in antimycin-treated hiPSC-CMs
Successful implementation of HCI assays depends on the ability to clearly distinguish between the negative (DMSO) and positive (treatment) controls for each measured cellular feature [20]. We investigated if different “blanket” or “sandwich” culturing conditions affected the detection of changes in cellular features and the scale of these changes. Treatment of hiPSC-CMs with antimycin (1 μM, 2 h) resulted in a decrease to complete fading of the mitochondrial dye TMRM (Fig. 3A), as expected for a compound that diminishes mitochondrial membrane potential. A decrease in size of nuclei and increase in Hoechst nuclear dye fluorescence intensity was also observed, consistent with changes occurring during the early apoptotic stage [21].
Fig. 3.
Changes in cellular features in response to treatment with antimycin vs control (DMSO) depend on cell culture substrate. Cells were plated on fibronectin (10 μg/mL) or MaxGel (25% and 10%), and a top layer of MaxGel was added 7 or 11d after cell seeding as specified below. A – Examples of representative images showing hiPSC-CMs in control and after treatment with antimycin (1 μM, 2 h). Representative images from cells grown in MaxGel/MaxGel 25% 7d are shown. Note smaller and brighter nuclei in cells treated with antimycin, green dots indicating apoptosis, and a red nucleus (arrow) indicating a necrotic cell. Scale bar is 50 μm. B – Groups of cellular features were measured using nuclear and mitochondrial dyes. Each data point is the mean of n = 4–8 wells. 1 – fibronectin/MaxGel 10% 11d; 2 – fibronectin/MaxGel 10% 7d; 3 – MaxGel/MaxGel 10% 7d; 4 – MaxGel/MaxGel 25% 7d. White – Min; Dark – Max. C – Cellular feature AUC fingerprints based on culturing condition after treatment with antimycin (1 μM, 2 h). D – Z-score fingerprints of four different culturing conditions after treatment with antimycin (1 μM, 2 h). The data point not shown for fibronectin/MaxGel 10% 7d is Mitoch_Haralick_ Correlation_2_px = -117. Each point is a median of n = 8 wells. Error bars are median absolute deviation (MAD).  fibronectin/MaxGel 10% 11d;
fibronectin/MaxGel 10% 11d;  fibronectin/MaxGel 10% 7d;
fibronectin/MaxGel 10% 7d;  MaxGel/MaxGel 10% 7d;
MaxGel/MaxGel 10% 7d;  MaxGel/MaxGel 25% 7d.
MaxGel/MaxGel 25% 7d.
Fig. 3B shows a heat map of the experiment described above and shown in Fig. 2. Each column represents a cellular feature measured using Hoechst (nuclear features) or TMRM (mitochondrial features) in control (DMSO) or after treatment with antimycin (1 μM, 2 h). Each row represents a different culturing condition. Only live cells were included in the analysis, as described in the Materials and Methods section. Culturing conditions clearly affected the detection of cellular features both in control and in the presence of antimycin. There were no statistical significant differences for all or most features in control cells when compared between the two “blanket” culturing conditions (58/58 for fibronectin/MaxGel 10% 11d and fibronectin/MaxGel 10% 7d) and between the two “sandwich” conditions (55/58 for MaxGel/MaxGel 10% 7d and MaxGel/MaxGel 25% 7d), as confirmed with the statistical analysis using ANOVA (Appendix B). However, when the features in DMSO-treated cells were compared between the “blanket” and the “sandwich” conditions, there were 12/58 cellular features significantly different between the two conditions (Appendix B, third table). Specifically, nuclear width (measured as Nucleus_Axial_Small_Length) and multiple mitochondrial features characterizing intensity and texture differed, suggesting similar culturing conditions produce similar results. This confirmed that the observed differences between the cellular features measured in different culturing conditions are not due to random experimental variability, but instead, to the effect of the culturing condition on the sensitivity of the assay. These observations are in line with the previously reported effects of both culturing medium and extracellular matrix composition on assay responsiveness and baseline measurements in different cell types such as breast cancer cells, neurons, hepatocytes [22], as well as hiPSC-CMs [23] in both imaging [22] and functional multi-electrode array [23] assays.
Some of the measured cellular features were significantly different when compared between DMSO and antimycin wells in cells grown on fibronectin/MaxGel 10% 7d (13/58 features significantly different) and fibronectin/MaxGel 10% 11d (3/58 features significantly different). The complete list of p-values illustrating the differences between DMSO and antimycin for all cellular features in the four culturing conditions is included in Appendix C. The conditions with the highest number of statistically significant cellular features between DMSO and antimycin were the MaxGel “sandwich” systems (20/58 in MaxGel/MaxGel 10% 7d and 25/58 in MaxGel/MaxGel 25% 7d). This classification is further illustrated in Fig. 3C and Table 3. Area under the curve (AUC) values were calculated, as described in the Materials and Methods section, for each cellular feature by culturing condition, grouped for each monitored cell region, and plotted in Fig. 3C. The AUC fingerprints of the complete set of cellular features for the four culturing conditions were superimposed for comparison. The AUC values were near or equal to 1 for a majority of mitochondrial features, with the exception of the fibronectin/MaxGel 10% 11d culturing condition. These profound changes in the mitochondrial region were expected since mitochondria are the primary target for antimycin.
Table 3.
Mean AUC values for cells treated with antimycin (1 μM, 2 h) grown in four culturing conditions.
| Bottom Coat: |
MaxGel 25% | MaxGel 10% | Fibronectin |
||
|---|---|---|---|---|---|
| Top Coat: |
MaxGel 25% 7d | MaxGel 10% 7d | MaxGel 10% 7d | MaxGel 10% 11d | |
| Organelle | Feature Group | ||||
| N/A | Overall | 0.77 | 0.77 | 0.76 | 0.68 |
| Mitochondria | Overall | 0.98 | 0.99 | 1.0 | 0.84 |
| Intensity | 0.99 | 0.99 | 1.0 | 0.79 | |
| Texture | 0.98 | 0.99 | 1.0 | 0.86 | |
| Nucleus | Overall | 0.66 | 0.65 | 0.63 | 0.60 |
| Morphology | 0.60 | 0.59 | 0.56 | 0.55 | |
| Intensity | 0.79 | 0.79 | 0.75 | 0.65 | |
| Texture | 0.69 | 0.68 | 0.68 | 0.63 | |
Certain nuclear features were also significantly different in antimycin-treated vs non-treated cells, specifically intensity features and features characterizing nuclear texture (Haralick and some SER features). Interestingly, although the nuclei in antimycin-treated cells were clearly smaller than those in DMSO-treated cells (Fig. 3A), the nuclear symmetry features were mostly indistinguishable between the controls in all culturing conditions (AUCs < 0.6).
When all features were taken into consideration, the MaxGel/MaxGel “sandwich” culturing conditions showed wider experimental windows than the “blanket” conditions. In particular, the MaxGel 25% “sandwich” produced the highest mean AUC out of all investigated culturing conditions. The AUC values, both aggregated and grouped by organelle (mitochondria, nucleus) and type of feature (morphology, intensity, texture) for each condition, are shown in Table 3. When only mitochondrial features were considered, the AUCs were very close to 1 (as described above and also illustrated in Fig. 3C) for both “sandwich” conditions and fibronectin/MaxGel 10% 7d. Both mitochondrial intensity and texture features yielded high AUCs. However, nuclear AUCs were overall lower than mitochondrial, with the nuclear intensity features providing the highest AUCs compared to morphology or texture. Therefore, the overall aggregated AUCs were lower than the AUCs considering mitochondrial features only, validating the selection of antimycin as a positive control for mitochondrial effects in hiPSC-CMs.
3.4. Effect of cell culture conditions on the scale of changes in cellular features and data variability in antimycin-treated hiPSC-CMs
In addition to the number of features that undergo detectable changes after treatment, the scale of feature value differences was also taken into consideration when comparing different culturing conditions. A bigger scale of differences with smaller data dispersion is imperative for a more reliable detection of changes in cellular features upon treatment with various compounds. As illustrated by the Z-score fingerprints for the four different culturing conditions (Fig. 3D), MaxGel/MaxGel 25% 7d “sandwich” yielded the largest overall scale of differences between DMSO and antimycin for most cellular features (a sum of absolute Z-score values for 58 individual features = 813) and the lowest variability of data (smallest MADs), as compared to MaxGel/MaxGel 10% 7d (Z-score sum = 635) and the “blanket” conditions (741 for fibronectin/MaxGel 10% 7d and 445 for fibronectin/MaxGel 10% 11d). The fibronectin/MaxGel 10% 11d condition had the biggest MADs.
Taking into account cell retention after treatment, separation between the values of cellular features measured in control and antimycin-treated cells, scale of compound-induced changes in cellular features, and data dispersion, MaxGel/MaxGel 25% 7d was superior to all other tested conditions when the cells were treated with 1 μM antimycin for 2 h, with MaxGel/MaxGel 10% 7d providing similar overall results.
3.5. Further optimization of MaxGel “sandwich” culturing conditions
Since the MaxGel 25% “sandwich” systems performed the best with antimycin, we investigated if higher gel concentration and shorter timing of top gel layer addition in this configuration could further improve results. We introduced three additional treatment compounds that differed in mechanism of action, as detailed below. MaxGel “sandwich” systems were prepared at 25% and 50% concentrations with top gel layer addition either 2 or 7d after seeding. Average numbers of cells/well in DMSO control wells were comparable among the four culturing conditions (Table 4). It is necessary to note that slight variations in cell numbers from well to well can also result from unequal number of cells picked up by the pipette during seeding.
Table 4.
Number of cells per well after treatment with DMSO for 2, 4, 24, and 48 h in four culturing conditions.
| Treatment Time | MaxGel Dilutions: |
|||
|---|---|---|---|---|
| 50% 2d | 50% 7d | 25% 2d | 25% 7d | |
| 2 h | 1259 ± 48 n = 4 |
1210 ± 40 n = 4 |
1449 ± 86 n = 4 |
1431 ± 63 n = 4 |
| 4 h | 1226 ± 63 n = 12 |
1212 ± 39 n = 12 |
1379 ± 47 n = 12 |
1214 ± 72 n = 12 |
| 24 h | 1243 ± 25 n = 3 |
1136 ± 18 n = 4 |
1374 ± 153 n = 4 |
1348 ± 54 n = 4 |
| 48 h | 1245 ± 88 n = 8 |
1183 ± 59 n = 7 |
1322 ± 94 n = 7 |
1134 ± 49 n = 6 |
n = number of wells. Numbers are mean ± SD.
3.5.1. Cell retention in “sandwich” culturing conditions after treatment with antimycin, staurosporine, amiodarone, and crizotinib
The following compounds (concentration; incubation time) were used for treatment: antimycin (0.003, 0.01, 0.03, 0.1 and 0.3 μM; 2 h), staurosporine (0.1, 0.3, 1, 3 and 10 μM; 2 and 24 h), a positive control for apoptosis and cell death, and two test compounds amiodarone (0.3, 1, 3, 10 and 30 μM; 4 and 48 h), a class III antiarrhythmic, and crizotinib (0.3, 1, 3, 10 and 30 μM; 4 and 48 h), a non-selective receptor tyrosine kinase inhibitor.
Summary data containing number of nuclei per well for antimycin, staurosporine, amiodarone, and crizotinib are shown in Appendix D. At 2 and 4 h treatment, MaxGel 50% 2d, 25% 2d, and 25% 7d showed excellent cell retention up to the highest concentration tested. In MaxGel 50% 7d, treatment with four compounds for 2 or 4 h resulted in concentration-dependent cell loss when compared to DMSO wells. Specifically, there was 52% loss with 30 μM amiodarone, 61% loss with 30 μM crizotinib, 45% loss with 10 μM staurosporine, and 32% loss with 0.3 μM antimycin.
Treatment with staurosporine for 24 h led to concentration-dependent cell loss in all four culturing conditions, with the lowest loss in MaxGel 25% 2d (18% loss at 10 μM) and highest loss in MaxGel 25% 7d (73% loss at 10 μM), similar to MaxGel 50% 7d (65% loss at 10 μM). Treatment with amiodarone for 48 h resulted in high cell retention in MaxGel 25% 2d and MaxGel 50% 2d, with minimal cell loss at the highest tested concentration, 30 μM. Moderate cell loss at this concentration was observed in MaxGel 25% 7d (52% loss), with more pronounced concentration-dependent cell loss in MaxGel 50% 7d (82% loss).Treatment with crizotinib at 48 h also resulted in concentration-dependent cell loss in all four culturing conditions, with the highest loss in MaxGel 50% 7d.
Thus, based on cell retention during treatment with three different compounds, MaxGel 25% dilution showed slightly better results than MaxGel 50% dilution, especially at longer treatment time points (24 h and 48 h). It is important to note that the observed decrease in cell numbers during longer term treatment with compounds can be attributed to at least two processes: cell loss due to detachment from the substrate, as mentioned above, and cell death as a result of compound treatment. Given enough time, the dead cells would disintegrate, making them undetectable with fluorescent dyes. Since it’s difficult to distinguish between these two processes with long-term treatments, results from short-term treatments (2 h and 4 h) were considered to be more reliable measures for cell retention.
3.5.2. Cell survival in different “sandwich” culturing conditions
Since cellular feature data was only collected from live cells, it was important to establish whether or not the culturing conditions affected cell survival in control conditions. There was a slight overall decrease in the percentage of live cells with increasing DMSO incubation time. Specifically, the average percentage of cells at 2 h time point was 96–98% compared to the 48 h average of 84–90%. However, there were no statistically significant differences between the percentages of live cells in control wells between the four different culturing conditions (p > 0.05).
3.5.3. The number of cellular features with significant changes in hiPSC-CMs treated with antimycin, staurosporine, amiodarone, and crizotinib
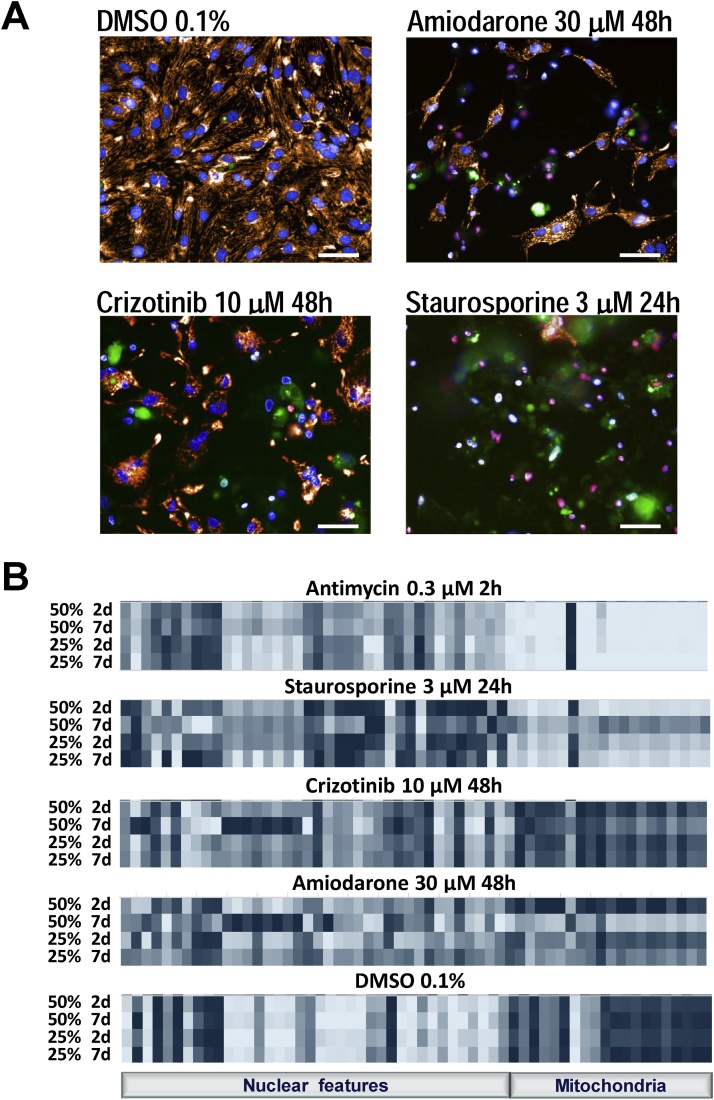
Fig. 4A shows sample images in control (DMSO) and after treatment with the compounds, as indicated. Treatments resulted in a notable decrease in nuclei size, an increase in fluorescence intensity of Hoechst dye, and a decrease to complete fading of TMRM fluorescence. Pink nuclei indicate necrotic cells (excluded from analysis). Amiodarone and crizotinib also resulted in visible changes to mitochondrial texture. All three compounds caused an increase in apoptosis, as evident from the increase in green fluorescence. A summary of changes for all measured cellular features in the four different culturing conditions are shown in Fig.4B. In DMSO-treated wells, most cellular features were not statistically significantly different among the four culturing conditions (Appendix E), except for mitochondrial SER and Haralick features and a few nuclear features at 48 h treatment between MaxGel 25% 2d and 7d.
Fig. 4.
Changes in cellular features in response to treatment with antimycin, staurosporine, amiodarone, and crizotinib, as compared to DMSO 0.1%, are highly dependent on MaxGel dilutions and timing of top gel layer addition. A – Representative examples of composite images showing hiPSC-CMs in control (DMSO 0.1%) and after treatment with compounds as indicated. Images obtained from cells grown on MaxGel 25% with top gel layer added 7d after cell seeding. Scale bar is 50 μm. B – Groups of features measured using nuclear (Hoechst) and mitochondrial (TMRM) dyes. Cells were plated on MaxGel at 25% and 50% concentration with top gel layer addition either 2 or 7d after seeding. Each data point is a mean of live cells in n = 4 wells. Light blue – Min; Dark blue – Max.
The highest number of cellular features with statistically significant differences between DMSO and staurosporine was obtained in the MaxGel 25% culturing conditions (13/58 in MaxGel 25%7d, and 9/58 in MaxGel 25% 2d). The complete list of p-values illustrating the differences between DMSO and staurosporine for all cellular features in the four culturing conditions is included in Appendix F and further illustrated in Fig. 5A.
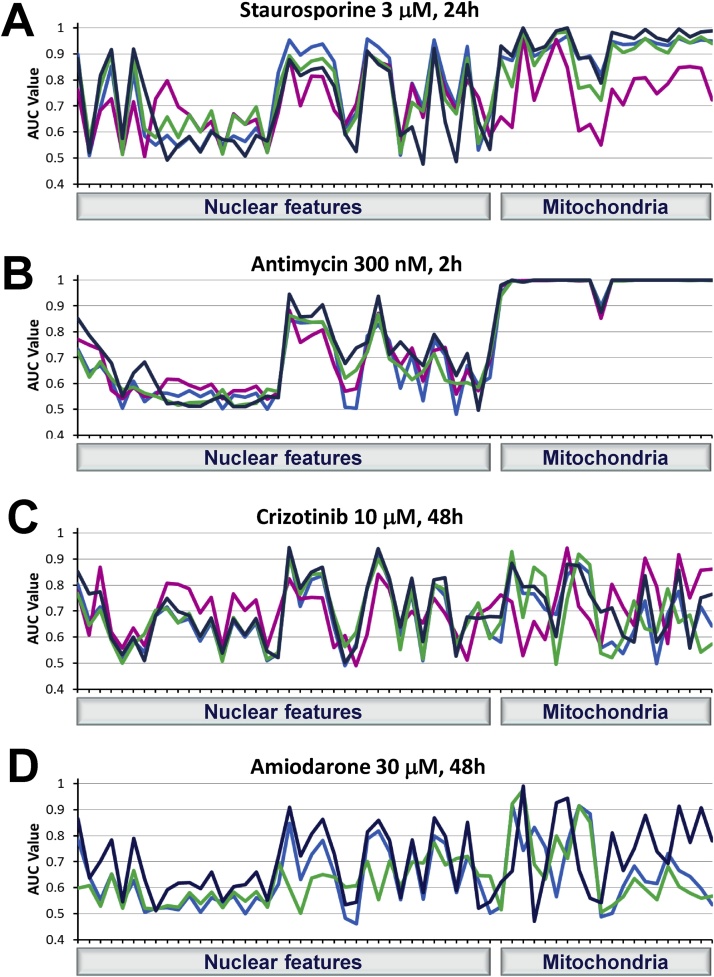
Fig. 5.
AUC fingerprints of cellular features in response to treatment with four compounds are affected by cell culturing conditions. AUC values were calculated for each cellular feature as described in the Materials and Methods section. Groups of cellular features were measured using nuclear and mitochondrial dyes. The fingerprints of the complete set of cellular features in four experimental conditions for each compound were superimposed for comparison. MaxGel 50% 7d AUCs for amiodarone was not calculated due to insufficient number of cells captured in the imaging fields after treatment.  MaxGel 50% 2d;
MaxGel 50% 2d;  MaxGel 50% 7d;
MaxGel 50% 7d;  MaxGel 25% 2d;
MaxGel 25% 2d;  MaxGel 25% 7d.
MaxGel 25% 7d.
3.5.4. The AUC values for cellular features measured in hiPSC-CMs treated with antimycin, staurosporine, amiodarone, and crizotinib in four culturing conditions
Specific groups of features underwent profound changes in both the nuclear and mitochondrial regions after treatment with staurosporine at 3 μM for 24 h in all four culturing conditions. The experimental windows characterized by the AUC values for the nuclear intensity, certain nuclear morphology (profile 5/5, radial relative deviation, radial mean), and certain nuclear texture (Haralick, SER saddle and edge) features were all greater than or near 0.8. The majority of mitochondrial features were also noticeably affected by staurosporine treatment. AUCs were above 0.9 for most of these features in all culturing conditions, with the exception of MaxGel 50% 7d (Fig. 5A). Interestingly, nuclear symmetry features were not significantly affected by staurosporine, with AUCs below 0.7 in all culturing conditions.
The results from treatment with antimycin 300 nM for 2 h (Fig. 5B) are comparable to the ones shown in Fig. 3C (1 μM for 2 h), with strong effects (AUCs ˜ 1) on a majority of mitochondrial features, as well as notable effects (AUC > 0.7) on nuclear intensity and certain nuclear morphology (Profile_5/5) and texture (Haralick homogeneity, Haralick sum variance, and SER saddle) features in all culturing conditions.
Treatment with the two test compounds, crizotinib at 10 μM for 48 h (Fig. 5C) and amiodarone at 30 μM for 48 h (Fig. 5D), resulted in different AUC profiles. A lower number of features was clearly affected (AUCs > 0.7) compared to the positive control compounds, as expected. Interestingly, nuclear compactness and symmetry features had higher AUC values with crizotinib compared to staurosporine, especially in MaxGel 50% 7d.
When the AUCs for 58 individual cellular features were analyzed for four compounds combined, MaxGel 25% 7d yielded the highest values (P < 0.05) among all tested culturing conditions. While MaxGel 50% 7d yielded the lowest AUCs with staurosporine treatment (P < 0.05), MaxGel 25% 2d and 7d and MaxGel 50% 2d were comparable in value (P > 0.05) (Table 5). The similar mean overall mitochondrial AUCs obtained from all four culturing conditions with antimycin (P > 0.05) suggest that mitochondrial responses are independent of culturing condition with this experimental setup. When considering only mitochondrial features, results were similar in nature to those obtained in the previous experiment (Table 3).
Table 5.
Mean AUC values for cells treated with four compounds as indicated and grown in four different experimental conditions.
| Compound | Organelle | Feature Group | MaxGel 50% 2d | MaxGel 50% 7d | MaxGel 25% 2d | MaxGel 25% 7d |
|---|---|---|---|---|---|---|
|
Antimycin, 300 nM, 2 h |
N/A | Overall | 0.76 | 0.77 | 0.77 | 0.79 |
| Mitochondria | Overall | 0.99 | 0.99 | 0.99 | 0.99 | |
| Intensity | 0.99 | 0.99 | 0.99 | 0.99 | ||
| Texture | 0.99 | 0.99 | 0.99 | 0.99 | ||
| Nucleus | Overall | 0.63 | 0.65 | 0.64 | 0.68 | |
| Morphology | 0.58 | 0.60 | 0.57 | 0.60 | ||
| Intensity | 0.78 | 0.75 | 0.78 | 0.81 | ||
| Texture | 0.65 | 0.68 | 0.67 | 0.72 | ||
|
Staurosporine, 3 μM, 24 h |
N/A | Overall | 0.79 | 0.72 | 0.78 | 0.77 |
| Mitochondria | Overall | 0.93 | 0.76 | 0.9 | 0.95 | |
| Intensity | 0.91 | 0.80 | 0.9 | 0.94 | ||
| Texture | 0.93 | 0.75 | 0.9 | 0.95 | ||
| Nucleus | Overall | 0.72 | 0.70 | 0.72 | 0.68 | |
| Morphology | 0.62 | 0.64 | 0.66 | 0.64 | ||
| Intensity | 0.90 | 0.76 | 0.85 | 0.81 | ||
| Texture | 0.76 | 0.74 | 0.73 | 0.68 | ||
|
Crizotinib, 10 μM, 48 h |
N/A | Overall | 0.68 | 0.7 | 0.68 | 0.71 |
| Mitochondria | Overall | 0.7 | 0.74 | 0.69 | 0.73 | |
| Intensity | 0.73 | 0.67 | 0.74 | 0.75 | ||
| Texture | 0.68 | 0.77 | 0.67 | 0.72 | ||
| Nucleus | Overall | 0.67 | 0.69 | 0.69 | 0.69 | |
| Morphology | 0.63 | 0.69 | 0.63 | 0.65 | ||
| Intensity | 0.75 | 0.72 | 0.76 | 0.77 | ||
| Texture | 0.68 | 0.66 | 0.69 | 0.71 | ||
|
Amiodarone, 30 μM, 48 h |
N/A | Overall | 0.64 | N/A | 0.63 | 0.71 |
| Mitochondria | Overall | 0.68 | N/A | 0.67 | 0.75 | |
| Intensity | 0.72 | N/A | 0.76 | 0.72 | ||
| Texture | 0.66 | N/A | 0.63 | 0.76 | ||
| Nucleus | Overall | 0.62 | N/A | 0.61 | 0.68 | |
| Morphology | 0.57 | N/A | 0.56 | 0.64 | ||
| Intensity | 0.71 | N/A | 0.62 | 0.79 | ||
| Texture | 0.66 | N/A | 0.67 | 0.69 |
In the case of staurosporine, the AUCs in the mitochondrial region were higher than those in the nuclear region (P < 0.05) in all four culturing conditions, also evident in Fig. 5A, possibly due to the mode of action of TMRM dye, the intensity (and therefore, the ability to collect data from the mitochondrial region) of which depends on mitochondrial membrane potential. The nuclear intensity and texture features provided the best separation between staurosporine and DMSO-treated cells in this region. Similar results were obtained with crizotinib and amiodarone treatments, with high AUCs in the mitochondrial region. Generally, nuclear AUCs were lower than mitochondrial for all drugs in all four culturing conditions (P < 0.05). Among the nuclear features, intensity features showed the best response for four drugs in all the culturing conditions (P < 0.05), with the exception of MaxGel 25% 2d with amiodarone, where nuclear texture was not statistically significantly different from morphology or intensity (P > 0.05).
Overall, the MaxGel 25% 7d culturing condition showed the widest experimental windows for all treatment compounds with mitochondrial features showing more pronounced effects than nuclear.
3.5.5. The scale of changes in cellular features and data variability in hiPSC-CMs treated with antimycin, staurosporine, amiodarone, and crizotinib
Based on the Z-score fingerprints for the four different culturing conditions with staurosporine (3 μM, 24 h) (Fig.6), the MaxGel 50% 7d “sandwich” yielded the largest overall scale of differences between control and treatment, mostly due to several features having a very high absolute Z-score value and a sum of absolute Z-score values for the 58 individual features = 1643. The MADs were also quite large for 50% 7d matrix. MaxGel 25% 7d provided the second highest sum of absolute Z-score values ( = 1396) with very small MADs for most cellular features. The lowest Z-score sum was obtained for MaxGel 25% 2d ( = 965).
Fig. 6.
Z-score fingerprints of staurosporine (3μM, 24h) in four different cell culturing conditions. Each data point is a median of n = 3–12 wells. Error bars are median absolute deviation (MAD).  MaxGel 50% 2d;
MaxGel 50% 2d;  MaxGel 50% 7d;
MaxGel 50% 7d;  MaxGel 25% 2d;
MaxGel 25% 2d;  MaxGel 25% 7d.
MaxGel 25% 7d.
With respect to cell retention after short-term treatment, separation between the values of cellular features measured in control and treated cells, scale of compound-induced changes in cellular features, and data dispersion, MaxGel/MaxGel 25% 7d was superior to all other tested conditions when the cells were treated with 300 nM antimycin for 2 h, 10 μM crizotinib for 48 h, and 30 μM amiodarone for 48 h. For 3 μM staurosporine treatment at 24 h, the best culturing condition varied depending on which parameter was under consideration. Specifically, the maximal cell retention and lowest MADs were achieved in MaxGel 50% 2d, highest AUC values - in MaxGel 25% 2d and 7d and MaxGel 50% 2d, whereas the maximal number of features significantly different between DMSO and treatment and highest composite Z-score were obtained in MaxGel 50% 7d.
4. Conclusions
The ability to detect maximal changes in cellular features in response to compound treatment is dependent on cell culture substrate composition and timing of top gel layer addition during hiPSC-CM culturing. These conditions need to be carefully optimized depending on which intracellular structures, corresponding features, and biological characteristics are of interest.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Transparency document
Acknowledgements
We would like to sincerely thank Drs. Matthew Kuhls and Amy Aslamkhan for providing access to the imaging equipment and for scientific guidance and generous technical assistance during assay establishment and validation stages. We would also like to thank Edward Lis for preparing the matrices and culturing the cells, Michelle Laussen of Perkin Elmer for invaluable help with imaging data analysis, and Dr. A. Lagrutta for critical reading of the manuscript.
Appendix A.
Intracellular compartments with the corresponding parameters and cellular features analyzed in Columbus™.
| Level | Cellular Region | Fluorescent Dye Used | Parameter | Cellular Feature |
|---|---|---|---|---|
| Cell based | Nucleus | Hoechst | Morphology | Nucleus Profile 5/5 |
| Nucleus Profile 4/5 | ||||
| Nucleus Radial Relative Deviation | ||||
| Nucleus Radial Mean | ||||
| Nucleus Axial Length Ratio | ||||
| Nucleus Axial Small Length | ||||
| Nucleus Threshold Compactness 60% | ||||
| Nucleus Threshold Compactness 50% | ||||
| Nucleus Threshold Compactness 40% | ||||
| Nucleus Threshold Compactness 30% | ||||
| Nucleus Symmetry 15 | ||||
| Nucleus Symmetry 14 | ||||
| Nucleus Symmetry 13 | ||||
| Nucleus Symmetry 12 | ||||
| Nucleus Symmetry 05 | ||||
| Nucleus Symmetry 04 | ||||
| Nucleus Symmetry 03 | ||||
| Nucleus Symmetry 02 | ||||
| Intensity | Intensity Nucleus Contrast | |||
| Intensity Nucleus CV [%] | ||||
| Intensity Nucleus Minimum | ||||
| Intensity Nucleus Maximum | ||||
| Intensity Nucleus StdDev | ||||
| Intensity Nucleus Mean | ||||
| Texture | Nucleus Gabor Max 2 px w4 | |||
| Nucleus Gabor Min 2 px w4 | ||||
| Nucleus Haralick Homogeneity 2 px | ||||
| Nucleus Haralick Sum Variance 2 px | ||||
| Nucleus Haralick Contrast 2 px | ||||
| Nucleus Haralick Correlation 2 px | ||||
| Nucleus SER Dark 2 px | ||||
| Nucleus SER Bright 2 px | ||||
| Nucleus SER Saddle 2 px | ||||
| Nucleus SER Valley 2 px | ||||
| Nucleus SER Ridge 2 px | ||||
| Nucleus SER Edge 2 px | ||||
| Nucleus SER Hole 2 px | ||||
| Nucleus SER Spot 2 px | ||||
| Mitochondria | TMRM | Intensity | Intensity Cytoplasm Contrast | |
| Intensity Cytoplasm CV [%] | ||||
| Intensity Cytoplasm Minimum | ||||
| Intensity Cytoplasm Maximum | ||||
| Intensity Cytoplasm StdDev | ||||
| Intensity Cytoplasm Mean | ||||
| Texture | Mitoch Haralick Homogeneity 2 px | |||
| Mitoch Haralick Sum Variance 2 px | ||||
| Mitoch Haralick Contrast 2 px | ||||
| Mitoch Haralick Correlation 2 px | ||||
| Mitoch Gabor Max 2 px w4 | ||||
| Mitoch Gabor Min 2 px w4 | ||||
| Mitoch SER Dark 2 px | ||||
| Mitoch SER Bright 2 px | ||||
| Mitoch SER Saddle 2 px | ||||
| Mitoch SER Valley 2 px | ||||
| Mitoch SER Ridge 2 px | ||||
| Mitoch SER Edge 2 px | ||||
| Mitoch SER Hole 2 px | ||||
| Mitoch SER Spot 2 px | ||||
| Well based | N/A | Hoechst | N/A | Number of Objects - Nuclei |
| RedDot | % Dead Cells | |||
| NucView | % Apoptotic Cells | |||
Appendix B.
Statistical significance (t-test, p-value < 0.05, adjusted by Bonferroni correction) of differences in cellular features measured from control cells (DMSO-treated) cultured in four different culturing conditions as follows:
-
–
Blanket (fibronectin/MaxGel 10% 11d and fibronectin/MaxGel 10% 7d).
-
–
Sandwich (MaxGel/MaxGel 10% 7d and MaxGel/MaxGel 25% 7d).
The assumptions of homogeneity of variances and normality were tested by Bartlett and Shapiro-Wilk tests, respectively.
| Culturing conditions | Number of significantly different features |
|---|---|
| Blanket: fibronectin/MaxGel 10% 11d vs. fibronectin/MaxGel 10% 7d | 0 |
| Sandwich: MaxGel/MaxGel 10% 7d vs. MaxGel/MaxGel 25% 7d | 3 |
| Sandwich vs. Blanket | 12 |
List of statistically significantly different cellular features between sandwich culturing conditions:
| Cellular feature | p-value |
|---|---|
| Nucleus_Symmetry_13 | 4.69e-02 |
| Intensity_Cytoplasm_CV_pcts | 1.46e-02 |
| Mitoch_Haralick_Sum_Variance_2_px | 1.38e-02 |
List of statistically significantly different cellular features between sandwich and blanket culturing conditions:
| Cellular feature | p-value |
|---|---|
| Nucleus_Axial_Small_Length | 8.01e-04 |
| Intensity_Cytoplasm_Contrast | 4.26e-02 |
| Intensity_Cytoplasm_CV_pcts | 3.41e-03 |
| Intensity_Cytoplasm_Minimum | 5.40e-05 |
| Intensity_Cytoplasm_Maximum | 1.12e-05 |
| Intensity_Nucleus_Contrast | 1.78e-04 |
| Nucleus_Haralick_Homogeneity_2_px | 9.19e-05 |
| Mitoch_Haralick_Sum_Variance_2_px | 6.73e-04 |
| Mitoch_Haralick_Contrast_2_px | 9.18e-06 |
| Mitoch_Gabor_Min_2_px_w4 | 1.67e-05 |
| Mitoch_SER_Dark_2_px | 2.48e-04 |
| Mitoch_SER_Valley_2_px | 2.77e-04 |
Appendix C.
The counts of the significantly (α = 0.05) different features between control (DMSO) and antimycin (1 μM, 2 h) in four different culturing conditions. A t-test was conducted for each feature under each condition (i.e., a combination of bottom and top coats) and the resulting p-values were adjusted by Bonferroni correction to control the family-wise error rate within each condition. The assumptions of homogeneity of variances and normality were tested by Bartlett and Shapiro-Wilk tests, respectively. The adjusted p-values are listed in the table below.
| Bottom coat | Top coat | Count of significantly different features |
|---|---|---|
| Fibronectin | MaxGel 10% 11d | 3 |
| Fibronectin | MaxGel 10% 7d | 13 |
| MaxGel 10% | MaxGel 10% 7d | 20 |
| MaxGel 25% | MaxGel 25% 7d | 25 |
| Culturing condition | Cellular feature | p-value |
|---|---|---|
| Fibronectin+MaxGel 10% 11d | Mitoch_Haralick_Contrast_2_px | 1.25e-02 |
| Fibronectin+MaxGel 10% 11d | Mitoch_Gabor_Max_2_px_w4 | 3.09e-02 |
| Fibronectin+MaxGel 10% 11d | Mitoch_Gabor_Min_2_px_w4 | 4.00e-02 |
| Fibronectin+MaxGel 10% 7d | Nucleus_Radial_Relative_Deviation | 2.57e-02 |
| Fibronectin+MaxGel 10% 7d | Intensity_Nucleus_CV_pcts | 1.70e-02 |
| Fibronectin+MaxGel 10% 7d | Intensity_Nucleus_Minimum | 2.32e-03 |
| Fibronectin+MaxGel 10% 7d | Intensity_Nucleus_Maximum | 2.78e-04 |
| Fibronectin+MaxGel 10% 7d | Intensity_Nucleus_StdDev | 1.83e-05 |
| Fibronectin+MaxGel 10% 7d | Intensity_Nucleus_Mean | 1.23e-02 |
| Fibronectin+MaxGel 10% 7d | Nucleus_Haralick_Homogeneity_2_px | 9.30e-03 |
| Fibronectin+MaxGel 10% 7d | Nucleus_Haralick_Sum_Variance_2_px | 1.71e-03 |
| Fibronectin+MaxGel 10% 7d | Nucleus_Haralick_Contrast_2_px | 1.43e-02 |
| Fibronectin+MaxGel 10% 7d | Nucleus_SER_Dark_2_px | 1.07e-02 |
| Fibronectin+MaxGel 10% 7d | Nucleus_SER_Saddle_2_px | 1.38e-02 |
| Fibronectin+MaxGel 10% 7d | Nucleus_SER_Valley_2_px | 2.51e-03 |
| Fibronectin+MaxGel 10% 7d | Nucleus_SER_Edge_2_px | 4.70e-03 |
| MaxGel 10%+MaxGel 10% 7d | Nucleus_Profile_5/5 | 3.64e-09 |
| MaxGel 10%+MaxGel 10% 7d | Nucleus_Radial_Relative_Deviation | 4.65e-08 |
| MaxGel 10%+MaxGel 10% 7d | Nucleus_Radial_Mean | 3.94e-04 |
| MaxGel 10%+MaxGel 10% 7d | Nucleus_Axial_Small_Length | 1.80e-05 |
| MaxGel 10%+MaxGel 10% 7d | Nucleus_Threshold_Compactness_30_pc | 7.61e-04 |
| MaxGel 10%+MaxGel 10% 7d | Nucleus_Symmetry_15 | 3.50e-02 |
| MaxGel 10%+MaxGel 10% 7d | Nucleus_Symmetry_14 | 2.41e-03 |
| MaxGel 10%+MaxGel 10% 7d | Nucleus_Symmetry_13 | 6.80e-03 |
| MaxGel 10%+MaxGel 10% 7d | Nucleus_Symmetry_12 | 4.37e-02 |
| MaxGel 10%+MaxGel 10% 7d | Nucleus_Symmetry_04 | 1.60e-02 |
| MaxGel 10%+MaxGel 10% 7d | Intensity_Cytoplasm_Minimum | 7.06e-10 |
| MaxGel 10%+MaxGel 10% 7d | Intensity_Nucleus_CV_pcts | 9.72e-10 |
| MaxGel 10%+MaxGel 10% 7d | Intensity_Nucleus_Mean | 3.55e-07 |
| MaxGel 10%+MaxGel 10% 7d | Nucleus_Haralick_Sum_Variance_2_px | 2.27e-10 |
| MaxGel 10%+MaxGel 10% 7d | Nucleus_Haralick_Contrast_2_px | 2.39e-05 |
| MaxGel 10%+MaxGel 10% 7d | Nucleus_SER_Dark_2_px | 3.44e-06 |
| MaxGel 10%+MaxGel 10% 7d | Nucleus_SER_Saddle_2_px | 8.25e-07 |
| MaxGel 10%+MaxGel 10% 7d | Nucleus_SER_Valley_2_px | 3.03e-06 |
| MaxGel 10%+MaxGel 10% 7d | Nucleus_SER_Edge_2_px | 3.17e-04 |
| MaxGel 10%+MaxGel 10% 7d | Nucleus_SER_Spot_2_px | 1.79e-03 |
| MaxGel 25%+MaxGel 25% 7d | Nucleus_Profile_5/5 | 1.77e-10 |
| MaxGel 25%+MaxGel 25% 7d | Nucleus_Axial_Length_Ratio | 4.23e-02 |
| MaxGel 25%+MaxGel 25% 7d | Nucleus_Axial_Small_Length | 1.28e-04 |
| MaxGel 25%+MaxGel 25% 7d | Nucleus_Symmetry_15 | 1.21e-04 |
| MaxGel 25%+MaxGel 25% 7d | Nucleus_Symmetry_13 | 3.99e-05 |
| MaxGel 25%+MaxGel 25% 7d | Nucleus_Symmetry_05 | 1.59e-02 |
| MaxGel 25%+MaxGel 25% 7d | Nucleus_Symmetry_04 | 7.61e-05 |
| MaxGel 25%+MaxGel 25% 7d | Nucleus_Symmetry_03 | 5.07e-05 |
| MaxGel 25%+MaxGel 25% 7d | Intensity_Cytoplasm_Mean | 5.03e-12 |
| MaxGel 25%+MaxGel 25% 7d | Intensity_Nucleus_Contrast | 8.94e-03 |
| MaxGel 25%+MaxGel 25% 7d | Intensity_Nucleus_CV_pcts | 4.90e-11 |
| MaxGel 25%+MaxGel 25% 7d | Intensity_Nucleus_Mean | 3.49e-05 |
| MaxGel 25%+MaxGel 25% 7d | Nucleus_Gabor_Max_2_px_w4 | 1.10e-04 |
| MaxGel 25%+MaxGel 25% 7d | Nucleus_Gabor_Min_2_px_w4 | 5.27e-04 |
| MaxGel 25%+MaxGel 25% 7d | Nucleus_Haralick_Homogeneity_2_px | 3.29e-09 |
| MaxGel 25%+MaxGel 25% 7d | Nucleus_Haralick_Sum_Variance_2_px | 1.87e-08 |
| MaxGel 25%+MaxGel 25% 7d | Nucleus_Haralick_Contrast_2_px | 8.31e-06 |
| MaxGel 25%+MaxGel 25% 7d | Nucleus_Haralick_Correlation_2_px | 6.69e-05 |
| MaxGel 25%+MaxGel 25% 7d | Nucleus_SER_Dark_2_px | 5.28e-07 |
| MaxGel 25%+MaxGel 25% 7d | Nucleus_SER_Bright_2_px | 3.54e-04 |
| MaxGel 25%+MaxGel 25% 7d | Nucleus_SER_Saddle_2_px | 4.56e-11 |
| MaxGel 25%+MaxGel 25% 7d | Nucleus_SER_Valley_2_px | 4.25e-08 |
| MaxGel 25%+MaxGel 25% 7d | Nucleus_SER_Ridge_2_px | 9.31e-03 |
| MaxGel 25%+MaxGel 25% 7d | Nucleus_SER_Edge_2_px | 4.42e-07 |
| MaxGel 25%+MaxGel 25% 7d | Nucleus_SER_Spot_2_px | 3.81e-06 |
Appendix D.
Number of nuclei (average of n = 4 wells) in control (DMSO) and in the presence of different concentrations of staurosporine (at 2 and 24 h) and amiodarone and crizotinib (at 4 and 48 h) in four different culturing conditions. Error bars are SD. * P < 0.05 vs. DMSO at the corresponding treatment time.
Appendix E.
Statistical significance (one-way ANOVA p-value < 0.05, adjusted by Bonferroni correction) of differences in mean cellular features measured in control cells (DMSO) in the four culturing “sandwich” conditions (i.e. the four factors were MaxGel 1:2 2 d; MaxGel 1:2 7 d; MaxGel 1:4 2 d; and MaxGel 1:4 7 d) at 24 and 48 h treatment. Note that none of the differences was statistically significant for the 24 h treatment. The ANOVA assumptions of homogeneity of variances and normality of residuals were tested by Bartlett and Shapiro-Wilk tests, respectively.
| Treatment time | Cellular feature | p-value |
|---|---|---|
| 48 h | Nucleus_Profile_4/5 | 3.60e-02 |
| 48 h | Nucleus_Threshold_Compactness_60_pc | 2.71e-02 |
| 48 h | Intensity_Cytoplasm_TexasRed_CV_pcts | 1.74e-03 |
| 48 h | Mitoch_Haralick_Haralick_Contrast_2_px | 3.00e-04 |
| 48 h | Mitoch_Haralick_Haralick_Correlation_2_px | 6.89e-04 |
| 48 h | Mitoch_Gabor_Gabor_Max_2_px_w4 | 1.30e-04 |
| 48 h | Mitoch_Gabor_Gabor_Min_2_px_w4 | 3.51e-04 |
| 48 h | Mitoch_SER_SER_Dark_2_px | 2.24e-03 |
| 48 h | Mitoch_SER_SER_Bright_2_px | 4.30e-04 |
| 48 h | Mitoch_SER_SER_Saddle_2_px | 2.42e-03 |
| 48 h | Mitoch_SER_SER_Valley_2_px | 2.56e-03 |
| 48 h | Mitoch_SER_SER_Ridge_2_px | 2.95e-04 |
| 48 h | Mitoch_SER_SER_Edge_2_px | 1.89e-02 |
| 48 h | Mitoch_SER_SER_Hole_2_px | 2.31e-03 |
| 48 h | Mitoch_SER_SER_Spot_2_px | 1.38e-03 |
| 48 h | Nucleus_DAPI_SER_Bright_2_px | 8.95e-03 |
| 48 h | Nucleus_DAPI_SER_Ridge_2_px | 1.16e-02 |
| 48 h | Nucleus_DAPI_SER_Hole_2_px | 1.25e-03 |
Appendix F.
The counts of the significantly (α = 0.05) different features between control (DMSO) and staurosporine (3 μM; 24 h) treatment in four different culturing conditions. A t-test was conducted for each feature under each condition and the resulting p values were adjusted by Bonferroni correction to control the family-wise error rate within each condition. The adjusted p values are listed in the table below. The assumptions of homogeneity of variances and normality were tested by Bartlett and Shapiro-Wilk tests, respectively.
| Top coat | Count of significantly different features |
|---|---|
| MaxGel 50% 2d | 3 |
| MaxGel 50% 7d | 7 |
| MaxGel 25% 2d | 9 |
| MaxGel 25% 7d | 13 |
| Top coat | Cellular feature | p-value |
|---|---|---|
| MaxGel 50% 2d | Nucleus_Haralick_Homogeneity_2_px | 2.00e-04 |
| MaxGel 50% 2d | Nucleus_Haralick_Sum_Variance_2_px | 2.97e-02 |
| MaxGel 50% 2d | Nucleus_Haralick_Contrast_2_px | 9.47e-03 |
| MaxGel 50% 7d | Nucleus_Radial_Relative_Deviation | 9.92e-05 |
| MaxGel 50% 7d | Nucleus_Threshold_Compactness_50_pc | 1.02e-02 |
| MaxGel 50% 7d | Nucleus_Symmetry_04 | 2.30e-02 |
| MaxGel 50% 7d | Intensity_Cytoplasm_Minimum | 1.03e-02 |
| MaxGel 50% 7d | Intensity_Nucleus_CV_pcts | 4.64e-02 |
| MaxGel 50% 7d | Nucleus_Haralick_Homogeneity_2_px | 3.40e-02 |
| MaxGel 50% 7d | Nucleus_Haralick_Sum_Variance_2_px | 4.06e-02 |
| MaxGel 25% 2d | Nucleus_Profile_5/5 | 1.80e-03 |
| MaxGel 25% 2d | Intensity_Cytoplasm_CV_pcts | 1.54e-05 |
| MaxGel 25% 2d | Intensity_Cytoplasm_Minimum | 7.00e-04 |
| MaxGel 25% 2d | Intensity_Cytoplasm_Maximum | 1.29e-02 |
| MaxGel 25% 2d | Nucleus_Haralick_Homogeneity_2_px | 2.17e-05 |
| MaxGel 25% 2d | Mitoch_Haralick_Homogeneity_2_px | 2.29e-04 |
| MaxGel 25% 2d | Mitoch_SER_Saddle_2_px | 9.31e-05 |
| MaxGel 25% 2d | Mitoch_SER_Edge_2_px | 1.12e-06 |
| MaxGel 25% 2d | Nucleus_SER_Saddle_2_px | 2.60e-05 |
| MaxGel 25% 7d | Nucleus_Profile_5/5 | 6.58e-03 |
| MaxGel 25% 7d | Nucleus_Radial_Mean | 1.08e-02 |
| MaxGel 25% 7d | Nucleus_Axial_Small_Length | 9.70e-04 |
| MaxGel 25% 7d | Nucleus_Threshold_Compactness_60_pc | 1.67e-03 |
| MaxGel 25% 7d | Intensity_Cytoplasm_Minimum | 6.59e-05 |
| MaxGel 25% 7d | Intensity_Cytoplasm_Mean | 1.25e-04 |
| MaxGel 25% 7d | Intensity_Nucleus_Contrast | 2.26e-02 |
| MaxGel 25% 7d | Intensity_Nucleus_CV_pcts | 3.90e-03 |
| MaxGel 25% 7d | Intensity_Nucleus_Minimum | 4.13e-02 |
| MaxGel 25% 7d | Intensity_Nucleus_Mean | 9.57e-04 |
| MaxGel 25% 7d | Nucleus_Haralick_Homogeneity_2_px | 1.32e-05 |
| MaxGel 25% 7d | Nucleus_Haralick_Contrast_2_px | 1.01e-03 |
| MaxGel 25% 7d | Mitoch_Haralick_Homogeneity_2_px | 1.30e-07 |
References
- 1.Guo L., Abrams R.M., Babiarz J.E., Cohen J.D., Kameoka S., Sanders M.J. Estimating the risk of drug-induced proarrhythmia using human induced pluripotent stem cell-derived cardiomyocytes. Toxicol. Sci. 2011;123:281–289. doi: 10.1093/toxsci/kfr158. [DOI] [PubMed] [Google Scholar]
- 2.Guo L., Coyle L., Abrams R.M., Kemper R., Chiao E.T., Kolaja K.L. Refining the human iPSC-cardiomyocyte arrhythmic risk assessment model. Toxicol. Sci. 2013;136:581–594. doi: 10.1093/toxsci/kft205. [DOI] [PubMed] [Google Scholar]
- 3.Zeng H., Roman M.I., Lis T., Lagrutta A., Sannajust F. Use of FDSS/μCell imaging platform for preclinical cardiac electrophysiology safety screening of compounds in human induced pluripotent stem cell-derived cardiomyocytes. J. Pharmacol. Toxicol. Methods. 2016;81:217–222. doi: 10.1016/j.vascn.2016.05.009. [DOI] [PubMed] [Google Scholar]
- 4.Abassi Y.A., Xi B., Li N., Ouyang W., Seiler A., Watzele M. Dynamic monitoring of beating periodicity of stem cell-derived cardiomyocytes as a predictive tool for preclinical safety assessment. Br. J. Pharmacol. 2012;165:1424–1441. doi: 10.1111/j.1476-5381.2011.01623.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doherty K.R., Talbert D.R., Trusk P.B., Moran D.M., Shell S.A., Bacus S. Structural and functional screening in human induced-pluripotent stem cell-derived cardiomyocytes accurately identifies cardiotoxicity of multiple drug types. Toxicol. Appl. Pharmacol. 2015;285:51–60. doi: 10.1016/j.taap.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 6.Grimm F.A., Iwata Y., Sirenko O., Bittner M., Rusyn I. High-content assay multiplexing for toxicity screening in induced pluripotent stem cell-derived cardiomyocytes and hepatocytes. Assay Drug Dev. Technol. 2015;13:529–546. doi: 10.1089/adt.2015.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roquemore L., Kauss M.A., Hather C., Thomas N., Uppal H. In vitro cardiotoxicity investigation using high content analysis and human stem cell-derived models. In: Clements M., Roquemore L., editors. Stem Cell-Derived Models in Toxicology. Methods in Pharmacology and Toxicology. Humana Press; New York, NY: 2017. pp. 247–269. [Google Scholar]
- 8.Ju H., Dixon I.M.C. Cardiac extracellular matrix and its role in the development of heart failure. In: Singal P.K., Dixon I.M.C., Beamish R.E., Dhalla N.S., editors. Vol. 167. Springer; Boston, MA: 1995. pp. 75–90. (Mechanisms of Heart Failure. Developments in Cardiovascular Medicine). [Google Scholar]
- 9.Rodin S., Domogatskaya A., Strom S., Hansson E.M., Chien K.R., Inzunza J. Long-term self-renewal of human pluripotent stem cells on human recombinant laminin-511. Nat. Biotechnol. 2010;28:611–615. doi: 10.1038/nbt.1620. [DOI] [PubMed] [Google Scholar]
- 10.Zhang J., Klos M., Wilson G.F., Herman A.M., Lian X., Raval K.K. Extracellular matrix promotes highly efficient cardiac differentiation of human pluripotent stem cells: the matrix sandwich method. Circ. Res. 2012;111:1125–1136. doi: 10.1161/CIRCRESAHA.112.273144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herron T.J., Rocha A.M., Campbell K.F., Ponce-Balbuena D., Willis B.C., Guerrero-Serna G. Extracellular matrix-mediated maturation of human pluripotent stem cell-derived cardiac monolayer structure and electrophysiological function. Circ. Arrhythm. Electrophysiol. 2016;9:1–12. doi: 10.1161/CIRCEP.113.003638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fong A.H., Romero-López M., Heylman C.M., Keating M., Tran D., Sobrino A. Three-dimensional adult cardiac extracellular matrix promotes maturation of human induced pluripotent stem cell-derived cardiomyocytes. Tissue Eng. Part A. 2016;22:1016–1025. doi: 10.1089/ten.tea.2016.0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.HydroMatrixTM Peptide Hydrogel . 2019. Sigma-Aldrich Product Information.https://www.sigmaaldrich.com/life-science/stem-cell-biology/3d-stem-cell-culture/hydromatrix-peptide.html Available from: [Google Scholar]
- 14.Geltrex® Matrix Products . 2019. Thermo Fisher Product Information.https://www.thermofisher.com/sg/en/home/life-science/cell-culture/3d-cell-culture/3d-cell-culture-misc/geltrex.html Available from: [Google Scholar]
- 15.MaxGelTM Human ECM . 2019. Sigma-Aldrich Product Information.https://www.sigmaaldrich.com/life-science/stem-cell-biology/3d-stem-cell-culture/maxgel-human-ecm.html Available from: [Google Scholar]
- 16.Perkin Elmer Inc; 2014. Columbus© 2.5.0 User Manual. [Google Scholar]
- 17.Lazarevic A., Kumar V. Feature bagging for outlier detection. Proc. 11th ACM SIGKDD International Conference on Knowledge Discovery in Data Mining. 2005:157–166. [Google Scholar]
- 18.Breunig M.M., Kriegel H.-P., Ng R.T., Sander J. LOF: identifying density-based local outliers. Proceedings of the 2000 ACM SIGMOD International Conference on Management of Data. SIGMOD. 2000:93–104. [Google Scholar]
- 19.Zou K.H., O’Malley A.J., Mauri L. Receiver-operating characteristic analysis for evaluating diagnostic tests and predictive models. Circulation. 2007;115:654–657. doi: 10.1161/CIRCULATIONAHA.105.594929. [DOI] [PubMed] [Google Scholar]
- 20.Bray M.A., Carpenter A. Advanced assay development guidelines for image-based high content screening and analysis. In: Sittampalam G.S., Coussens N.P., Brimacombe K., editors. Assay Guidance Manual. 2012. pp. 3–7. [Google Scholar]
- 21.Ziegler U., Groscurth P. Morphological features of cell death. News Physiol. Sci. 2004;19:124–128. doi: 10.1152/nips.01519.2004. [DOI] [PubMed] [Google Scholar]
- 22.Buscher W., Collins M., Garyantes T., Guha R., Haney S., Lemmon V. Assay development guide for image-based high content screening, high content analysis and high content imaging. In: Sittampalam G.S., Coussens N.P., Brimacombe K., editors. Assay Guidance Manual. 2012. pp. 21–22. [Google Scholar]
- 23.Schocken D., Stohlman J., Vicente J., Chan D., Patel D., Matta M.K. Comparative analysis of media effects on human induced pluripotent stem cell-derived cardiomyocytes in proarrhythmia risk assessment. J. Pharmacol. Toxicol. Methods. 2018;90:39–47. doi: 10.1016/j.vascn.2017.11.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.