Abstract
Staphylococcus argenteus, a novel species of the genus Staphylococcus or a member of the S. aureus complex, is closely related to S. aureus and is usually misidentified. In this study, the presence of S. argenteus in isolated S. aureus was investigated in 67 rabbits with abscess lesions during 2014–2016. Among 19 S. aureus complex isolates, three were confirmed to be S. argenteus by matrix‐assisted laser desorption/ionization time‐of‐flight mass spectrometry, nonribosomal peptide synthetase gene amplification, and multilocus sequence type. All S. aureus complex isolates, including the S. aureus isolates, were examined for their antimicrobial resistance phenotype by disk diffusion and for their resistance genotype by PCR assays. Among the S. argenteus isolates, one was susceptible to all antimicrobial drugs and the other two were resistant to penicillin and doxycycline. In contrast, most S. aureus isolates were resistant to penicillin (37.5%), and gentamicin (12.5%). Moreover, S. aureus isolates harbored the blaZ, mecA, aacA‐aphD, and mrs(A) as well as mutations of gyrA and grlA, but S. argenteus isolates carried solely the blaZ. S. argenteus isolates were investigated for enterotoxin (sea‐sed) and virulence genes by PCR. One isolate carried sea, sec, and sed, whereas the other two isolates carried only sea or sed. No isolate carried seb and see. All three S. argenteus isolates carried hla, hlb, and clfA, followed by pvl, whereas coa, spa (IgG‐binding region), and spa (x region) were not detected in the three isolates. This paper presents the first identification of S. argenteus from rabbits in Thailand. S. argenteus might be pathogenic because the isolates carried virulence genes. Moreover, antimicrobial resistance was observed. Investigations of this new bacterial species should be conducted in other animal species as well as in humans.
Keywords: antimicrobial resistance, antimicrobial resistance genes, rabbit, Staphylococcus argenteus, Staphylococcus aureus, Staphylococcus aureus complex
1. INTRODUCTION
The Staphylococcus aureus complex consists of opportunistic pathogens that can cause a wide spectrum of diseases in both humans and animals (Corpa et al., 2009). These pathogens are nonspore‐forming, nonmotile, spherical organisms, appearing as grapelike clusters under a microscope. They are facultatively anaerobic, catalase‐positive (Foster & Geoghegan, 2015), coagulase‐positive and can produce protein A. In rabbits, infection with the S. aureus complex usually results in small dermal lesions; the invasion of subcutaneous tissue and the development of pododermatitis, subcutaneous abscesses, and mastitis. Abscesses in internal organs are sometimes observed, such as in the lungs, liver, and uterus. This gives rise to poor reproductive results, infertility, and death (Corpa et al., 2009; Vancraeynest et al., 2004; Viana et al., 2007).
Recently, a novel coagulase‐positive Staphylococcus species, S. argenteus (S. aureus complex), was identified from clinical human and animal sources (Argudín et al., 2016; Chantratita et al., 2016; Schuster et al., 2017; Thaipadungpanit et al., 2015; Tong et al., 2015). Bacterial colonies were characterised by a nonpigmented, creamy white appearance and showed β‐hemolysis on blood agar. Moreover, the bacteria were shown to be gram‐positive cocci in clusters and gave positive results in the catalase and coagulase tests, which are characteristic findings for S. aureus (Tong et al., 2015). Therefore, routine diagnostic analyses can lead to S. argenteus being misidentified as S. aureus. Moreover, identification by molecular methods, such as 16S rRNA sequencing, cannot differentiate S. argenteus from S. aureus (Tong et al., 2015). Other molecular techniques, such as matrix‐assisted laser desorption/ionization time‐of‐flight mass spectrometry (MALDI‐TOF MS), nonribosomal peptide synthetase (NRPS) gene amplification, and multilocus sequence type (MLST) determination, were thus recommended in many publications as tools for the identification of S. argenteus (Chantratita et al., 2016; Schuster et al., 2017; Zhang et al., 2016). Some sequencing types of S. aureus were previously confirmed to be S. argenteus, such as ST2793, ST1223, and ST2250 (Chantratita et al., 2016; Schuster et al., 2017; Thaipadungpanit et al., 2015; Tong et al., 2015).
The aim of this study was to characterize S. argenteus and S. aureus isolated from rabbits with clinical abscesses.
2. MATERIALS AND METHODS
2.1. Specimen collection and bacterial isolation
Sixty‐seven pus samples were collected from rabbits with clinical abscesses by a veterinarian at Prasu‐Arthorn Animal Hospital, Thailand, during 2014–2016. They were transported to a microbiological laboratory within 24 hr after collection. Individual samples were inoculated on sheep blood and mannitol salt agar (Oxoid, Basingstoke, UK) and incubated at 37°C for 24–48 hr. After incubation, the suspected bacterial colonies were selected and identified by conventional methods, including Gram staining, catalase, mannitol fermentation, coagulase (Ramel; Oxoid), and commercial latex agglutination (Dryspot Staphytect Plus; Oxoid), to detect protein A for S. aureus identification. This study was approved by The Faculty of Veterinary Science Animal Care and Use Committee, Mahidol University (protocol number MUVS‐2013‐35).
2.2. S. argenteus identification
2.2.1. MALDI‐TOF MS
Mass spectra were generated using a MALDI Biotyper 3.0 Ultraflex platform (Bruker Daltonics, Massachusetts, USA). For individual suspected S. argenteus isolates, with white colonies, 1 ml of crude protein extract or one colony was deposited on a 96‐spot polished steel target plate (Bruker Daltonics), air‐dried and covered with 1 ml of HCCA matrix solution (Bruker Daltonics) (Kolecka et al., 2013). As a positive control and calibration reference, 1 ml of Bacterial Test Standard (Bruker Daltonics) was used. The main spectrum was acquired using the MALDI Biotyper Automated Flex Control software v.3.0 (Bruker Daltonics). The identification of isolates was performed using the Bruker database and in‐house databases from Chantratita et al. (2016) and Moradigaravand et al. (2017).
2.2.2. NRPS gene amplification
Primer sequences and the PCR protocol for NRPS gene indels were in accordance with those of Zhang et al. (2016). The NRPS gene was amplified in a total reaction volume of 25 μl. The PCR reaction was performed using the thermal cycles (Bio‐Rad, California, USA) with initial denaturation at 94°C for 4 min, followed by 35 cycles of 94°C for 30 s, 53°C for 30 s, and 72°C for 40 s and then final extension at 72°C for 10 min. Individual PCR amplicons were purified using the GenepHlow™ Gel/PCR purification kit (Geneaid, New Taipei, Taiwan) and submitted for sequencing. DNA sequences were analyzed for similarity with the GenBank database.
2.2.3. MLST
MLST was used to analyze the suspected S. argenteus isolates through the amplification of seven S. aureus housekeeping genes, by a method developed by Enright et al. (2000) at Imperial College London for analyzing a query profile for MLST. Before sequencing, individual PCR amplicons were purified using the GenepHlow™ Gel/PCR purification kit (Geneaid). The allelic number queries and sequence types (STs) obtained from trimmed DNA sequencing results for seven genes were determined using the online S. aureus MLST database (https://pubmlst.org/saureus/). The suspected novel alleles or queried allelic profiles of novel STs that did not match the database were submitted to the curator of PubMLST (https://pubmlst.org/saureus/) to check and assign a novel allele or novel ST number.
2.3. Antimicrobial susceptibility testing
Antimicrobial susceptibility testing and interpretation were performed by the disk diffusion in according to the Clinical and Laboratory Standards Institute (2012). A total of 13 antimicrobial drugs were tested: amikacin (30 μg), azithromycin (15 μg), cefazolin (30 μg), cefoxitin (30 μg), ceftriaxone (30 μg), chloramphenicol (30 μg), ciprofloxacin (5 μg), doxycycline (30 μg), gentamicin (10 μg), moxifloxacin (5 μg), norfloxacin (10 μg), penicillin (10 units), and trimethoprim/sulfamethoxazole (1.25 μg/23.75 μg). S. aureus ATTC®25923 was used as the control strain.
2.4. Detection of 16s rRNA, antimicrobial resistance, and enterotoxin and virulence genes
All isolated S. aureus samples were prepared for genomic DNA extraction using a DNA extraction kit (Geneaid). The extraction protocol involved the measurement of the OD 260/280 nm ratio using a spectrophotometer. Amplification with specific oligonucleotide primers was performed for 16s rRNA of the S. aureus complex group (McClure et al., 2006) and eight antimicrobial resistance genes representative of each antibiotic drug group: blaZ (Gómez‐Sanz et al., 2010) and mecA (Vancraeynest et al., 2004) for β‐lactam resistance, aacA‐aphD (Laplana et al., 2007) for aminoglycoside resistance, mrs(A) (Gómez‐Sanz et al., 2010) for macrolide resistance, tet(K) (Vancraeynest et al., 2004) for tetracycline resistance, dfrG (Lagier et al., 2009) for trimethoprim resistance, and cfr (Kehrenberg & Schwarz, 2006) for chloramphenicol resistance. PCR amplification for determined fluoroquinolone resistance, namely gyrA and grlA (Iihara et al., 2006) was performed. The PCR amplicon, namely the QRDR region, was sequenced and analyzed for resistance determining mutations. Deduced amino acid sequences of the PCR amplicons were analyzed using the GenBank database with accession numbers AAC31138.1 for gyrA and WP075108737.1 for grlA. The PCR reaction mixture was subjected to the following thermal cycling conditions using Flexcycler2 (Analytik Jena, Überlingen, Germany): 5 min of 95°C; then 30 cycles of amplification with denaturing at 95°C for 30 s, annealing at a temperature specific for each primer for 30 s and extension at 72°C for 60 s; followed by a final extension at 72°C for 10 min.
In S. argenteus isolates, further identification of the virulence genes was performed, including classical enterotoxin (sea, seb, sec, sed, and see) (Wu et al., 2011), hemolysin (hla and hlb) (Jarraud et al., 2002), clumping factor (clfA) (Tristan et al., 2003), protein A [spa x region (Frénay et al., 1996) and spa IgG‐binding region (Seki et al., 1998)], coagulase (coa) (Aslantas et al., 2007), and Panton‐Valentine leukocidin (pvl) (Jarraud et al., 2002), with specific oligonucleotide primers. PCR reactions were performed, involving initial denaturation at 95°C for 10 min; 35 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s and a final extension at 72°C for 10 min.
PCR amplicons were analyzed using 1.5% agarose gel electrophoresis and SYBR safe (Invitrogen, New York, USA) staining. The DNA bands were observed under a UV transilluminator (UVP Bioimaging System; Invitrogen). Control bacteria for the PCR included the strains S. aureus ATCC 19095 (sea and sec), ATCC 14458 (seb), ATCC 23235 (sed), ATCC 27664 (see), ATCC 13565 (coa, clfA, hla, hlb, spa x region, and spa IgG‐binding region) and laboratory control strains, which were sequenced and analyzed as accession numbers KX371630.1 (pvl). For gene amplification with no reference control, the PCR product from positive samples was subjected to nucleotide sequencing and sequence analysis for gene confirmation.
3. RESULTS
3.1. Detection of S. argenteus by MALDI‐TOF MS
From 67 samples (total of 19 bacterial isolates), we obtained 11 S. aureus isolates and 8 suspected S. argenteus isolates. These suspected S. argenteus isolates were analyzed by MALDI‐TOF MS. After visual inspection and obtaining the spectral results of their ionizable cell surface components, which were compared for similarity with the spectral data in the Bruker and in‐house databases. The results showed that there are five isolates that match with the S. aureus database and three isolates that match with the S. argenteus database, with a high score (>2.3) (isolate no. U19 T10.1, U43 S18.1, and U43 S18.3; Table 1).
Table 1.
Prevalence of enterotoxin and virulence genes, MLST and MALDI‐TOF MS for suspected Staphylococcus argenteus identification
| Isolate no. | Enterotoxin and virulence genes detection | MLST identification | Sequence results of NRPS PCR amplicon | MALDI‐TOF MS identification | |||
|---|---|---|---|---|---|---|---|
| ST | CC | Size (bp) | BLAST species | Sequence identities (%) | |||
| U27 OC2.1 | ND | ST4209a | Singleton | 160 | S. aureus strain K5 (CP020656.1) | 97 | S. aureus |
| U27 OC2.2 | ND | ST4209a | Singleton | 160 | S. aureus strain FDAARGOS (CP007539.3) | 99 | S. aureus |
| U27 OC2.3 | ND | ST4209a | Singleton | 160 | S. aureus strain K5 (CP020656.1) | 96 | S. aureus |
| U19 T10.1 | sed, hla, hlb, clfA, pvl | ST4210a, b | Singleton | 340 | S. argenteus strain XNO106 (CP025023.1) | 99 | S. argenteus |
| U43 S18.1 | sea, hla, hlb, clfA | ST4211a, b | Singleton | 340 | S. argenteus strain XNO106 (CP025023.1) | 100 | S. argenteus |
| U43 S18.3 | sea, sec, sed, hla, hlb, clfA, pvl | ST4211a, b | Singleton | 340 | S. argenteus strain XNO106 (CP025023.1) | 100 | S. argenteus |
| U14 T6.2 | ND | ST4212a | Singleton | 160 | S. aureus strain FDAARGOS_159 (CP014064.2) | 95 | S. aureus |
| U65 S3 | ND | ST4213a | Singleton | 160 | S. aureus strain NRS137 (CP026080.1) | 92 | S. aureus |
bp: base pair; BLAST: basic local alignment search tool; CC: clonal complex; MALDI‐TOF MS: matrix‐assisted laser desorption/ionization time‐of‐flight mass spectrometry; ND: not determine; MLST: multilocus sequence type; NRPS: nonribosomal peptide synthetase.
Novel STs from this study.
S. argenteus ST.
3.2. NRPS gene amplification of S. argenteus
The NRPS gene was amplified for eight suspected S. argenteus isolates, as shown in Figure 1. The results showed that three isolates (U19 T10.1, U43 S18.1, and U43 S18.3) have a PCR product of approximately 340 bp, which was correlated with S. argenteus as reported by Zhang et al. (2016). In contrast, the five other suspected isolates had a PCR product of nearly 160 bp, which was reported to correlate with S. aureus (Zhang et al., 2016). The sequences of NRPS amplicons were analyzed for similarity using the GenBank database, which showed that bacterial isolate no. U19 T10.1, U43 S18.1, and U43 S18.3 have 99% identity with S. argenteus strain XNO106 (accession number: CP025023.1). In contrast, the other five isolates showed high identity with S. aureus strain K5 (accession number: CP020656.1), S. aureus strain FDAARGOS (accession number: CP007539.3), S. aureus strain FDAARGOS_159 (accession number: CP014064.2), and S. aureus strain NRS137 (accession number: CP026080.1). These results correlated with the MALDI‐TOF MS results (Table 1).
Figure 1.
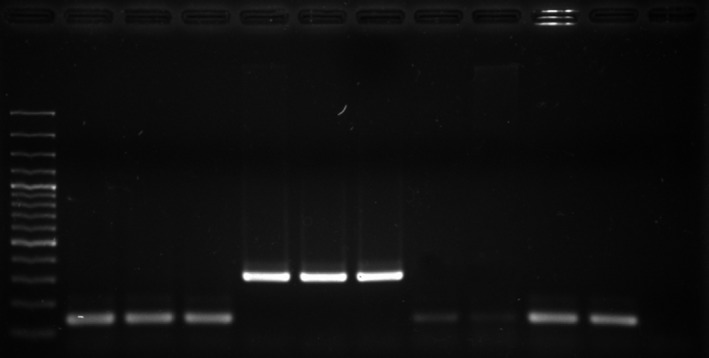
Non ribosomal peptide synthetase amplicon of eight suspected Staphylococcus argenteus. Lane M: DNA marker; lane 1: isolate no. U27 OC2.1; lane 2: isolate no. U27 OC2.2; lane 3: isolate no. U27 OC2.3; lane 4: isolate no. U19 T10.1; lane 5: isolate no. U43 S18.1; lane 6: isolate no. U43 S18.3; lane 7: isolate no. U14 T6.2; lane 8: isolate no. U65 S3; lane 9: S. aureus ATCC13565; lane 10: S. aureus ATCC25923; lane 11: negative control
3.3. MLST of S. argenteus
All eight suspected S. argenteus isolates were further analyzed by MLST to define STs. Several isolates were shown to be novel STs of the Staphylococcus complex, which included ST4209 (isolate no. U27 OC2.1, U27 OC2.2, and U27 OC2.3), ST4210 (isolate no. U19 T10.1), ST4211 (isolate no. U43 S18.1 and U43 S18.3), ST4212 (isolate no. U14 T6.2), and ST4213 (isolate no. U65 S3), derived from the curator of the PubMLST S. aureus database (https://pubmlst.org), University of Oxford, UK, and the Wellcome Trust fund. The results showed that three isolates (isolate no. U19 T10.1, ST4210; isolates no. U43 S18.1 and U43 S18.3, ST4211) were identified as S. argenteus, which correlated with the results of MALDI‐TOF MS and the NRPS gene. The neighbor‐joining and maximum likelihood analyses yielded similar phylogenetic trees. Based on arcC, aroE, gmk, and pta, three bacterial isolates, ST4210 (isolate no. U19 T10.1) and ST4211 (isolates no. U43 S18.1 and U43 S18.3), showed close similarity to the S. argenteus group (ST1223, ST2250, ST2854, and ST2198) (Figure 2).
Figure 2.
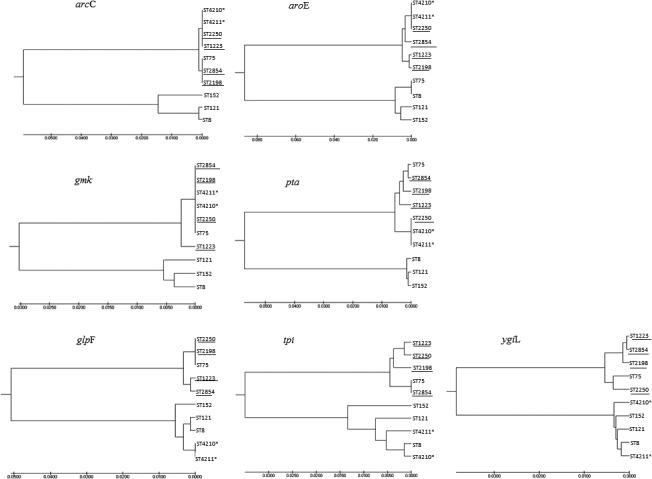
Phylogenetic neighbor‐joining tree of suspected Staphylococcus argenteus. The tree is constructed from arcC, aroE, gmk, glpF, pta, tpi, and ygiL gene sequences from the suspected S. argenteus (ST4210*, ST4211*), S. aureus reference strain (ST75, ST152, ST121, ST8) and ST of published S. argenteus group (ST1223, ST2250, ST2854, ST2198). The phylogenetic analysis was performed using MEGA7
3.4. Enterotoxin and virulence genes
Five classical enterotoxin genes and seven virulence genes, as mentioned in the Materials and Methods section, were investigated in the three S. argenteus isolates by PCR, the results of which are shown in Table 1. The detected enterotoxin genes were sea, sec, and sed. No isolate carried seb and see. Regarding the virulence genes, the most prevalent ones were hla, hlb, and clfA present at a rate of 100%, followed by pvl at 66.67%, whereas coa, spa (IgG‐binding region) and spa (x region) were not detected in the S. argenteus isolates.
3.5. Antimicrobial susceptibility testing
From bacterial identification, we obtained 16 S. aureus isolates and three S. argenteus isolates. Among the three S. argenteus isolates, one (isolate no. U19 T10.1) was susceptible to all the tested antimicrobial drugs, whereas the remaining two (isolate no. U43 S18.1 and U43 S18.3) were resistant to penicillin and doxycycline. Meanwhile, among the S. aureus isolates, six were resistant to penicillin and two were resistant to gentamicin. One S. aureus isolate (isolate no. U14 T6.2) was resistant to several antimicrobial classes, namely, β‐lactams, aminoglycosides, macrolides, tetracyclines, and fluoroquinolones.
3.6. Antimicrobial resistance genes
In the S. aureus and S. argenteus isolates, the presence of antimicrobial resistance genes was tested. We found that all S. aureus isolates carried blaZ, mecA, aacA‐aphD, and mrs(A), but none carried dfrG, tet(K), and cfr, whereas all three S. argenteus isolates carried only the blaZ (isolate no. U43 S18.1 and U43 S18.3). Mutations of gyrA and grlA were found in the S. aureus isolate no. U14 T6.2. The mutations of gyrA occurred at codons 88 [GAA (Glu) → GAT (Asn)] and 96 [GAT (Asp) → ACA (Thr)], whereas the mutation of grlA occurred at codon 80 [TCC (Ser) → TTA (Leu)]. The presence of antimicrobial resistance genes in S. aureus is shown in Table 2.
Table 2.
Antimicrobial drug resistance phenotypes and antimicrobial resistance genes of Staphylococcus aureus isolated from rabbit
| Antimicrobial susceptibility testing | Antimicrobial resistance genes amplification | ||||
|---|---|---|---|---|---|
| Drug | S (%) | I (%) | R (%) | Gene | No. of isolates (%) |
| β‐lactams | |||||
| Penicillin | 62.5 | 0 | 37.5 | blaZ | 1 (6.25) |
| Cefoxitin | 93.75 | 0 | 6.25 | mecA | 1 (6.25) |
| Cefazolin | 93.75 | 0 | 6.25 | ||
| Ceftriaxone | 75.0 | 18.75 | 6.25 | ||
| Aminoglycosides | |||||
| Gentamicin | 87.5 | 0 | 12.5 | aacA‐ | 1 (6.25) |
| Amikacin | 93.75 | 0 | 6.25 | aphD | |
| Macrolides | |||||
| Azithromycin | 93.75 | 0 | 6.25 | mrs(A) | 1 (6.25) |
| Tetracycline | |||||
| Doxycycline | 93.75 | 0 | 6.25 | tet(K) | 0 (0) |
| Fluoroquinolones | |||||
| Ciprofloxacin | 93.75 | 0 | 6.25 | gyrA | 1 (6.25) |
| Moxifloxacin | 93.75 | 0 | 6.25 | grlA | 1 (6.25) |
| Norfloxacin | 93.75 | 0 | 6.25 | ||
| Folate pathway inhibitors | |||||
| Trimethoprim/sulfamethoxazole | 100 | 0 | 0 | dfrG | 0 (0) |
| Phenicols | |||||
| Chloramphenicol | 100 | 0 | 0 | cfr | 0 (0) |
S: susceptible; I: intermediate; R: resistance.
4. DISCUSSION
Staphylococci are considered the most important veterinary bacterial pathogens because they cause a multiplicity of infections and a wide range of diseases in many host species, including humans and animals (Holmes et al., 2016). This virulent group of pathogens is not only important for livestock, causing conditions such as bovine mastitis or lameness in chickens, but also for causing skin infections resulting in abscesses in pets, such as dogs, cats, and rabbits (Drougka et al., 2016; Goñi et al., 2004; Loncaric et al., 2014; Youn et al., 2014). Pathogenic strains of staphylococci that cause skin infections have been well studied and characterized. Moreover, in farm rabbits, abscesses in the lungs, liver, and uterus lead to poor production, infertility, and death (Corpa et al., 2009). This study revealed the presence of S. aureus, including a new member of the S. aureus complex, S. argenteus, which caused skin infections producing abscesses in rabbits.
Intriguingly, from the 19 S. aureus isolates, eight had white colonies, which differs from the normal colony color of S. aureus, which is golden or yellowish. The suspected eight isolates were further analyzed using MALDI‐TOF MS, and bacterial diversity discriminated by NRPS amplification and MLST. The MALDI‐TOF MS results revealed three S. argenteus isolates, which were investigated for enterotoxin and virulence genes. The enterotoxins detected were sea and sed. All three isolates carried the common virulence hla, hlb, and clfA. Surprisingly, previous studies have reported that S. argenteus is negative for the pvl (Thaipadungpanit et al., 2015), but we found the pvl in two S. argenteus isolates in this study. These three S. argenteus isolates, isolated from rabbits, had the ability to cause severe illness in these animals, particularly via the presence of clfA, which usually contributes to abscess formation in rabbits, as previously reported (Malachowa et al., 2016). Although S. argenteus is regionally distributed in animals other than humans, they have been misidentified as S. aureus; this has been suggested in several previous publications (Chantratita et al., 2016; Schuster et al., 2017; Thaipadungpanit et al., 2015; Tong et al., 2015).
The NRPS gene has been identified in studies of a diverse array of related S. aureus and S. argenteus. This study found that NRPS amplification can differentiate S. argenteus from S. aureus, as also reported previously (Zhang et al., 2016). The sequences of NRPS amplicons analyzed using the GenBank database also corresponded to the MALDI‐TOF MS results. Furthermore, when we performed molecular identification using MLST, all eight isolates were found to have diverse novel STs belonging to S. aureus (ST4209, ST4212, and ST4213) and S. argenteus (ST4210 and ST4211). The results showed high heterogeneity among these pathogenic bacterial isolates from rabbits. Moreover, this study revealed a new pathogenic member of the S. aureus complex, S. argenteus, for the first time. These bacteria form a genetically diverse lineage from S. aureus (Tong et al., 2015), being recently discovered in humans, in 2014. This study may be the first to report on S. argenteus originating from rabbits. We found that one S. argenteus isolate was susceptible to all the tested antibiotic agents and the other two S. argenteus isolates were resistant to penicillin and doxycycline. These results could be useful for veterinarians who have difficulty treating rabbits successfully with penicillin. In this study, we found one S. aureus isolate with mutations of gyrA and grlA, which indicated the possibility of quinolone resistance. Mutations in these genes have typically been found at codon 88 [GAA (Glu) → AAA (Lys)] in gyrA (Griggs et al., 2003; Iihara et al., 2006) and codon 80 [TCC (Ser) → TTC (Phe)] in grlA (Aligholi et al., 2011; Iihara et al., 2006). Interestingly, the mutation of grlA at codon 80 found in our study was TCC (Ser) → TTA (Leu), and the mutation of gyrA at codon 88 was GAA (Glu) → GAT (Asp). Comparing the antibiotic resistance pattern, the isolated S. argenteus showed higher susceptibility to antibiotic agents than the isolated S. aureus. However, it is necessary to monitor the development of drug resistance in S. argenteus in the future. From the discovery of S. argenteus in rabbits, further study of its virulence factors, pathogenesis, clinical manifestations, antimicrobial resistance, and severity or outcome should be performed to improve our knowledge for treating, controlling, or preventing this novel pathogen in exotic pets.
CONFLICT OF INTEREST
The authors state that there are no conflicts of interest.
ACKNOWLEDGMENTS
We thank Prasu‐Arthorn Animal Hospital for help with specimen collection. We appreciative Mr. Paul Adams for correcting grammatical and typing errors in this manuscript. This work was supported by the Annual Research Fund of the Faculty of Veterinary Science, Mahidol University 2014; and Thailand Research Fund (RSA5980048).
Indrawattana N, Pumipuntu N, Suriyakhun N, et al. Staphylococcus argenteus from rabbits in Thailand. MicrobiologyOpen. 2019;8:e665 10.1002/mbo3.665
REFERENCES
- Aligholi, M. , Mirsalehian, A. , Halimi, S. , Imaneini, H. , Taherikalani, M. , Jabalameli, F. , … Emaneini, M. (2011). Phenotypic and genotypic evaluation of fluoroquinolone resistance in clinical isolates of Staphylococcus aureus in Tehran. Medical Science Monitor, 17, PH71–PH74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argudín, M. A. , Dodémont, M. , Vandendriessche, S. , Rottiers, S. , Tribes, C. , Roisin, S. , … Denis, O. (2016). Low occurrence of the new species Staphylococcus argenteus in a Staphylococcus aureus collection of human isolates from Belgium. European Journal of Clinical Microbiology and Infectious Diseases, 35, 1017–1022. 10.1007/s10096-016-2632-x [DOI] [PubMed] [Google Scholar]
- Aslantas, O. , Demir, C. , Turutoglu, H. , Cantekin, Z. , Ergun, Y. , & Dogruer, G. (2007). Coagulase gene polymorphism of Staphylococcus aureus isolated form subclinical mastitis. Turkish Journal of Veterinary and Animal Science, 31, 253–257. [Google Scholar]
- Chantratita, N. , Wikraiphat, C. , Tandhavanant, S. , Wongsuvan, G. , Ariyaprasert, P. , Suntornsut, P. , … Chaimanee, P. (2016). Comparison of community‐onset Staphylococcus argenteus and Staphylococcus aureus sepsis in Thailand: A prospective multicenter observational study. Clinical Microbiology and Infection, 22, 458e11–458e19. 10.1016/j.cmi.2016.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute . (2012). Performance standards for antimicrobial susceptibility testing; twentieth informational supplement. CLSI document M100‐S20, Wayne, PA. [Google Scholar]
- Corpa, J. M. , Hermans, K. , & Haesebrouck, F. (2009). Main pathologies associated with Staphylococcus aureus infections in rabbits: A review. World Rabbit Science, 17, 115–125. [Google Scholar]
- Drougka, E. , Fokaa, A. , Koutinas, C. K. , Jelastopulu, E. , Giormezis, N. , Farmaki, O. , … Spiliopoulou, I. (2016). Interspecies spread of Staphylococcus aureus clones among companion animals and human close contacts in a veterinary teaching hospital. Across‐sectional study in Greece. Preventive Veterinary Medicine, 126, 190–198. 10.1016/j.prevetmed.2016.02.004 [DOI] [PubMed] [Google Scholar]
- Enright, M. C. , Day, N. P. J. , Davies, C. E. , Peacock, S. J. , & Spratt, B. G. (2000). Multilocus sequence typing for characterization of methicillin‐resistant and methicillin‐susceptible clones of Staphylococcus aureus . Journal of Clinical Microbiology, 38, 1008–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster, T. J. , & Geoghegan, J. A. (2015). Staphylococcus aureus . Medical Microbiology and Immunology. 10.1016/B978-0-12-397169-2.00037-8 [DOI] [Google Scholar]
- Frénay, H. M. , Bunschoten, A. E. , Schouls, L. M. , Van Leeuwen, W. J. , Vandenbrouke‐Grauls, C. M. , Verhoef, J. , & Mooi, F. R. (1996). Molecular typing of methicillin‐resistant Staphylococcus aureus on the basis of protein a gene polymorphism. European Journal of Clinical Microbiology and Infectious Diseases, 15, 60–64. 10.1007/BF01586186 [DOI] [PubMed] [Google Scholar]
- Gómez‐Sanz, E. , Torres, C. , Lozano, C. , Fernández‐Pérez, R. , Aspiroz, C. , Ruiz‐Larrea, F. , & Zarazaga, M. (2010). Detection, molecular characterization, and clonal diversity of methicillin‐resistant Staphylococcus aureus CC398 and CC97 in Spanish slaughter pigs of different age groups. Foodborne Pathogens and Disease, 7, 1269–1277. 10.1089/fpd.2010.0610 [DOI] [PubMed] [Google Scholar]
- Goñi, P. , Vergara, Y. , Ruiz, J. , Albizu, I. , Vila, J. , & Gómez‐Lus, R. (2004). Antibiotic resistance and epidemiological typing of Staphylococcus aureus strains from ovine and rabbit mastitis. International Journal of Antimicrobial Agents, 23, 268–272. [DOI] [PubMed] [Google Scholar]
- Griggs, D. J. , Marona, H. , & Piddock, L. J. V. (2003). Selection of moxifloxacin‐resistant Staphylococcus aureus compared with five other fluoroquinolones. Journal of Antimicrobial Chemotherapy, 51, 1403–1407. 10.1093/jac/dkg241 [DOI] [PubMed] [Google Scholar]
- Holmes, M. A. , Harrison, E. M. , Fisher, E. A. , Graham, E. M. , Parkhill, J. , Foster, G. , & Paterson, G. K. (2016). Genomic analysis of companion rabbit Staphylococcus aureus . PLoS ONE, 11, e0151458 10.1371/journal.pone.0151458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iihara, H. , Suzuki, T. , Kawamura, Y. , Ohkusu, K. , Inoue, Y. , Zhang, W. , … Ezaki, T. (2006). Emerging multiple mutations and high‐level fluoroquinolone resistance in methicillin‐resistant Staphylococcus aureus isolated from ocular infections. Diagnostic Microbiology and Infectious Disease, 56, 297–303. 10.1016/j.diagmicrobio.2006.04.017 [DOI] [PubMed] [Google Scholar]
- Jarraud, S. , Mougel, C. , Thioulouse, J. , Lina, G. , Meugnier, H. , Forey, F. , … Vandenesch, F. (2002). Relationships between Staphylococcus aureus genetic background, virulence factors, agr groups (alleles), and human disease. Infection and Immunity, 70, 631–641. 10.1128/IAI.70.2.631-641.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehrenberg, C. , & Schwarz, S. (2006). Distribution of florfenicol resistance genes fexA and cfr among chloramphenicol‐resistant Staphylococcus isolates. Antimicrobial Agents and Chemotherapy, 50, 1156–1163. 10.1128/AAC.50.4.1156-1163.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolecka, A. , Khayhan, K. , Groenewald, M. , Theelen, B. , Arabatzis, M. , Velegraki, A. , … Boekhout, T. (2013). Identification of medically relevant species of arthroconidial yeasts by use of matrix‐assisted laser desorption ionization‐time offlight mass spectrometry. Journal of Clinical Microbiology, 51, 2491–2500. 10.1128/JCM.00470-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagier, J. , Fenollar, F. , Hallé, O. , Lepidi, H. , & Raoult, D. (2009). Letters to the editor: Trimethoprim resistance genes in vancomycin‐resistant Enterococcus faecium clinical isolates from France. International Journal of Antimicrobial Agents, 34, 390–391. [DOI] [PubMed] [Google Scholar]
- Laplana, L. M. , Cepero, P. G. , Ruiz, J. , Zolezzi, P. C. , Calvo, C. R. , Erazo, M. C. , & Gómez‐Lus, R. (2007). Molecular typing of Staphylococcus aureus clinical isolates by pulsed‐field gel electrophoresis, staphylococcal cassette chromosome mec type determination and dissemination of antibiotic resistance genes. International Journal of Antimicrobial Agents, 30, 505–513. 10.1016/j.ijantimicag.2007.06.020 [DOI] [PubMed] [Google Scholar]
- Loncaric, I. , Künzel, F. , Licka, T. , Simhofer, H. , Spergser, J. , & Rosengarten, R. (2014). Identification and characterization of methicillin‐resistant Staphylococcus aureus (MRSA) from Austrian companion animals and horses. Veterinary Microbiology, 168, 381–387. 10.1016/j.vetmic.2013.11.022 [DOI] [PubMed] [Google Scholar]
- Malachowa, N. , Kobayashi, S. D. , Porter, A. R. , Braughton, K. R. , Scott, D. P. , & Gardner, D. J. (2016). Contribution of Staphylococcus aureus coagulases and clumping factor A to abscess formation in a rabbit model of skin and soft tissue infection. PLoS ONE, 11, e0158293 10.1371/journal.pone.0158293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure, J. , Conly, J. M. , Lau, V. , Elsayed, S. , Louie, T. , Hutchins, W. , & Zhang, K. (2006). Novel multiplex PCR assay for detection of the Staphylococcal virulence marker panton‐valentine leukocidin genes and simultaneous discrimination of methicillin‐susceptible from ‐resistant Staphylococci . Journal of Clinical Microbiology, 44, 1141–1144. 10.1128/JCM.44.3.1141-1144.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moradigaravand, D. , Jamrozy, D. , Mostowy, R. , Anderson, A. , Nickerson, E. K. , Thaipadungpanit, J. , … Wongsuvan, G. (2017). Evolution of the Staphylococcus argenteus ST2250 clone in Northeastern Thailand is linked with the acquisition of livestock‐associated Staphylococcal genes. MBio, 8, e00802‐17 10.1128/mbio.00802-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster, D. , Rickmeyer, J. , Gajdiss, M. , Thye, T. , Lorenzen, S. , Reif, M. , … Liégeois, F. (2017). Differentiation of Staphylococcus argenteus (formerly: Staphylococcus aureus complex 75) by mass spectrometry from S. aureus using the first strain isolated from a wild African great ape. International Journal of Medical Microbiology, 307, 57–63. 10.1016/j.ijmm.2016.11.003 [DOI] [PubMed] [Google Scholar]
- Seki, K. , Sakurada, J. , Seong, H. K. , Murai, M. , Tachi, H. , Ishii, H. , & Masuda, S. (1998). Occurrence of coagulase serotype among Staphylococcus aureus strains isolated from healthy individuals‐special reference to correlation with size of protein‐A gene. Microbiology and Immunology, 42, 407–409. [DOI] [PubMed] [Google Scholar]
- Thaipadungpanit, J. , Amronchai, P. , Nickerson, E. K. , Wongsuvan, G. , Wuthiekanun, V. , Limmathurotsakul, D. , … Peacock, S. J. (2015). Clinical and molecular epidemiology of Staphylococcus argenteus infections in Thailand. Journal of Clinical Microbiology, 53, 1005–1008. 10.1128/JCM.03049-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong, S. Y. C. , Schaumburg, F. , Ellington, M. J. , Corander, J. , Pichon, B. , Leendertz, F. , … Giffard, P. M. (2015). Novel staphylococcal species that form part of a Staphylococcus aureus‐related complex: The non‐pigmented Staphylococcus argenteus sp. nov. and Staphylococcus schweitzeri sp. nov. International Journal of Systematic and Evolutionary Microbiology, 65, 15–22. 10.1099/ijs.0.062752-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tristan, A. , Ying, L. , Bes, M. , Etienne, J. , Vandenesch, F. , & Lina, G. (2003). Use of multiplex PCR to identify Staphylococcus aureus adhesins involved in human hematogenous infections. Journal of Clinical Microbiology, 41, 4465–4467. 10.1128/JCM.41.9.4465-4467.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vancraeynest, D. , Hermans, K. , Martel, A. , Vaneechoutte, M. , Devriese, L. A. , & Haesebrouck, F. (2004). Antimicrobial resistance and resistance genes in Staphylococcus aureus strains from rabbits. Veterinary Microbiology, 101, 245–251. 10.1016/j.vetmic.2004.03.021 [DOI] [PubMed] [Google Scholar]
- Viana, D. , Selva, L. , Segura, P. , Penades, J. R. , & Corpa, J. M. (2007). Genotypic characterization of Staphylococcus aureus strains isolated from rabbit lesions. Veterinary Microbiology, 121, 288–298. 10.1016/j.vetmic.2006.12.003 [DOI] [PubMed] [Google Scholar]
- Wu, D. , Li, X. , Yang, Y. , Zheng, Y. , Wang, C. , Deng, L. , … Yu, S. (2011). Superantigen gene profiles and presence of exfoliative toxin genes in community‐acquired meticillin‐resistant Staphylococcus aureus isolated from Chinese children. Journal of Medical Microbiology, 60, 35–45. 10.1099/jmm.0.023465-0 [DOI] [PubMed] [Google Scholar]
- Youn, J. , Park, Y. H. , Hang'ombe, B. , & Sugimoto, C. (2014). Prevalence and characterization of Staphylococcus aureus and Staphylococcus pseudintermedius isolated from companion animals and environment in the veterinary teaching hospital in Zambia, Africa. Comparative Immunology, Microbiology and Infectious Diseases, 37, 123–130. 10.1016/j.cimid.2014.01.003 [DOI] [PubMed] [Google Scholar]
- Zhang, D. , Xu, X. , Song, Q. , Bai, Y. , Zhang, Y. , Song, M. , & Shi, X. (2016). Identification of Staphylococcus argenteus in Eastern China based on nonribosomal peptide synthetase (NRPS) gene. Future Microbiology, 11, 1113–1121. [DOI] [PubMed] [Google Scholar]


