Abstract
Autophagy is a conserved intracellular degradation pathway essential for protein homeostasis, survival and development. Defects in autophagic pathways have been connected to a variety of human diseases, including cancer and neurodegeneration. In the process of macroautophagy, cytoplasmic cargo is enclosed in a double-membrane structure and fused to the lysosome to allow for digestion and recycling of material. Autophagosome formation is primed by the ULK complex, which enables the downstream production of PI(3)P, a key lipid signalling molecule, on the phagophore membrane. The PI(3)P is generated by the PI3 kinase (PI3K) complex, consisting of the core components VPS34, VPS15 and Beclin 1. Beclin 1 is a central player in autophagy and constitutes a molecular platform for the regulation of autophagosome formation and maturation. Post-translational modifications of Beclin 1 affect its stability, interactions and ability to regulate PI3K activity, providing the cell with a plethora of strategies to fine-tune the levels of autophagy. Being such an important regulator, Beclin 1 is a potential target for therapeutic intervention and interfering with the post-translational regulation of Beclin 1 could be one way of manipulating the levels of autophagy. In this review, we provide an overview of the known post-translational modifications of Beclin 1 that govern its role in autophagy and how these modifications are maintained by input from several upstream signalling pathways.
▓
Subject terms: Macroautophagy, Molecular biology
Facts
Beclin 1 is a core component of the PI3K complex, important for producing PI(3)P on the phagophore membrane in order to recruit downstream autophagy effectors.
Beclin 1 is a multi-domain protein, with a large interactome that facilitates regulation of autophagy in both a positive and negative manner.
Post-translational modifications of Beclin 1 affect protein stability, confirmation, activity and its interactome and can be used as a molecular rheostat to fine-tune autophagic activity.
Targeting Beclin 1 modifiers to regulate Beclin 1 post-translational modifications could provide a possible therapeutic intervention for upregulating autophagy.
Open Questions
Understand the full repertoire of Beclin 1 post-translational modifications relevant to autophagy
Understand the relevance of Beclin 1 post-translational modifications to the activities of the different Beclin 1 complexes
Can regulation of Beclin 1 post-translational modifications be exploited as a therapeutic strategy to counteract accumulation of aggregate-prone proteins in neurodegenerative diseases?
Introduction
Autophagy is an essential and widely conserved cellular degradation process that plays a crucial role in housekeeping and stress survival. In the process of macroautophagy (henceforth referred to as autophagy), a portion of cytoplasmic content, including proteins and organelles, is sequestered into a double-membrane structure called the phagophore. The phagophore membrane expands to fully enclose its cargo, forming the autophagosome, and the autophagosome is trafficked along microtubules to ultimately fuse with the lysosome [1, 2]. Fusion with this lytic compartment ensures the enzymatic degradation of the cargo and subsequent release of recycled material. Autophagy was initially believed to be a non-selective bulk degradation process, but recent data indicate that it is often selective using specific receptors for cargo recognition [3]. Autophagy has been shown to aid in clearing intracellular aggregate-prone proteins underlying neurodegenerative diseases, including Alzheimer and Parkinson disease, and upregulating autophagy induction may be a promising therapeutic approach for these types of diseases [4].
PI(3)P production on the phagophore membrane
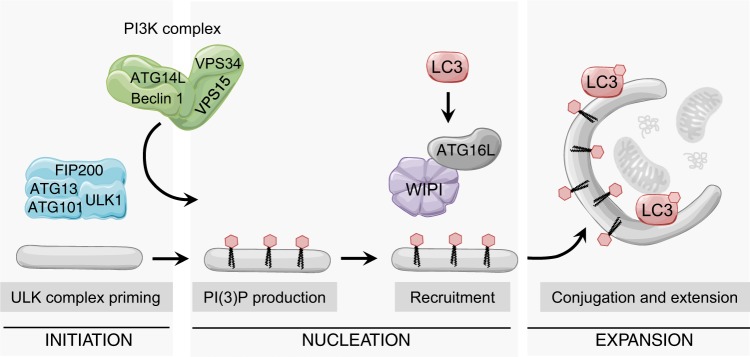
Under normal conditions, cells sustain basal levels of autophagy to maintain homeostasis. In addition, autophagy can be triggered by a variety of stimuli, such as nutrient deprivation, metabolic imbalance, protein aggregation and oxidative stress. The process of autophagosome biogenesis includes three main stages: initiation, nucleation, and expansion of the isolation membrane (Fig. 1). Key initiating events in autophagosome biogenesis are dictated by the Unc-51-like autophagy-activating kinase (ULK) complex, which consists of ULK1 (or ULK2), ATG13, ATG101 and FIP200 [5]. The ULK complex facilitates the production of phosphatidylinositol-3-phosphate (PI(3)P) by recruiting and activating the kinase VPS34, which is found in the class III PI3 kinase (PI3K) complex together with a number of proteins, including VPS15, ATG14L and Beclin 1, the mammalian orthologue of yeast Atg6 [6] (Fig. 1). The pool of PI(3)P produced at the site of the phagophore creates a platform for the recruitment of subsequent autophagy machinery effectors, like WIPI family proteins and ATG16L. These effectors facilitate conjugation of ATG8 proteins, such as LC3, to the phagophore membrane, which in turn mediates cargo recruitment, and membrane extension of the phagophore [1, 7, 8].
Fig. 1.
Autophagy initiation depends on ULK1 priming and PI(3)P production. Upon autophagy initiation signals, the ULK1 complex (containing ULK1 or ULK2) activates the PI3K complex, which directs PI(3)P production to a membrane site. The PI(3)P-rich membrane recruits downstream autophagy effectors, like WIPI proteins and ATG16L, which mediates the membrane conjugation of LC3. LC3 then stimulates membrane expansion and cargo recruitment into the forming phagophore.
The main signalling cues for autophagy initiation converge on PI3K regulation
The early initiation steps of autophagy are regulated by diverse signal-sensing proteins, including four protein kinases: mammalian target of rapamycin complex 1 (mTORC1), ULK1, AMP-activated protein kinase (AMPK), and AKT (Fig. 2). Nutrient starvation is one of the most studied autophagy inducers and the serine/threonine mTOR kinase plays a major role in sensing nutrient availability [9–11]. Lack of nutrients, mainly amino acids, triggers an intracellular signalling cascade, which inhibits mTORC1 activity [12]. Upon starvation, inactivated mTORC1 dissociates from ULK1, resulting in dephosphorylation and activation of the ULK1 complex [13, 14]. Activated ULK1 phosphorylates several proteins involved in autophagy initiation and progression, including the ULK complex components ATG13, ATG101 and FIP200 [15], and downstream effectors like the VPS34, ATG14L, Beclin 1 and AMBRA1 components of the PI3K complex [16–18].
Fig. 2.
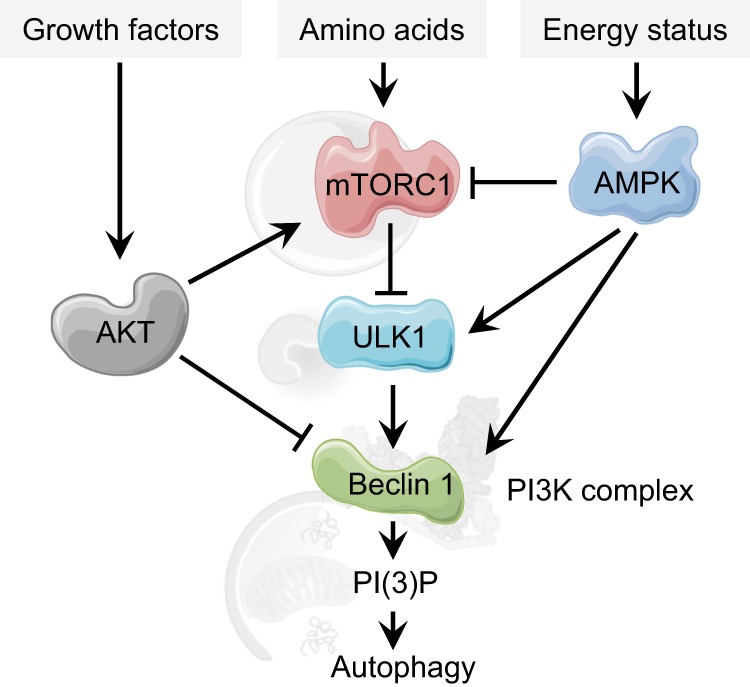
Upstream modulation of autophagy responds to nutritional cues and converges on regulation of the Beclin 1-containing PI3K complex. When amino acids are abundant, mTORC1 is active and phosphorylates and inhibits the autophagy-priming complex ULK1. This prevents ULK1 from activating the PI3K complex to initiate PI(3)P production. During amino acid starvation, the inhibitory effect of mTORC1 is released and autophagy can be induced. The presence of growth factors is sensed by the AKT kinase, which activates mTORC1 and inhibits PI3K activity. Cellular energy status can also be detected via the AMP/ATP ratio, where increase in AMP activates the kinase AMPK, which activates autophagy via mTORC1 inhibition, ULK1 activation, and PI3K activation
Autophagy can also be triggered by low energy conditions through activation of AMPK. AMPK responds to low energy conditions, detected as changes in the ATP-to-ADP or ATP-to-AMP ratios, by inhibiting ATP-consuming anabolic processes and by promoting catabolic processes to restore the proper metabolic balance [19]. Activated AMPK triggers autophagy by coordinated activation of ULK1 and negative regulation of the mTOR kinase [20–22]. As described in the following sections, AMPK also exerts a direct role in regulating components of the PI3K complex.
Another important input for nutrient status is the presence of extracellular growth factors. The serine threonine kinase AKT (also called protein kinase B (PKB)) is recruited to active growth factor receptors at the plasma membrane, and binding initiates its downstream signalling cascade [23]. AKT phosphorylation activates mTORC1 and inhibits autophagy, and AKT also suppresses autophagy via mTOR-independent pathways by direct phosphorylation of key autophagy proteins, including components of the PI3K complex [24–26].
Beclin 1 is a key regulator of autophagy
Beclin 1 was one of the first mammalian autophagy proteins identified [27], and BECN1 is an essential gene required for embryonic survival and development [28]. BECN1 also functions as a tumour-suppressor gene and has been found to be monoallelically deleted in many cancer types, and Beclin 1 deficiency is also associated with several neurodegenerative diseases [6, 28–32]. Beclin 1 regulates both autophagosome synthesis and autophagosome maturation, by forming three distinct PI3K complexes together with the core lipid kinase VPS34 and the regulatory component VPS15 [33–35]. Complex I includes the core proteins together with ATG14L and is involved in autophagy initiation and autophagosome formation, whereas in complex II ATG14L is replaced by UVRAG and this complex regulates autophagosome maturation and endocytosis [33, 36]. In the third PI3K complex, RUBICON interacts with Beclin 1, VPS34 and UVRAG of complex II to inhibit lipid kinase activity and reduce autophagic flux [34, 35, 37].
Beclin 1 interacts with the PI3K core component VPS34 and lipid membranes via its C-terminal β/α-repeated, autophagy-related (BARA) domain (previously denoted evolutionary conserved domain) [38–40] (Fig. 3a, b) and binds ATG14L or UVRAG in a mutually exclusive manner via its coiled-coiled domain (CCD) [36]. Additionally, Beclin 1 has an unstructured N-terminal domain, followed by a BCL2-homology-3 (BH3) domain and a flexible helical domain (HD) (Fig. 3a, b) [41, 42]. The many domains of Beclin 1 mediate communications with multiple interaction partners that can alter its conformation and binding accessibility, thus making Beclin 1 an important molecular platform for the regulation of PI3K activity and autophagy [43]. For example, the BH3 domain of Beclin 1 mediates its interaction with the antiapoptotic protein BCL2 and this interaction causes a steric block inhibiting PI3K complex formation [27, 44]. Thus, BCL2 binding to Beclin 1 inhibits autophagy, and nutrient-dependent regulation of this interaction modifies the pool of Beclin 1 molecules available for VPS34 interaction [44, 45]. Another example of how the Beclin 1 interactome can be utilised as a way of fine-tuning autophagic activity is demonstrated by the interaction with AMBRA1. AMBRA1 is a positive regulator of autophagy and competes with BCL2 for binding to the Beclin 1 BH3 domain [46]. Binding of AMBRA1 promotes Beclin 1 interaction with VPS34 and regulates the positioning of the PI3K complex to the site of phagophore formation, thereby mediating autophagy initiation [47, 48]. Consequently, modifications that alter the interaction between Beclin 1 and the PI3K components, or the interaction with AMBRA1 and BCL2, allow cells to modulate autophagic activity in response to a multitude of internal and external cues and could also provide potential targets for pharmaceutical interventions.
Fig. 3.
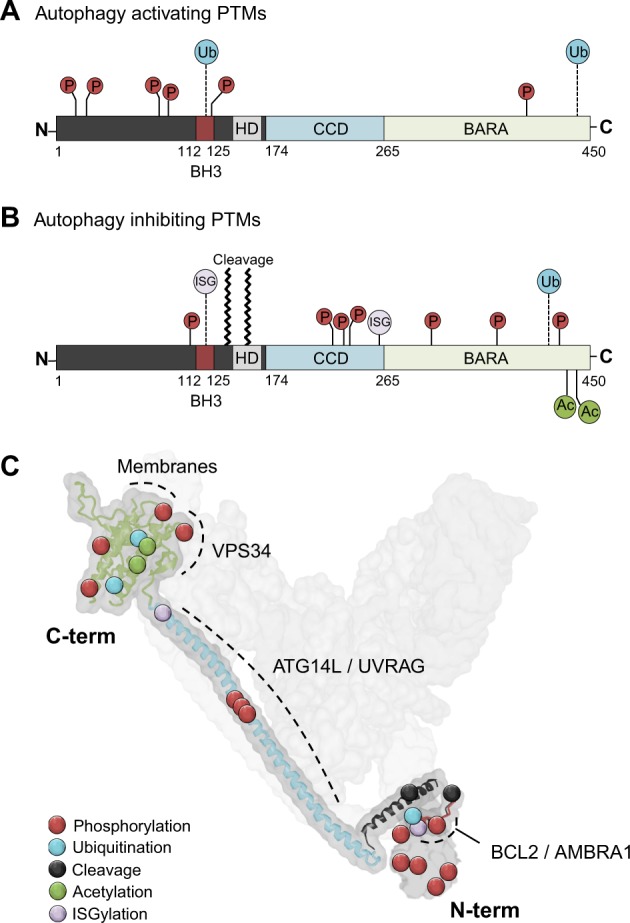
Positions of Beclin 1 post-translational modifications. a, b Linear domain structure of Beclin 1 indicating the BCL2 homology 3 domain (BH3), helical domain (HD), coiled-coiled domain (CCD), and β/α-repeated, autophagy-related (BARA) domain. Known post-translational modifications are indicated by position and type: P phosphorylation, Ub ubiquitination, Ac acetylation, ISG ISGylation. Modifications known to induce autophagy are illustrated in a and modifications known to inhibit autophagy are shown in b. Further details on modifications can be found in Table 1. c Positions of post-translational modifications of Beclin 1. Beclin 1 is visualised oriented in the PI3K complex II (in light grey) in a structure model based on the crystal structure of the yeast PI3K complex, with the Beclin 1 homologue Atg6 [113]. Domains important for Beclin 1 interactors are denoted in dashed lines with labels for interactors. The orientation of Beclin 1 in the complex is indicated by denotation of the C- and N-terminals. The N-terminal of Beclin 1 is intrinsically disordered and has no determined crystal structure; therefore, this part is represented only by a grey surface representation without the overlay of a ribbon model. Protein structure downloaded from iCn3D web viewer [114]
The functions of many proteins directly involved in autophagosome biogenesis are regulated by their post-translational modifications. These modifications include phosphorylation, ubiquitination, acetylation, lipidation and redox regulation. The initiation of autophagy relies on the coordinated spatio-temporal regulation of the levels of modification of key players constituting the core machinery. In this review, we provide an overview of the known post-translational modifications of Beclin 1 that govern its role in autophagy, and how these modifications are maintained by inputs from upstream signalling pathways. The modifications of Beclin 1 mentioned in this paper are listed in Table 1 together with information on supporting experimental evidence, and the physiological relevance in terms of effect on autophagy. Post-translational modifications of other autophagy-related proteins are reviewed elsewhere [49].
Table 1.
Post-translational modifications of Beclin 1
| Site | Modification | Modifier | Consequence | Conditions, cell system | Reference |
|---|---|---|---|---|---|
| Ser15 | Phosphorylation | ULK1 | Activation | Amino acid starvation, MEFs, HEK293, C elegans, in vitro | [18] |
| Ser90, Ser93 | Phosphorylation | AMPK | Activation | Glucose starvation, HeLa, MEFs, in vitro | [53] [36] |
| Thr388 | Phosphorylation | AMPK | Activation | Glucose starvation, HEK293T, MEFs, treatment with AMPK stimulator/inhibitor, in vitro | [55] |
| Ser90 | Phosphorylation | MAPKAPK2/3 | Activation | starvation (HBSS), HeLa, MEFs, in vitro | [54] |
| Ser90 | Phosphorylation | DAPK3 | Activation | Serum starvation, mouse tissues, HeLa, MCF7, in vitro | [57] |
| Ser90 | Dephosphorylation | PP2A | Inhibition | Insulin addition, okadic acid treatment, mouse tissues, HeLa, MCF7, in vitro | [57] |
| Ser90 | Phosphorylation | CaMKII | Activation | Ionomycin treatment, HEK293, in vitro | [58] |
| Ser30 | Phosphorylation | PGK1 | Activation | Glutamine starvation, Hypoxia, U87, BxPC-3, MDA-MB-231, in vitro | [60] |
| Ser234, Ser295 | Phosphorylation | AKT1 | Inhibition | Starvation (EBSS), HeLa, in vitro | [26] |
| Tyr229, Tyr233, Tyr352 | Phosphorylation | EGFR | Inhibition | Serum starvation, HeLa, A549 | [61] |
| Tyr233 | Phosphorylation | FAK | Inhibition | Phenylephrine treatment, Cardiomyocytes, Cos | [62] |
| Tyr233 | Phosphorylation | HER2 | Inhibition | Basal conditions, serum starvation, HER2-positive breast cancer cell lines (BT-474, SK-BR3, and MDA-MB-361) | [63] |
| Thr119 | Phosphorylation | DAPK | Activation | Basal condition, HEK293, in vitro | [64] |
| Thr119 | Phosphorylation | ROCK1 | Activation | starvation (HBSS), HeLa, MEFs, in vitro | [65] |
| Thr108 | Phosphorylation | MST1 | Inhibition | Glucose starvation, cardiomyocytes, in vitro | [68] |
| — | Lys11- and Lys63-linked ubiquitination | NEDD4 | Inhibition? | Basal conditions, HeLa. Role in autophagy not established | [70] |
| — | Lys6- and Lys27-linked ubiquitination | NEDD4 | Activation | Basal conditions, Torin treatment, HeLa, HEK293, MEFs | [71] |
| Lys117 | Lys63-linked ubiquitination | TRAF6 | Activation | LPS and IFN-γ treatment, starvation (HBSS), mouse macrophages (RW264.7), primary human monocytes | [59] |
| Lys437 | Lys63-linked ubiquitination | AMBRA1 | Activation | Starvation (HBSS), MEFs, HeLa | [75] |
| — | Lys63-linked ubiquitination | TRIM50 | Activation | Basal condition, starvation (EBSS), rapamycin treatment, HEK293, HeLa, MEFs | [77] |
| — | Lys63-linked deubiquitination | USP14 | Inhibition | Serum starvation, H4 | [78] |
| — | Lys48-linked ubiquitination | RNF216 | Inhibition | LPS treatment, starvation (HBSS), mouse macrophages (RW264.7), HEK293T | [79] |
| — | Lys48-linked ubiquitination | KLHL20 | Inhibition | Starvation (HBSS), HeLa, mouse muscle cells, HEK293T | [80] |
| — | Lys48-linked deubiquitination | USP10, USP13 | Activation | Basal conditions, MEFs, HeLa, HEK293T, H4, Bcap-37 | [81] |
| — | Deubiquitination | USP9X | Activation | Basal conditions, HEK293T | [83] |
| Lys402 | Lys48-linked deubiquitination | ATAXIN3 | Activation | Starvation (HBSS), HeLa, primary neurons, mouse striatal-derived cells | [31] |
| — | Cleavage | CASP3,6,9,10 | Basal conditions, HeLa | [84] | |
| TDVD133, DQLD149 | Cleavage | CASP3,7 and 8 | Inhibition | Growth-factor withdrawal (interleukin-3), pro-apoptotic compounds, mouse (Ba/F3, FDCP1) and human (U937) cell lines, HeLa | [86] |
| DQLD149 | Cleavage | CASP3 | Inhibition | BAX overexpression, pro-apoptotic compounds, HeLa, SK-N-SH, MEFs | [85] |
| — | Cleavage | Calpain | Inhibition | Ischaemia injury, Rat retina | [89] |
| Lys430, Lys437 | Acetylation | EP300 | Inhibition | Basal conditions, HEK293T, MCF7, HeLa | [90] |
| Ser409 | Phosphorylation | CK1γ2 | Inhibition | Serum starvation, HEK293 | [90] |
| Lys430, Lys437 | Deacetylation | SIRT1 | Activation | Basal conditions, HEK293T, MCF7 | [90] |
| Lys117, Lys263 | ISGylation | HERC5/USP18 | Inhibition | Type I interferons treatment, H4 cells, HepG2, HEK293T | [91] |
| — | O-GlcNAc | — | — | Cardiomyocytes | [92] |
Known residues of Beclin 1 targeted for modification, the modifying enzyme and the relevance of that modification in terms of autophagy (activation/inhibition). The table also lists the autophagy-inducing conditions used and what systems/cell models were used to establish the modification of Beclin 1, and whether the modification was confirmed in vitro
Activational phosphorylation of Beclin 1
Phosphorylation is the most intensively studied post-translational modification in the autophagy process, where a phosphate group is reversibly attached onto serine, threonine or tyrosine residues in the substrate protein [50]. This post-translational modification changes protein conformation and can affect accessibility for interactions and enzymatic activity, and many of the events in autophagy initiation rely on kinase signalling. Upon amino acid depletion, mTORC1 is inactivated and its inhibitory effect on the ULK1 complex is relieved [13]. ULK1 then phosphorylates Beclin 1 on Ser15 (Ser14 in mouse) to induce the activity of ATG14L-containing VPS34 complexes and stimulate autophagy initiation [18]. This phosphorylation site is in the N-terminal domain of Beclin 1 (Fig. 3a, c; Table 1), which is dispensable for interaction with the PI3K components. Indeed, mutating the ULK1 phosphorylation site did not affect the Beclin 1 interactions with PI3K components, indicating that this phosphorylation site is important for the regulatory role of Beclin 1 on VPS34 kinase activity, rather than affecting complex assembly [18, 51].
The activating phosphorylation of Ser15 of Beclin 1 in autophagy initiation is dependent on ATG14L, which acts as an adaptor for recruiting ULK1 to Beclin 1 at the phagophore by binding Beclin 1 and ATG13 of the ULK1 complex [17]. Indeed, expression of a mutant ATG14L (ΔBATS) that binds Beclin 1 but is unable to localise to the phagophore severely compromised ULK1-mediated phosphorylation of Beclin 1 [18, 52]. In addition, the recruitment of Beclin 1 to the phagophore is dependent on ULK1-mediated phosphorylation of AMBRA1 [48]. Thus ULK1-dependent phosphorylation activates autophagy via two steps: Beclin 1 recruitment and activation. ULK1-dependent phosphorylation of Beclin 1 Ser15 is also thought to be important during later steps of autophagosome formation, where UVRAG has been suggested to play a role similar to ATG14L in stimulating ULK1 activity [18].
The activity of ULK1 is also governed by AMPK, another major nutrient-sensing kinase. During glucose depletion and in low energy states, AMPK phosphorylates ULK1 and stimulates ULK1-dependent autophagy initiation (Fig. 2) [21]. AMPK also directly phosphorylates Beclin 1 on Ser90 and Ser93 (Ser91 and Ser94 in mouse) (Table 1) [53]. Similar to the activating phosphorylation by ULK1, phosphorylation at these sites do not alter PI3K complex formation or membrane association but rather regulate PI3K kinase activity [39, 54]. Interestingly, AMPK exerts a dual regulation of the PI3K activity governed by ATG14L, where AMPK mediates an inhibitory phosphorylation on VPS34 unless ATG14L is present to inhibit this phosphorylation and instead stimulates the activational phosphorylation of Beclin 1 [53]. In vitro studies show that UVRAG might play a similar role in directing AMPK activity to the PI3K complex II, but further investigations of this are required [53]. Remarkably, the regulation of AMPK activity by ATG14L seems independent of phagophore localisation of ATG14L, as phosphorylation was still stimulated by an ATG14L ΔBATS mutant [34, 53]. Consequently, the timing and location of AMPK-mediated phosphorylation of Beclin 1 with respect to autophagy initiation remains unclear. Another study confirmed that AMPK phosphorylates Beclin 1 but identified Thr388 as the target residue [55]. As opposed to Ser90 phosphorylation, this modification affected PI3K complex formation by shifting Beclin 1 affinity, disrupting the Beclin 1–BCL2 interaction and favouring the Beclin 1–VPS34 interaction [55]. While the importance of AMPK phosphorylation for inducing autophagy during glucose-deprivation was evident in the two separate studies, it is not known if and how phosphorylation at the two different sites are co-regulated. In addition, an autophagy-independent role for Beclin 1 phosphorylation by AMPK has been shown, where phosphorylation of Ser90, Ser93 and Ser96 enables Beclin 1 to interact with SLC7A11 and initiate ferroptosis, a type of programmed cell death [56].
Phosphorylation at Ser90 seems to be a key event for Beclin 1 activation and is considered to be the initial event that mediates further phosphorylation at Ser93 [39]. Monophosphorylation at Ser90 is considered sufficient to have an activating effect upon autophagy induction, and this modification appears to be mediated by different kinases, depending on cellular context and tissue type (Fig. 3; Table 1). In addition to AMPK-mediated phosphorylation upon glucose starvation, Ser90 is phosphorylated by the stress-responsive kinases MAPKAPK2/3 upon amino acid starvation, and by death-associated protein kinase 3 (DAPK3) during serum starvation in mouse skeletal muscle tissue [54, 57]. Upon insulin addition, the same site is dephosphorylated by the protein phosphatase 2A (PP2A). When autophagy is induced by treatment with ionomycin, the calcium/calmodulin-dependent protein kinase II (CamKII) phosphorylates Beclin 1 at Ser90 in a manner that increases interaction with VPS34 and activates Beclin 1 via TRAF6-mediated K63-linked ubiquitination [58, 59]. Furthermore, the Ser90 phosphorylation site is located within the BCL2-binding domain of Beclin 1, where phosphorylation and BCL2 binding occurs in a competitive manner, providing another important point of regulation [54, 58]. The importance of the Ser90 phosphorylation site is further emphasised by studies showing that this site is important for the tumour-suppression function of Beclin 1 [54].
PI3K activity can also be enhanced by phosphorylation of Beclin 1 Ser30, and this seems to be the preferred mechanism for autophagy activation upon glutamine deprivation and hypoxia in cancer cell lines, where Ser15/Ser90/Ser96 phosphorylation by ULK1 and AMPK seem less essential [60]. This phosphorylation is accomplished by phosphoglycerate kinase 1 (PGK1) and contributes to cancer cell proliferation during hypoxic conditions.
Phosphorylation of Beclin 1 is not only a way to positively regulate autophagy, it can also be used to reduce PI3K activity and decelerate autophagic flux. In the presence of extracellular growth factors, AKT phosphorylates Beclin 1 Ser295 and Ser234 [26]. This phosphorylation bridges Beclin 1 and 14-3-3 proteins and sequesters the protein complex to intermediate filaments, inhibiting the activation of Beclin 1 in nutrient-rich conditions to suppress autophagy.
Phosphorylation regulates the Beclin 1 interactome
Several phosphorylation events can directly affect protein interactions and shift the Beclin 1 interactome to induce or inhibit autophagy. In growth-factor rich conditions, active epidermal growth factor receptor (EGFR) binds Beclin 1, leading to multisite phosphorylation on residues Tyr229, Tyr233 and Tyr352, which decreases the Beclin 1 interaction with VPS34 and increases its interaction with negative regulators, such as RUBICON and BCL2 [61]. Inhibitory phosphorylation of Beclin 1 Tyr233 occurs also during phenylephrine-stimulated repression of autophagy, where focal adhesion kinase (FAK) seems to be the responsible modifier [62]. Another growth factor receptor that regulates autophagy by tyrosine phosphorylation on Beclin 1 was identified in human breast cancer cells [63]. Here the activated human epidermal growth factor receptor 2 (HER2) was shown to bind Beclin 1 and phosphorylate Tyr233, a modification that leads to a decrease in both basal and starvation-induced autophagy [63]. However, the direct phosphorylation of these sites has not been verified in vitro, and therefore the possible involvement of other kinases and regulators cannot be excluded.
As mentioned previously, the antiapoptotic protein BCL2 affects the interaction between Beclin 1 and VPS34 and also sterically hinders accessibility for activating kinases [54, 58]. The interaction between Beclin 1 and BCL2 is itself highly regulated by phosphorylation and can be prevented via direct phosphorylation of the BH3 domain in either Beclin 1 or BCL2 to induce autophagy. Beclin 1 can be phosphorylated at Thr119 by DAPK or Rho kinase 1 (ROCK1) [64, 65]. ROCK1 may also prevent the inhibitory Beclin 1/BCL2 dimer formation upon nutrient stress by activating the c-Jun amino-terminal kinase 1 (JNK1) pathway, where JNK1 phosphorylates BCL2 [65, 66]. However, it is not clear which kinase is directly phosphorylating Thr119 of Beclin 1. There may be differences in this process between cell types and conditions, and it should be noted that Thr119 phosphorylation was not observed upon starvation in an MCF7 cell line [57]. The phosphorylation that inhibits the Beclin 1-BCL2 interaction does not seem to interfere with Beclin 1 binding to AMBRA1, even though this positive autophagy regulator also binds Beclin 1 via the BH3 domain. This might be due to slightly different binding motifs, and it is possible that AMBRA1 interacts with Beclin 1 also via other domains [46].
Phosphorylation can also stabilise the interaction of Beclin 1 and BCL2 to inhibit autophagy and possibly initiate apoptosis by limiting the amount of free BCL2 that can interact with and inactivate the pro-apoptotic protein BAX [67]. This is the case in cardiomyocytes, where stress-induced mammalian sterile 20-like kinase 1 (MST1) phosphorylates Beclin 1 in its BH3 domain at Thr108, promoting binding of Beclin 1 to BCL2 [68].
Activational ubiquitination of Beclin 1
Ubiquitination of protein is not only a key regulatory step targeting proteins for degradation but also could alter its localisation or composition and activity of multiprotein complexes. [69]. In the process of ubiquitination, ubiquitin is covalently attached to a lysine residue of a target protein by a cascade of enzymatic reactions involving E1, E2 and E3 enzymes. The action of ubiquitin ligases can be reversed by deubiquitinating enzymes (DUBs), a family of proteases that remove ubiquitin chains from substrate proteins by peptide cleavage.
Beclin 1 is regulated by ubiquitination in numerous ways that affect its stability and function in the autophagy initiation process. The E3 ubiquitin ligase NEDD4 (neural-precursor-cell-expressed developmentally downregulated 4) was reported to interact with Beclin 1 via a PY-motif located downstream of the BARA domain (aa 349–352), to mediate Beclin 1 ubiquitination and degradation (Fig. 3, Table 1) [70]. Levels of Beclin 1 correlate inversely with the levels of NEDD4, and enhanced expression of NEDD4 results in increased Lys11- and Lys63-linked polyubiquitination of Beclin 1 both in vivo and in vitro, further establishing the role of NEDD4 in Beclin 1 regulation. While the role of NEDD4-mediated Beclin 1 ubiquitination in autophagy was not established in the initial studies, it was assumed that it would inhibit autophagy by promoting Beclin 1 degradation [70]. Controversially, other studies found the opposite to be true, claiming that NEDD4 is a positive regulator of autophagy [71, 72]. The contradictory results suggest that NEDD4 mediates mainly Lys6- and Lys27-linked ubiquitination of Beclin 1, which functions to protect Beclin 1 against Lys48-linked ubiquitination and stabilise protein levels during autophagy induction [71]. This study further showed that NEDD4 also interacts with proteins of the ATG8 family and ULK1, extending the role of NEDD4 in autophagy regulation beyond its modification of Beclin 1.
Lys63-linked polyubiquitination of Beclin 1 has been linked to increased activity of the PI3K complex and is considered to be a modification that positively regulates autophagy. During the inflammatory response in murine macrophage cells stimulated with the Toll-like receptor 4 (TLR4) agonist lipopolysaccharide and interferon-γ, the E3 ligase TRAF6 modifies Beclin 1 with Lys63-linked ubiquitin chains [59]. This post-translational modification occurs on Beclin 1 Lys117, which is strategically located in the BH3 domain of Beclin 1 (Fig. 3a), and the modification was opposed by the activity of the DUB A20. Both TRAF6 and A20 were shown to bind Beclin 1 and the binding of TRAF6 was mapped to two sites, one in the Beclin 1 N-terminal domain (aa 54–58) and one in the C-terminal BARA domain (aa 297–301). The TRAF6-mediated polyubiquitination of Beclin 1 affected oligomerisation of Beclin 1, inhibiting homodimerisation and promoting complex formation with VPS34, thereby leading to autophagy induction [59, 73]. Additionally, Lys63-linked ubiquitination of Beclin 1 was enhanced by coexpression of the E3 ligase TRIM16, suggesting that this protein might be an additional regulator of Beclin 1 stability [74]. However, a direct interaction was not shown, and it is possible that this effect is mediated indirectly via other E3 ligases.
During mouse embryonic development, Lys63-linked ubiquitination of Beclin 1 is necessary to maintain autophagic flux and ensure embryonic survival [75]. In this scenario, Lys437 is the main target residue and ubiquitination is suggested to be mediated by the DDB1–Cullin 4 E3 ubiquitin ligase complex containing AMBRA1 as the substrate receptor to mediate the interaction with Beclin 1 (Fig. 3a, c, Table 1) [75, 76]. The WASH (Wiskott–Aldrich syndrome protein (WASP) and SCAR Homologue) protein was identified as a negative regulator of autophagy by binding Beclin 1 and inhibiting Beclin 1 Lys63-linked ubiquitination to suppress PI3K activity. WASH was shown to bind the central region of Beclin 1, competing with AMBRA1, and this interaction was decreased upon starvation-induced autophagy. Consistently, WASH deficiency increased Beclin 1 Lys63-linked ubiquitination and enhanced autophagy induction in mouse embryos [75].
A more recent study identified Tripartite motif protein 50 (TRIM50) as another E3 ligase responsible for Beclin 1 Lys63-linked ubiquitination, showing that TRIM50-dependent ubiquitination of Beclin 1 increases during autophagy-inducing conditions, such as starvation or treatment with rapamycin [77]. The ubiquitination of Beclin 1 did not affect interactions with VPS34 but did increase the interactions between the PI3K complex and ULK1, which could account for the activation of autophagy. In addition, TRIM50 activity was dependent on acetylation by the acetyltransferase EP300, placing Beclin 1 activating ubiquitination downstream of regulation governed by acetylation [77].
The activating effect of Beclin 1 Lys63-linked ubiquitination is counteracted by the DUB USP14 [78]. USP14 interacts directly with the Beclin 1 CCD, and depletion of USP14 or treatment with its inhibitor increases Beclin 1 Lys63-linked ubiquitination and promotes the Beclin 1 interaction with ATG14L and UVRAG but not with VPS34. Thus absence of USP14 increases activation of the VPS34 complex without changing the levels of the key PI3K complex components and promotes both autophagy initiation and autophagosome maturation by enhancing the activity of PI3K complexes I and II. Moreover, USP14 deubiquitinating activity is suggested to be regulated by AKT-mediated phosphorylation, providing evidence for the upstream signalling events governing Beclin 1 ubiquitination [78].
Ubiquitination and deubiquitination regulate Beclin 1 levels
Beclin 1 is marked for degradation via modification by Lys48-linked ubiquitin. Thus any proteins that increase this modification would be predicted to be negative regulators of autophagy. In macrophages, the E3 ligase RNF216 (ring finger protein 216) was identified as the protein responsible for Beclin 1 degradation [79], and overexpression of RNF216 was shown inhibit TLR-mediated autophagy induction in bacteria-infected mice, indicating that it is an important regulator of autophagy in the pathogen response [79].
The CUL3-KLHL20 ubiquitin ligase complex is another E3 ligase that governs autophagy by facilitating degradation of several autophagy proteins during prolonged starvation-induced autophagy [80]. KLHL20 was shown to directly bind and ubiquitinate ULK1, VPS34 and Beclin 1 at the phagophore to promote their degradation and prevent unrestrained autophagic activity during prolonged starvation [80]. Consequently, KLHL20 depletion caused an increase in the amplitude and duration of starvation-induced autophagy and reduced cell survival.
DUBs that remove Lys48-linked ubiquitin chains counteract the degradation signal from E3 ligases and rescue proteins from degradation. USP10 and USP13 were identified as two deubiquitinases that target and stabilise Beclin 1 [81]. Depletion of USP10 and USP13 reduced levels of VPS34, Beclin 1, ATG14L, VPS15 and UVRAG, replicating the effects observed when treating cells with Spautin-1, a compound known to inhibit the two DUBs. Intriguingly, the interaction between Beclin 1 and USP10/USP13 does not only stabilise Beclin 1 levels but also reciprocally stabilises the deubiquitinases. Thus Beclin 1 could be considered a regulator of other USP10 and USP13 targets, one of which is the tumour-suppressor protein p53 [82]. All together, these results provide one mechanism to explain the tumour-suppressor function of Beclin 1 and illustrate that PI3K components can regulate their own levels via feedback control of USP10 and USP13 [81]. Another mechanism that may contribute to the tumour-suppressor properties of Beclin 1, in addition to its role in autophagy, is mediated by the USP9X deubiquitinase, which is another modifier of Beclin 1 [83]. Beclin 1 competes with the antiapoptotic protein MCL1 for binding to USP9X and thus the levels of Beclin 1 and MCL1 are reciprocally regulated, providing cells with a way of upregulating autophagy at the same time as inducing apoptosis to prevent unrestricted cell growth [83].
Most recently, the deubiquitinase ATAXIN3 was also reported to control Beclin 1 stability upon starvation-induced autophagy [31]. Depletion of ATAXIN3 caused a decrease in autophagy flux as well as a decrease in the Beclin 1 levels. The drop in Beclin 1 levels was correlated with an increase in Lys48-linked ubiquitination and the Lys402 site within Beclin 1 BARA domain was shown to be a major site for ATAXIN3-mediated deubiquitination (Fig. 3, Table 1). Furthermore, these proteins were shown to directly interact through Beclin 1 BARA domain and ATAXIN3 polyQ region. Interestingly, this interaction was compromised in the presence of neurodegenerative disease-causing polyQ-expanded proteins, like mutant huntingtin, resulting in the decrease in Beclin 1 levels and impaired starvation-induced autophagy [31].
Proteolytic cleavage of Beclin 1
Specific proteolytic events often regulate protein activity and function. Beclin 1 can be regulated by a direct cleavage of its polypeptide chain into fragments, thereby providing additional means for regulating the autophagic response. Beclin 1 cleavage by caspases has been demonstrated and provides a direct link between autophagy and apoptosis [84–86]. Upon prolonged withdrawal of growth factors, treatment with pro-apoptotic compounds or overexpression of pro-apoptotic BAX, Beclin 1 was cleaved into three major fragments of 50, 37 and 35 kDa [85, 86]. Beclin 1 was specifically cleaved in the presence of Caspase 3, 7 and 8 and the generated fragments resulted from Caspase-3-mediated cleavage after TDVD133 and DQLD149 sites (Fig. 3, Table 1). Caspase-generated N-terminal fragments of Beclin 1 relocalise to the nucleus or mitochondria, enhancing cellular apoptotic response [85–87]. The caspase-3-mediated Beclin 1 cleavage can be inhibited by treatment with spermidine, revealing a neuroprotective effect through restoration of Beclin 1-dependent autophagy [88]. Additionally, Beclin 1 was shown to be a substrate for calpain-mediated cleavage following retinal ischaemic injury in rats, although the exact cleavage site was not identified [89].
Other post-translational modifications of Beclin 1
Acetylation is an additional important post-translational modification of proteins, and proteomic studies have identified thousands of acetylated proteins in mammalian cells, including Beclin 1, where this modification appears to be an important regulatory mechanism governing Beclin 1 function in autophagosome maturation [90]. Beclin 1 was found to be acetylated at Lys430 and 437 by EP300 and this modification was further shown to promote the binding of Beclin 1 to RUBICON, providing a mechanism for negative regulation of autophagy [90]. Beclin 1 interaction with EP300 and subsequent acetylation is enhanced when Beclin 1 is phosphorylated at Ser409 by CK1γ2 (Casein kinase 1 gamma 2). Finally, Beclin 1 acetylation and its increased interaction with RUBICON was shown to inhibit intracellular trafficking of autophagosomes and endocytic cargo to lysosomes for degradation [90].
The ubiquitin-like type modifier, ISG15, is one of the major interferon I-stimulated gene products (ISGs) and can be conjugated to the target protein (ISGylation) as part of the antiviral immune response. The ISG-conjugating enzyme, HERC5, and ISG-deconjugating enzyme, USP18, were shown to interact directly with the BH3 domain and CCD of Beclin 1 in cells stimulated with type I interferon [91]. The ISGylation of Beclin 1 was shown to decrease the activity of the PI3K complex by competing with Beclin 1 Lys63 ubiquitination that is essential for Beclin 1 activation, thereby decreasing autophagy response [91]. However, these findings have not been validated in other systems and might correspond to a cell-type-specific response.
The O-Linked β-N-acetylglucosamine (O-GlcNAc) modification is a dynamic, distinct form of protein glycosylation that can occur within the nucleus, cytoplasm or mitochondria as a response to cellular stress. Beclin 1 and BCL2 appear to be targets of O-GlcNAc modification in cardiomyocytes isolated from diabetic mice; however, the impact of this modification on autophagy process remains elusive [92].
Future perspectives
Post-translational modification of proteins has been revealed as an essential regulatory mechanism for controlling autophagy. By regulating the assembly and activity of the PI3K complex and recruitment of critical autophagy effectors, Beclin 1 has emerged as a critical regulatory node and a possible point for intervention to modify autophagic flux. Pharmacological stimulation of autophagy appears to be desirable, as an increase in autophagy flux has been shown to reduce levels of harmful aggregated protein species involved in neurodegeneration [93–96], and targeting Beclin 1 modifications could constitute a way of achieving this. The feasibility of this approach was indicated by mouse model studies where the inhibitory interaction of Beclin 1 and BCL2 was obstructed, and these mice showed signs of increased autophagic flux, increased neuroprotection and extended lifespan [45, 97]. While this study was based on a genetic disruption of the Beclin1–BCL2 complex, other studies are investigating the viability of BH3 mimetics for this approach [98, 99]. Additional studies have developed small peptides to alter Beclin 1 interactions, and these have been proved to be sufficient to regulate autophagy in in vivo settings relevant for disease [63, 100–103].
As phosphorylation of Beclin 1 Ser90 seems to be a key event for autophagy activation in many tissues, blocking or inhibiting this modification might provide a conceivable approach for developing an autophagy-modulating drug. Another approach to modify Beclin 1-dependent autophagy could be to alter levels of Beclin 1 by targeting E3 ligases or DUBs [104, 105]. The example of using the DUB inhibitor Spautin-1 have already been mentioned, and as another example, small molecule inhibitors targeting USP14 have been developed and are showing promising results in neuroprotection [81, 106]. However, specific targeting of modifying enzymes can be challenging as enzymes of a particular family often share similar domains and structure. Furthermore, as many enzymes have a wide range of substrates, inhibition may impact other cellular pathways. An alternative approach could be to specifically inhibit certain modifications by developing peptides or small molecules that target a specific site on the substrate, similar to what was recently reported for blocking the phosphorylation of the apoptotic protein BAD [107].
Future studies of Beclin 1 function are needed to uncover additional layers of regulation by post-translational modifications. There are several post-translational modifications such as methylation, SUMOylation and oxidation that have been identified as regulators of other key autophagy proteins [108–111], and prediction tools reveal that Beclin 1 might be a target for several of these modifications [112]. However, it remains to be determined whether any of these predicted sites are utilised in vivo. Another future direction in the research on post-translational regulation of autophagy would be to identify the proteins that are responsible for reversing the modifications, such as phosphatases, DUBs and deacetylases. It also remains to be determined how different modifications might be affecting each other and how regulation might diverge in different tissues and conditions.
Acknowledgements
We are grateful for funding from the UK Dementia Research Institute (funded by the MRC, Alzheimer’s Research UK and the Alzheimer’s Society) (to DCR), The Tau Consortium, Alzheimer’s Research UK, an anonymous donation to the Cambridge Centre for Parkinson-Plus, the Swedish Natural Research Council (VR) (to S.M.H; reference 2016–06605) and from the European Molecular Biology Organisation (EMBO long-term fellowships to SMH and LW; ALTF 1024-2016 and ALTF 135-2016, respectively).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by F. Pentimalli
Change history
5/9/2019
Reference 45 was incorrect in the original version of this article.
References
- 1.Bento CF, Renna M, Ghislat G, Puri C, Ashkenazi A, Vicinanza M, et al. Mammalian autophagy: how does it work? Annu Rev Biochem. 2016;85:685–713. doi: 10.1146/annurev-biochem-060815-014556. [DOI] [PubMed] [Google Scholar]
- 2.Dikic I, Elazar Z. Mechanism and medical implications of mammalian autophagy. Nat Rev Mol Cell Biol. 2018;19:349–64. doi: 10.1038/s41580-018-0003-4. [DOI] [PubMed] [Google Scholar]
- 3.Farre JC, Subramani S. Mechanistic insights into selective autophagy pathways: lessons from yeast. Nat Rev Mol Cell Biol. 2016;17:537–52. doi: 10.1038/nrm.2016.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rubinsztein DC, Bento CF, Deretic V. Therapeutic targeting of autophagy in neurodegenerative and infectious diseases. J Exp Med. 2015;212:979–90. doi: 10.1084/jem.20150956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lamb CA, Yoshimori T, Tooze SA. The autophagosome: origins unknown, biogenesis complex. Nat Rev Mol Cell Biol. 2013;14:759–74. doi: 10.1038/nrm3696. [DOI] [PubMed] [Google Scholar]
- 6.Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–6. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 7.Polson HE, de Lartigue J, Rigden DJ, Reedijk M, Urbe S, Clague MJ, et al. Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy. 2010;6:506–22. doi: 10.4161/auto.6.4.11863. [DOI] [PubMed] [Google Scholar]
- 8.Dooley HC, Razi M, Polson HE, Girardin SE, Wilson MI, Tooze SA. WIPI2 links LC3 conjugation with PI3P, autophagosome formation, and pathogen clearance by recruiting Atg12-5-16L1. Mol Cell. 2014;55:238–52. doi: 10.1016/j.molcel.2014.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gonzalez A, Hall MN. Nutrient sensing and TOR signaling in yeast and mammals. EMBO J. 2017;36:397–408. doi: 10.15252/embj.201696010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell. 2017;169:361–71. doi: 10.1016/j.cell.2017.03.035. [DOI] [PubMed] [Google Scholar]
- 11.Bar-Peled L, Sabatini DM. Regulation of mTORC1 by amino acids. Trends Cell Biol. 2014;24:400–6. doi: 10.1016/j.tcb.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Son Sung Min, Park So Jung, Lee Huikyong, Siddiqi Farah, Lee Jong Eun, Menzies Fiona M., Rubinsztein David C. Leucine Signals to mTORC1 via Its Metabolite Acetyl-Coenzyme A. Cell Metabolism. 2019;29(1):192-201.e7. doi: 10.1016/j.cmet.2018.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20:1981–91. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, et al. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Egan DF, Chun MG, Vamos M, Zou H, Rong J, Miller CJ, et al. Small molecule inhibition of the autophagy kinase ULK1 and identification of ULK1 substrates. Mol Cell. 2015;59:285–97. doi: 10.1016/j.molcel.2015.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou C, Ma K, Gao R, Mu C, Chen L, Liu Q, et al. Regulation of mATG9 trafficking by Src- and ULK1-mediated phosphorylation in basal and starvation-induced autophagy. Cell Res. 2017;27:184–201. doi: 10.1038/cr.2016.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park JM, Jung CH, Seo M, Otto NM, Grunwald D, Kim KH, et al. The ULK1 complex mediates MTORC1 signaling to the autophagy initiation machinery via binding and phosphorylating ATG14. Autophagy. 2016;12:547–64. doi: 10.1080/15548627.2016.1140293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Russell RC, Tian Y, Yuan H, Park HW, Chang YY, Kim J, et al. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol. 2013;15:741–50. doi: 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garcia D, Shaw RJ. AMPK: mechanisms of cellular energy sensing and restoration of metabolic balance. Mol Cell. 2017;66:789–800. doi: 10.1016/j.molcel.2017.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee JW, Park S, Takahashi Y, Wang HG. The association of AMPK with ULK1 regulates autophagy. PLoS ONE. 2010;5:e15394. doi: 10.1371/journal.pone.0015394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–41. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–61. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burgering BM, Coffer PJ. Protein kinase B (c-Akt) in phosphatidylinositol-3-OH kinase signal transduction. Nature. 1995;376:599–602. doi: 10.1038/376599a0. [DOI] [PubMed] [Google Scholar]
- 24.Inoki K, Li Y, Zhu T, Wu J, Guan KL. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol. 2002;4:648–57. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- 25.Bach M, Larance M, James DE, Ramm G. The serine/threonine kinase ULK1 is a target of multiple phosphorylation events. Biochem J. 2011;440:283–91. doi: 10.1042/BJ20101894. [DOI] [PubMed] [Google Scholar]
- 26.Wang RC, Wei Y, An Z, Zou Z, Xiao G, Bhagat G, et al. Akt-mediated regulation of autophagy and tumorigenesis through Beclin 1 phosphorylation. Science. 2012;338:956–9. doi: 10.1126/science.1225967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liang XH, Kleeman LK, Jiang HH, Gordon G, Goldman JE, Berry G, et al. Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J Virol. 1998;72:8586–96. doi: 10.1128/jvi.72.11.8586-8596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci USA. 2003;100:15077–82. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lipinski MM, Zheng B, Lu T, Yan Z, Py BF, Ng A, et al. Genome-wide analysis reveals mechanisms modulating autophagy in normal brain aging and in Alzheimer’s disease. Proc Natl Acad Sci USA. 2010;107:14164–9. doi: 10.1073/pnas.1009485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pickford F, Masliah E, Britschgi M, Lucin K, Narasimhan R, Jaeger PA, et al. The autophagy-related protein beclin 1 shows reduced expression in early Alzheimer disease and regulates amyloid beta accumulation in mice. J Clin Invest. 2008;118:2190–9. doi: 10.1172/JCI33585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ashkenazi A, Bento CF, Ricketts T, Vicinanza M, Siddiqi F, Pavel M, et al. Polyglutamine tracts regulate beclin 1-dependent autophagy. Nature. 2017;545:108–11. doi: 10.1038/nature22078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shibata M, Lu T, Furuya T, Degterev A, Mizushima N, Yoshimori T, et al. Regulation of intracellular accumulation of mutant Huntingtin by Beclin 1. J Biol Chem. 2006;281:14474–85. doi: 10.1074/jbc.M600364200. [DOI] [PubMed] [Google Scholar]
- 33.Liang C, Lee JS, Inn KS, Gack MU, Li Q, Roberts EA, et al. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat Cell Biol. 2008;10:776–87. doi: 10.1038/ncb1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, Kurotori N, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11:385–96. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- 35.Zhong Y, Wang QJ, Li X, Yan Y, Backer JM, Chait BT, et al. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol. 2009;11:468–76. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19:5360–72. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun Q, Zhang J, Fan W, Wong KN, Ding X, Chen S, et al. The RUN domain of rubicon is important for hVps34 binding, lipid kinase inhibition, and autophagy suppression. J Biol Chem. 2011;286:185–91. doi: 10.1074/jbc.M110.126425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Furuya N, Yu J, Byfield M, Pattingre S, Levine B. The evolutionarily conserved domain of Beclin 1 is required for Vps34 binding, autophagy and tumor suppressor function. Autophagy. 2005;1:46–52. doi: 10.4161/auto.1.1.1542. [DOI] [PubMed] [Google Scholar]
- 39.Fogel AI, Dlouhy BJ, Wang C, Ryu SW, Neutzner A, Hasson SA, et al. Role of membrane association and Atg14-dependent phosphorylation in beclin-1-mediated autophagy. Mol Cell Biol. 2013;33:3675–88. doi: 10.1128/MCB.00079-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ranaghan MJ, Durney MA, Mesleh MF, McCarren PR, Garvie CW, Daniels DS, et al. The autophagy-related Beclin-1 protein requires the coiled-coil and BARA domains to form a homodimer with submicromolar affinity. Biochemistry. 2017;56:6639–51. doi: 10.1021/acs.biochem.7b00936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee EF, Perugini MA, Pettikiriarachchi A, Evangelista M, Keizer DW, Yao S, et al. The BECN1 N-terminal domain is intrinsically disordered. Autophagy. 2016;12:460–71. doi: 10.1080/15548627.2016.1140292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mei Y, Su M, Soni G, Salem S, Colbert CL, Sinha SC. Intrinsically disordered regions in autophagy proteins. Proteins. 2014;82:565–78. doi: 10.1002/prot.24424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.He C, Levine B. The Beclin 1 interactome. Curr Opin Cell Biol. 2010;22:140–9. doi: 10.1016/j.ceb.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–39. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 45.Fernández Álvaro F., Sebti Salwa, Wei Yongjie, Zou Zhongju, Shi Mingjun, McMillan Kathryn L., He Congcong, Ting Tabitha, Liu Yang, Chiang Wei-Chung, Marciano Denise K., Schiattarella Gabriele G., Bhagat Govind, Moe Orson W., Hu Ming Chang, Levine Beth. Disruption of the beclin 1–BCL2 autophagy regulatory complex promotes longevity in mice. Nature. 2018;558(7708):136–140. doi: 10.1038/s41586-018-0162-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Strappazzon F, Vietri-Rudan M, Campello S, Nazio F, Florenzano F, Fimia GM, et al. Mitochondrial BCL-2 inhibits AMBRA1-induced autophagy. EMBO J. 2011;30:1195–208. doi: 10.1038/emboj.2011.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fimia GM, Stoykova A, Romagnoli A, Giunta L, Di Bartolomeo S, Nardacci R, et al. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007;447:1121–5. doi: 10.1038/nature05925. [DOI] [PubMed] [Google Scholar]
- 48.Di Bartolomeo S, Corazzari M, Nazio F, Oliverio S, Lisi G, Antonioli M, et al. The dynamic interaction of AMBRA1 with the dynein motor complex regulates mammalian autophagy. J Cell Biol. 2010;191:155–68. doi: 10.1083/jcb.201002100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wani WY, Boyer-Guittaut M, Dodson M, Chatham J, Darley-Usmar V, Zhang J. Regulation of autophagy by protein post-translational modification. Lab Invest. 2015;95:14–25. doi: 10.1038/labinvest.2014.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Humphrey SJ, James DE, Mann M. Protein phosphorylation: a major switch mechanism for metabolic regulation. Trends Endocrinol Metab. 2015;26:676–87. doi: 10.1016/j.tem.2015.09.013. [DOI] [PubMed] [Google Scholar]
- 51.Byfield MP, Murray JT, Backer JM. hVps34 is a nutrient-regulated lipid kinase required for activation of p70 S6 kinase. J Biol Chem. 2005;280:33076–82. doi: 10.1074/jbc.M507201200. [DOI] [PubMed] [Google Scholar]
- 52.Matsunaga K, Morita E, Saitoh T, Akira S, Ktistakis NT, Izumi T, et al. Autophagy requires endoplasmic reticulum targeting of the PI3-kinase complex via Atg14L. J Cell Biol. 2010;190:511–21. doi: 10.1083/jcb.200911141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim J, Kim YC, Fang C, Russell RC, Kim JH, Fan W, et al. Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress and autophagy. Cell. 2013;152:290–303. doi: 10.1016/j.cell.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wei Y, An Z, Zou Z, Sumpter R, Su M, Zang X, et al. The stress-responsive kinases MAPKAPK2/MAPKAPK3 activate starvation-induced autophagy through Beclin 1 phosphorylation. Elife. 2015. 10.7554/eLife.05289. [DOI] [PMC free article] [PubMed]
- 55.Zhang D, Wang W, Sun X, Xu D, Wang C, Zhang Q, et al. AMPK regulates autophagy by phosphorylating BECN1 at threonine 388. Autophagy. 2016;12:1447–59. doi: 10.1080/15548627.2016.1185576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Song X, Zhu S, Chen P, Hou W, Wen Q, Liu J, et al. AMPK-mediated BECN1 phosphorylation promotes ferroptosis by directly blocking system Xc(-) activity. Curr Biol. 2018;28:2388.e5–99.e5. doi: 10.1016/j.cub.2018.05.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fujiwara N, Usui T, Ohama T, Sato K. Regulation of Beclin 1 protein phosphorylation and autophagy by protein phosphatase 2A (PP2A) and death-associated protein kinase 3 (DAPK3) J Biol Chem. 2016;291:10858–66. doi: 10.1074/jbc.M115.704908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li X, Wu XQ, Deng R, Li DD, Tang J, Chen WD, et al. CaMKII-mediated Beclin 1 phosphorylation regulates autophagy that promotes degradation of Id and neuroblastoma cell differentiation. Nat Commun. 2017;8:1159. doi: 10.1038/s41467-017-01272-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shi CS, Kehrl JH. TRAF6 and A20 regulate lysine 63-linked ubiquitination of Beclin-1 to control TLR4-induced autophagy. Sci Signal. 2010;3:ra42. doi: 10.1126/scisignal.2000751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qian X, Li X, Cai Q, Zhang C, Yu Q, Jiang Y, et al. Phosphoglycerate kinase 1 phosphorylates Beclin1 to induce autophagy. Mol Cell. 2017;65:917.e6–31.e6. doi: 10.1016/j.molcel.2017.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wei Y, Zou Z, Becker N, Anderson M, Sumpter R, Xiao G, et al. EGFR-mediated Beclin 1 phosphorylation in autophagy suppression, tumor progression, and tumor chemoresistance. Cell. 2013;154:1269–84. doi: 10.1016/j.cell.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cheng Z, Zhu Q, Dee R, Opheim Z, Mack CP, Cyr DM, et al. Focal adhesion kinase-mediated phosphorylation of Beclin1 protein suppresses cardiomyocyte autophagy and initiates hypertrophic growth. J Biol Chem. 2017;292:2065–79. doi: 10.1074/jbc.M116.758268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vega-Rubin-de-Celis S, Zou Z, Fernandez AF, Ci B, Kim M, Xiao G, et al. Increased autophagy blocks HER2-mediated breast tumorigenesis. Proc Natl Acad Sci USA. 2018;115:4176–81. doi: 10.1073/pnas.1717800115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zalckvar E, Berissi H, Mizrachy L, Idelchuk Y, Koren I, Eisenstein M, et al. DAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-XL and induction of autophagy. EMBO Rep. 2009;10:285–92. doi: 10.1038/embor.2008.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gurkar AU, Chu K, Raj L, Bouley R, Lee SH, Kim YB, et al. Identification of ROCK1 kinase as a critical regulator of Beclin1-mediated autophagy during metabolic stress. Nat Commun. 2013;4:2189. doi: 10.1038/ncomms3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wei Y, Pattingre S, Sinha S, Bassik M, Levine B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell. 2008;30:678–88. doi: 10.1016/j.molcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Marquez RT, Xu L. Bcl-2:Beclin 1 complex: multiple, mechanisms regulating autophagy/apoptosis toggle switch. Am J Cancer Res. 2012;2:214–21. [PMC free article] [PubMed] [Google Scholar]
- 68.Maejima Y, Kyoi S, Zhai P, Liu T, Li H, Ivessa A, et al. Mst1 inhibits autophagy by promoting the interaction between Beclin1 and Bcl-2. Nat Med. 2013;19:1478–88. doi: 10.1038/nm.3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Grabbe C, Husnjak K, Dikic I. The spatial and temporal organization of ubiquitin networks. Nat Rev Mol Cell Biol. 2011;12:295–307. doi: 10.1038/nrm3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Platta HW, Abrahamsen H, Thoresen SB, Stenmark H. Nedd4-dependent lysine-11-linked polyubiquitination of the tumour suppressor Beclin 1. Biochem J. 2012;441:399–406. doi: 10.1042/BJ20111424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pei G, Buijze H, Liu H, Moura-Alves P, Goosmann C, Brinkmann V, et al. The E3 ubiquitin ligase NEDD4 enhances killing of membrane-perturbing intracellular bacteria by promoting autophagy. Autophagy. 2017;13:2041–55. doi: 10.1080/15548627.2017.1376160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li Y, Zhang L, Zhou J, Luo S, Huang R, Zhao C, et al. Nedd4 E3 ubiquitin ligase promotes cell proliferation and autophagy. Cell Prolif. 2015;48:338–47. doi: 10.1111/cpr.12184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Noble CG, Dong JM, Manser E, Song H. Bcl-xL and UVRAG cause a monomer-dimer switch in Beclin1. J Biol Chem. 2008;283:26274–82. doi: 10.1074/jbc.M804723200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chauhan S, Kumar S, Jain A, Ponpuak M, Mudd MH, Kimura T, et al. TRIMs and galectins globally cooperate and TRIM16 and Galectin-3 co-direct autophagy in endomembrane damage homeostasis. Dev Cell. 2016;39:13–27. doi: 10.1016/j.devcel.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xia P, Wang S, Du Y, Zhao Z, Shi L, Sun L, et al. WASH inhibits autophagy through suppression of Beclin 1 ubiquitination. EMBO J. 2013;32:2685–96. doi: 10.1038/emboj.2013.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jin J, Arias EE, Chen J, Harper JW, Walter JC. A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol Cell. 2006;23:709–21. doi: 10.1016/j.molcel.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 77.Fusco C, Mandriani B, Di Rienzo M, Micale L, Malerba N, Cocciadiferro D, et al. TRIM50 regulates Beclin 1 proautophagic activity. Biochim Biophys Acta. 2018;1865:908–19. doi: 10.1016/j.bbamcr.2018.03.011. [DOI] [PubMed] [Google Scholar]
- 78.Xu D, Shan B, Sun H, Xiao J, Zhu K, Xie X, et al. USP14 regulates autophagy by suppressing K63 ubiquitination of Beclin 1. Genes & Dev. 2016;30:1718–30. doi: 10.1101/gad.285122.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xu C, Feng K, Zhao X, Huang S, Cheng Y, Qian L, et al. Regulation of autophagy by E3 ubiquitin ligase RNF216 through BECN1 ubiquitination. Autophagy. 2014;10:2239–50. doi: 10.4161/15548627.2014.981792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu CC, Lin YC, Chen YH, Chen CM, Pang LY, Chen HA, et al. Cul3-KLHL20 ubiquitin ligase governs the turnover of ULK1 and VPS34 complexes to control autophagy termination. Mol Cell. 2016;61:84–97. doi: 10.1016/j.molcel.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 81.Liu J, Xia H, Kim M, Xu L, Li Y, Zhang L, et al. Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell. 2011;147:223–34. doi: 10.1016/j.cell.2011.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yuan J, Luo K, Zhang L, Cheville JC, Lou Z. USP10 regulates p53 localization and stability by deubiquitinating p53. Cell. 2010;140:384–96. doi: 10.1016/j.cell.2009.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Elgendy M, Ciro M, Abdel-Aziz AK, Belmonte G, Dal Zuffo R, Mercurio C, et al. Beclin 1 restrains tumorigenesis through Mcl-1 destabilization in an autophagy-independent reciprocal manner. Nat Commun. 2014;5:5637. doi: 10.1038/ncomms6637. [DOI] [PubMed] [Google Scholar]
- 84.Cho DH, Jo YK, Hwang JJ, Lee YM, Roh SA, Kim JC. Caspase-mediated cleavage of ATG6/Beclin-1 links apoptosis to autophagy in HeLa cells. Cancer Lett. 2009;274:95–100. doi: 10.1016/j.canlet.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 85.Luo S, Rubinsztein DC. Apoptosis blocks Beclin 1-dependent autophagosome synthesis: an effect rescued by Bcl-xL. Cell Death Differ. 2010;17:268–77. doi: 10.1038/cdd.2009.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wirawan E, Vande Walle L, Kersse K, Cornelis S, Claerhout S, Vanoverberghe I, et al. Caspase-mediated cleavage of Beclin-1 inactivates Beclin-1-induced autophagy and enhances apoptosis by promoting the release of proapoptotic factors from mitochondria. Cell Death Dis. 2010;1:e18. doi: 10.1038/cddis.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang L, Xu XB, You WW, Lin XX, Li CT, Qian HR, et al. The cytoplasmic nuclear shuttling of Beclin 1 in neurons with Alzheimer’s disease-like injury. Neurosci Lett. 2017;661:63–70. doi: 10.1016/j.neulet.2017.09.055. [DOI] [PubMed] [Google Scholar]
- 88.Yang Y, Chen S, Zhang Y, Lin X, Song Y, Xue Z, et al. Induction of autophagy by spermidine is neuroprotective via inhibition of caspase 3-mediated Beclin 1 cleavage. Cell Death Dis. 2017;8:e2738. doi: 10.1038/cddis.2017.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Russo R, Berliocchi L, Adornetto A, Varano GP, Cavaliere F, Nucci C, et al. Calpain-mediated cleavage of Beclin-1 and autophagy deregulation following retinal ischemic injury in vivo. Cell Death Dis. 2011;2:e144. doi: 10.1038/cddis.2011.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sun T, Li X, Zhang P, Chen WD, Zhang HL, Li DD, et al. Acetylation of Beclin 1 inhibits autophagosome maturation and promotes tumour growth. Nat Commun. 2015;6:7215. doi: 10.1038/ncomms8215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xu D, Zhang T, Xiao J, Zhu K, Wei R, Wu Z, et al. Modification of BECN1 by ISG15 plays a crucial role in autophagy regulation by type I IFN/interferon. Autophagy. 2015;11:617–28. doi: 10.1080/15548627.2015.1023982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Marsh SA, Powell PC, Dell’italia LJ, Chatham JC. Cardiac O-GlcNAcylation blunts autophagic signaling in the diabetic heart. Life Sci. 2013;92:648–56. doi: 10.1016/j.lfs.2012.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ravikumar B, Duden R, Rubinsztein DC. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum Mol Genet. 2002;11:1107–17. doi: 10.1093/hmg/11.9.1107. [DOI] [PubMed] [Google Scholar]
- 94.Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36:585–95. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 95.Sarkar S, Perlstein EO, Imarisio S, Pineau S, Cordenier A, Maglathlin RL, et al. Small molecules enhance autophagy and reduce toxicity in Huntington’s disease models. Nat Chem Biol. 2007;3:331–8. doi: 10.1038/nchembio883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Schaeffer V, Lavenir I, Ozcelik S, Tolnay M, Winkler DT, Goedert M. Stimulation of autophagy reduces neurodegeneration in a mouse model of human tauopathy. Brain. 2012;135(Pt 7):2169–77. doi: 10.1093/brain/aws143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rocchi A, Yamamoto S, Ting T, Fan Y, Sadleir K, Wang Y, et al. A Becn1 mutation mediates hyperactive autophagic sequestration of amyloid oligomers and improved cognition in Alzheimer’s disease. PLoS Genet. 2017;13:e1006962. doi: 10.1371/journal.pgen.1006962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Maiuri MC, Criollo A, Tasdemir E, Vicencio JM, Tajeddine N, Hickman JA, et al. BH3-only proteins and BH3 mimetics induce autophagy by competitively disrupting the interaction between Beclin 1 and Bcl-2/Bcl-X(L) Autophagy. 2007;3:374–6. doi: 10.4161/auto.4237. [DOI] [PubMed] [Google Scholar]
- 99.Lian J, Karnak D, Xu L. The Bcl-2-Beclin 1 interaction in (-)-gossypol-induced autophagy versus apoptosis in prostate cancer cells. Autophagy. 2010;6:1201–3. doi: 10.4161/auto.6.8.13549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shoji-Kawata S. [Identification of a candidate therapeutic autophagy-inducing peptide] Seikagaku. 2015;87:481–4. [PubMed] [Google Scholar]
- 101.Wu S, He Y, Qiu X, Yang W, Liu W, Li X, et al. Targeting the potent Beclin 1-UVRAG coiled-coil interaction with designed peptides enhances autophagy and endolysosomal trafficking. Proc Natl Acad Sci USA. 2018;115:E5669–78. doi: 10.1073/pnas.1721173115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bartolomeo R, Cinque L, De Leonibus C, Forrester A, Salzano AC, Monfregola J, et al. mTORC1 hyperactivation arrests bone growth in lysosomal storage disorders by suppressing autophagy. J Clin Invest. 2017;127:3717–29. doi: 10.1172/JCI94130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Soria LR, Allegri G, Melck D, Pastore N, Annunziata P, Paris D, et al. Enhancement of hepatic autophagy increases ureagenesis and protects against hyperammonemia. Proc Natl Acad Sci USA. 2018;115:391–6. doi: 10.1073/pnas.1714670115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jacomin AC, Taillebourg E, Fauvarque MO. Deubiquitinating enzymes related to autophagy: new therapeutic opportunities? Cells. 2018;7:8. doi: 10.3390/cells7080112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Harrigan JA, Jacq X, Martin NM, Jackson SP. Deubiquitylating enzymes and drug discovery: emerging opportunities. Nat Rev Drug Discov. 2018;17:57–78. doi: 10.1038/nrd.2017.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Boselli M, Lee BH, Robert J, Prado MA, Min SW, Cheng C, et al. An inhibitor of the proteasomal deubiquitinating enzyme USP14 induces tau elimination in cultured neurons. J Biol Chem. 2017;292:19209–25. doi: 10.1074/jbc.M117.815126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pandey V, Wang B, Mohan CD, Raquib AR, Rangappa S, Srinivasa V, et al. Discovery of a small-molecule inhibitor of specific serine residue BAD phosphorylation. Proc Natl Acad Sci USA. 2018;115:E10505–14. doi: 10.1073/pnas.1804897115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yang Y, Fiskus W, Yong B, Atadja P, Takahashi Y, Pandita TK, et al. Acetylated hsp70 and KAP1-mediated Vps34 SUMOylation is required for autophagosome creation in autophagy. Proc Natl Acad Sci USA. 2013;110:6841–6. doi: 10.1073/pnas.1217692110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Scherz-Shouval R, Shvets E, Elazar Z. Oxidation as a post-translational modification that regulates autophagy. Autophagy. 2007;3:371–3. doi: 10.4161/auto.4214. [DOI] [PubMed] [Google Scholar]
- 110.Azad N, Vallyathan V, Wang L, Tantishaiyakul V, Stehlik C, Leonard SS, et al. S-nitrosylation of Bcl-2 inhibits its ubiquitin-proteasomal degradation. A novel antiapoptotic mechanism that suppresses apoptosis. J Biol Chem. 2006;281:34124–34. doi: 10.1074/jbc.M602551200. [DOI] [PubMed] [Google Scholar]
- 111.Song H, Feng X, Zhang M, Jin X, Xu X, Wang L, et al. Crosstalk between lysine methylation and phosphorylation of ATG16L1 dictates the apoptosis of hypoxia/reoxygenation-induced cardiomyocytes. Autophagy. 2018;14:825–44. doi: 10.1080/15548627.2017.1389357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Audagnotto M, Dal Peraro M. Protein post-translational modifications: In silico prediction tools and molecular modeling. Comput Struct Biotechnol J. 2017;15:307–19. doi: 10.1016/j.csbj.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rostislavleva K, Soler N, Ohashi Y, Zhang L, Pardon E, Burke JE, et al. Structure and flexibility of the endosomal Vps34 complex reveals the basis of its function on membranes. Science. 2015;350:aac7365. doi: 10.1126/science.aac7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.National Center for Biotechnology; National Library of Medicine (US). iCn3D. 2016 [cited 19 Sep 2018]. Available from: https://www.ncbi.nlm.nih.gov/Structure/icn3d/icn3d.html