Abstract
The recent discovery of autosis as a variant of autophagy-dependent cell death has challenged the conventional understanding of cell death and programmed cell death in cellular decision making. In contrast to previous accounts of distinct cell death modalities, autosis occurs with high autophagic activity, in the absence of apoptotic and necrotic markers and yet is not fully regulated by typical autophagy markers. Given the metabolic importance of autophagic responses and the extensive cross-talk with both apoptosis and necrosis signalling, the classical and morphotype-driven characterization of cell death as pre-determined subroutines is being increasingly called into question. Furthermore, the conflicting evidence with regards to cell death induction through autophagy modulation in various cancer models highlights the lack of consensus over the extent to which autophagy assists in cell death ontrol and whether it is capable of being a bona fide lethal process. This review evaluates the evidence and context of autophagy-dependent cell death and delineates the role of an autophagic flux threshold associated with ‘lethal’ and ‘non-lethal’ autophagy and its role in autosis control. In doing so, cancer treatment avenues will be explored with regards to precision modulation of tumour autophagic flux to ascertain whether autosis induction may present a novel therapeutic strategy.
Subject terms: Macroautophagy, Lysosomes
Facts
Autosis has been shown to occur independently of apoptotic and necrotic markers.
Inhibition of the Na+, K+ ATPase pump with cardiac glycosides can prevent autosis onset.
A high level of autophagic activity is required to induce autosis, as it has been shown to occur after treatment with autophagy inducing peptides and under conditions of starvation and hypoxia-ischemia.
Open questions
What is the exact level of autophagic activity necessary to accomplish autosis?
Can an adaptive and efficient autophagic response become, flux dependently, autophagy-dependent cell death?
If autosis is mediated solely through autophagy, does a threshold exist that defines lethal and non-lethal autophagic flux?
Can autosis be considered as a druggable target to induce cell death in tumours resistant to apoptosis?
Introduction
Finding alternative, more effective means of cell death control through autophagy modulation remains a major challenge. Given recent advances in gene editing techniques, it seems to be more feasible than ever to not only target which cells to kill, but also to choose the mechanism by which they die [1–4]. Hence, it appears that future enquiries will not necessarily be guided by more precise gene editing approaches, but rather a strong consensus over which regulatory mechanisms to target for desirable cell death control. The elaborate regulatory overlap between cell death subroutines, including the historically employed cell death modalities—apoptosis (type I), autophagy (type II) and necrosis (type III)—complicates this implementation to the extent of delaying clinical translation.
The highly conserved cellular self-consumption process known as autophagy, which delivers cytoplasmic material to lysosomes, has been implicated in many processes crucial to cellular survival, ranging from proteostasis and metabolism to bacterial sequestration and immunity [5–7]. Given its intricate involvement in cellular homeostasis, it is not surprising that much controversy exists surrounding the functionality of autophagosomes. Following the discovery of lysosomal association with phagophores, the observation of dying cells presenting with an abundance of De Duve’s autophagosomes [8] led to the classification of autophagy as a specific cell death modality, historically referred to as type II cell death [8]. This classification was, however, largely based on morphological data, and subsequent molecular characterization revealed major molecular overlap between autophagy, apoptosis and necrosis [9]. Further evidence of the metabolic importance of autophagy has led many to view it as an initiator rather than an effector of cell death, questioning its validity as a cell death modality alltogether. Although it is now clear that cells are characterized by an inherent autophagic proficiency, ranging from baseline autophagic degradation that can dynamically and rapidly change from efficient to new adaptive levels, the role of autophagic activity in autophagy-dependent cell death (ADCD) remains unclear [10, 11]. Yet, studies by Levine et al. suggest the existence of a specific autophagy-mediated variant of cell death referred to as autosis [12]. In parallel to these advances, our understanding of the network-like nature of the autophagic system and its molecular regulation has grown tremendously [13], enabling a better approach to control autophagy in context of disease progression and autophagy defect [14]. Supported by novel tools [15] and approaches [16, 17] to measure autophagic flux, i.e., the rate of protein degradation through autophagy [11] in vitro and in vivo, it is now possible to quantify autophagy with a high degree of precision, with great promise in the treatment of key pathologies [14, 18].
This review will evaluate the evidence for autosis in light of the recent developments in autophagic flux determination to provide insights into the mechanisms that may govern an autophagic flux threshold which separates lethal from non-lethal autophagy. In doing so, the positioning of autophagy as a response mechanism at the interface of adaptation, dysfunction and form of regulated cell death (RCD) becomes clear, as we evaluate the potential of autosis induction as a therapeutic intervention in cancer.
The dynamic nature of cell death
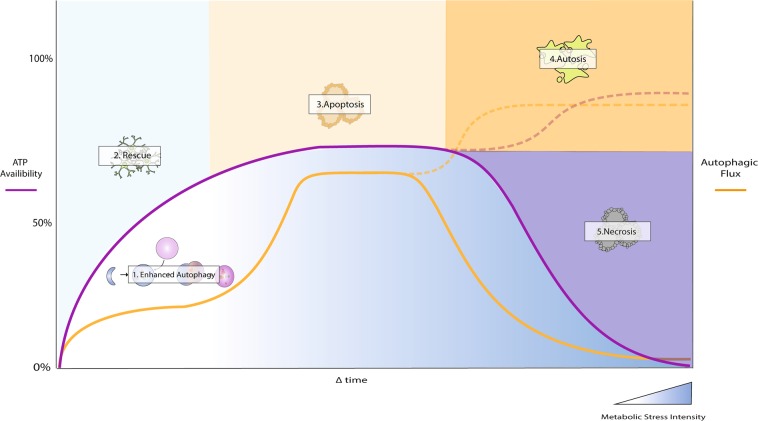
The historic and morphotypic driven description of the three major modes of cell death as ‘programmed’ has often led to the general assumption that cells die in a static, predetermined fashion. However, a growing body of evidence suggests that the degree of cell death control is largely dependent on the initial cellular energetic state and that ATP availability plays a central role in determining the final mode of cell death execution (Fig. 1) [19–23]. Only when factoring in the molecular mechanisms that govern energetic sensing and the degree of regulatory overlap that exists between apoptosis, autophagy, and necrosis, does the dynamic nature of RCD become fully apparent.
Fig. 1.
The dynamic nature of cell death. The specific cell death modality to be initiated is highly dependent on the intensity of the stressor and cellular ATP availability. Under conditions of nutrient deprivation or short-term cellular stress, autophagy is initiated to enhance ATP availability, thereby assisting in the cellular stress response. Prolonged stressors, such as chemotherapy or radiation, activate the apoptotic machinery, which makes use of the ATP provided by the initial autophagic response. Excessively prolonged damage will result in necrosis onset after a rapid decrease in ATP availability
Centrality of autophagy in cell death regulation
Metabolic sensing and the autophagic machinery
The importance of autophagy in cell death onset stems from its intricate involvement in nutrient sensing and cellular proteostasis. Under nutrient limiting conditions, increased ADP levels activate AMP kinase (AMPK), thereby inhibiting the mammalian target of rapamycin complex 1 (mTORC1) [24]. Autophagy is initiated once mTORC1 dissociates from the ULK-complex, dephosphorylating ULK1 and allowing for kinase-dependent activation of ATG13 and FIP2000, which form the pre-initiation complex [24]. The activated ULK-complex elicits its kinase activity on key components of the class III PI3K complex, otherwise known as the initiation complex. Composed of Beclin 1, Vps34, Vps13, and ATG14, the formation of this multi-domain complex is necessary for the allosteric activation of Vps34, which targets phosphatidylinositol to generate phosphatidylinositol-3-phosphate (PI3P) at the isolation membrane [25]. Attachment of PI3P is necessary for the recruitment of proteins involved in the elongation of the phagophore membrane. Elongation is mediated by two ubiquitin-like protein (UBL) conjugation systems that generate LC3-II, a proteolipid molecule necessary for the fusion of autophagosomes to lysosomes [26]. Importantly, light chain -3 (LC3) is lipidated by the UBL cascade which involves the E1 (ATG7) and E2 (ATG3) ligases as well as ATG4 protease activity. Formation of the ATG12-ATG5-ATG16L complex facilitates conjugation of LC3-I to phosphatidylethanolamine (PE), producing LC3-II to conclude the elongation reaction [27]. Both LC3-II and the ATG12-ATG5-ATG16L1 complex associate with the elongating membrane, although only LC3-II remains attached to the fully matured autophagosome. Fusion with acidic lysosomes is now possible, allowing for the degradation of cytoplasmic proteins, lipids and carbohydrates into their respective biosynthetic substrates. The rate at which this process operates, is metabolically critical, and can be assessed by measuring the autophagosome flux [11, 16, 17].
Dance of the dead: crosstalk between autophagy and apoptosis
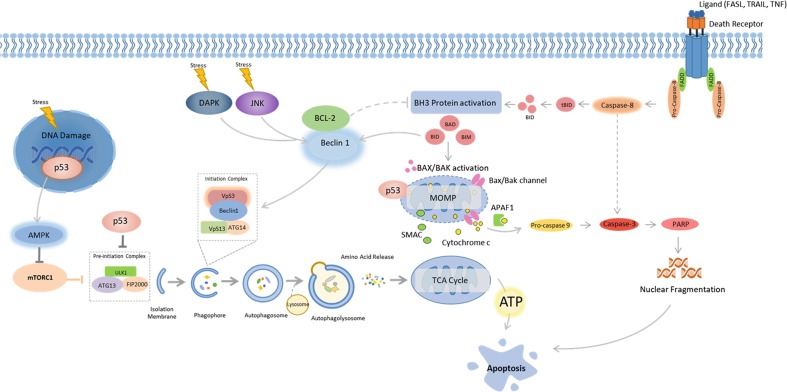
One of the main functions of autophagosomal catabolism—besides the degradation of long-lived proteins at basal levels—is to provide energetic substrates and ATP to rescue cells from starvation through adaptive autophagy (Fig. 1). However, under conditions of prolonged metabolic stress, this energy reservoir can be used to facilitate ATP dependent apoptosis [28]. In this instance, the stressor/insult severity as well as its duration plays a critical role in driving this process (Fig. 1). While autophagy can protect a cell from starvation, it can also be initiated through p53 signalling when cells are subjected to DNA damaging agents, such as chemotherapeutics or radiation [29]. Under basal conditions, tumour suppressor p53 is located in the cytosol and exerts an inhibitory effect on FIP200, thereby impairing the autophagy initiation complex [29, 30]. In response to excessive stress however, p53 translocates to the nucleus where it coordinates the expression of pro-autophagic proteins including AMPK, damage-regulated autophagy modulator 1 (DRAM1) and both Sestrin 1 and 2, a process that promotes cellular survival [31, 32]. An intriguing phenomenon occurs under prolonged stress, when p53 translocates to cyclophin D on the outer mitochondrial membrane, resulting in opening of the permeability transition pore (PTP), thereby promoting apoptosis [33]. In terms of cellular survival, it might seem surprising that p53 would first activate autophagy, which provides metabolic substrates to the mitochondria, only to later impair its function (Fig. 2). However, a low degree of PTP opening can still be counteracted by engaging mitochondrial specific degradation through autophagy (referred to as mitophagy) thereby clearing damaged mitochondria and circumventing apoptosis onset [34, 35]. PTP opening can reach a defined threshold, a ‘point of no return’ (PONR), after which mitochondrial outer membrane permeabilization (MOMP) becomes unavoidable and wide-scale apoptosis is evident [28]. Therefore, in terms of the intrinsic (mitochondrial) apoptotic pathway, a fine balance exists between autophagic inhibition of apoptosis though mitophagy and global mitochondrial failure (Fig. 2). The level of autophagic activity required for driving and controlling either process however, remains to be elucidated.
Fig. 2.
Cross-talk between autophagy and apoptosis. Autophagy and apoptosis share several inducers; p53, death-associated protein kinase (DAPK), JUN N-terminal kinase (JNK), BH3-only proteins BAD (BCL-2 antagonist of cell death), and BID (BH3-interacting domain death agonist). p53 translocates to the nucleus in response to excessive cellular stress and coordinates the expression of pro-autophagic proteins including AMPK. However, under prolonged stress p53 translocates to cyclophin D on the outer mitochondrial membrane, resulting in opening of the permeability transition pore (PTP) which promotes apoptosis. BH3-only proteins are conventional regulators of apoptosis, but are also capable of impairing the interaction between BCL-2 and Beclin 1. Unbound Beclin 1 is able to interact with VPS34 which initiates autophagy. BCL-2 loses its ability to repress apoptosis when not activated by Beclin 1. The Ser/Thr kinases DAPK and JNK contribute to a similar outcome on autophagy and apoptosis, where the BH3 binding domain of Beclin 1 is phosphorylated by DAPK, making binding of Beclin 1 to VPS34 possible, while JNK phosphorylates and inhibits BCL2, which activates Beclin 1
Consequently, apoptosis and autophagy share several inducers in addition to p53. These include the kinases death-associated protein kinase (DAPK) and JUN N-terminal kinase (JNK) as well as the BH3-only proteins BAD (BCL-2 antagonist of cell death), BID (BH3-interacting domain death agonist), NIX (NIP3-like protein X), NOXA and PUMA (Fig. 2). Although BH3-only proteins are commonly associated with apoptosis mediation, they are also capable of impairing the interaction between BCL-2 and Beclin 1. As a result, unbound Beclin 1 is able to interact with VPS34, which initiates autophagosome synthesis [36]. BCL-2 also loses its ability to repress apoptosis when not activated by Beclin 1 [37]. The Ser/Thr kinases DAPK and JNK contribute to a similar scenario with a resulting outcome on autophagy and apoptosis onset in response to stress. The BH3 binding domain of Beclin 1 is phosphorylated by DAPK, which makes binding of Beclin 1 to VPS34 possible, whereas JNK phosphorylates and inhibits BCL2 [38–40]. Given these examples, it appears likely that autophagy is activated concomitantly with apoptosis to act as an initial cytoprotective mechanism during low levels of cellular stress. Should this insult continue to increase in intensity, the cell would already have all apoptotic regulators, as well as the energetic capacity available to successfully carry out apoptosis to its completion.
The controversy surrounding autophagy-dependent cell death (ADCD)
Given the major interplay between autophagy and apoptosis signalling, what is the likelihood of autophagic cell death occurring independently? This has been a topic of heated debate since the term autophagic cell death (ACD) was first coined and to date strong evidence exists to support both arguments [8, 41–43]. Initially, the presence of accumulated autophagosomes in dying cells prompted the description of a newly defined mode of cell death, as it was morphologically distinct from that of apoptotic or necrotic cells. More recently, studies have shown that in BAX/BAK or caspase knock-out cells, these distinct features of ACD are present during cell death induction [44–46]. It is important to note that, although an increased number of autophagosomes is observed in these specific scenarios, their mere presence observed at a single time point is no indication of the autophagy pathway function. When quantifying the abundance of autophagosomes present through electron microscopy or LC3-II expression through immunoblotting, the only valid conclusion to be made is that either an increased or decreased autophagosomal synthesis or degradation took place [16, 17]. The increased presence or autophagosome pool size however, could either be as a result of their accumulation, indicating degradative dysfunction, or increased autophagosomal turnover due to a heightened flux through the pathway, indicative of a survival response or adaptive autophagy. Therefore, relying solely on morphological features to attribute cause of cell death to autophagy only can be highly misleading, which, as recently highlighted by the nomenclature committee on cell death (NCCD), demands for a molecularly oriented definition that includes genetic silencing of at least two autophagy markers to validate whether death would still occur in the absence of autophagy [17]. It is now largely accepted that cellular demise that relies on the autophagy machinery or key components thereof, can be defined as ADCD [10]. It is however less clear which level of autophagic activity is required to transit heightened and adaptive autophagy to this position of lethality leading to the induction of this subroutine. However, given a severe enough stimulus, necrosis would still be able to operate independently of autophagy or apoptosis signalling, highlighting the importance of the time required by which death is delayed due to autophagy inhibition [9].
Therefore, when examining the evidence for ADCD, very few examples exist where both the flux dependent nature of the pathway as well as genetic characterisation are combined to define its existence. For example, the silencing of specific Atg genes during the developmental stages of D. melanogaster resulted in inadequate removal of specific tissues [47, 48]. Although it was concluded that ADCD is solely responsible for these actions, the caspase-independent removal of the larval midgut during Drosophila development, on the other hand, has been shown not to be dependent on all components of the macroautophagic machinery required during nutrient deprivation [49]. It is important to note that the most compelling examples for ADCD are mainly derived from developmental studies and highlights that the dependence on autophagy in these scenarios is related to energy requirements as opposed to true cell death mediation. Although there is ample evidence supporting the involvement of autophagy in cell death onset [12, 50–53], very little data exist on the exact mechanism through which autophagy acts as an independent cell death modality. The recent identification of a Na+, K+-ATPase dependent form of ADCD, referred to as autosis, shed much light onto these discrepancies, providing evidence that lethal autophagy is regulated somewhat differently than ‘non-lethal’ housekeeping autophagy associated with efficient autophagic responses [12].
The search for an autophagic flux threshold: how much is too much?
It is well established that basal autophagic activity is necessary to promote homeostasis and that its upregulation to enhanced levels promotes cellular survival under nutrient limiting conditions. It is now clear that the degree of efficient and adaptive autophagic responses is cell specific and defines the cellular and tissue autophagy proficiency. In this regard, the initial experimental approaches to assess autophagic activity focussed primarily on measuring the presence of autophagy-related markers in the presence and absence of a stressor/inducer as well as saturating concentrations of lysosomal inhibitors [17]. Subsequently, quantifying the number of autophagic vacuoles combined with measuring the protein expression of autophagosome related proteins has become the gold standard for evaluating autophagic activity. Advances in live cell imaging have since complemented these methods substantially, as autophagosomal degradation has been observed to be a highly dynamic process [15–17]. Furthermore, although the multi-step nature of autophagy, progressing from initiation to degradation, has been characterised extensively, it has only recently been viewed using minimal models of phase transitions [54]. In light of the findings that targeted autophagy induction can result in autosis, it is intriguing to speculate to what degree autophagosomal degradation is required to achieve such outcome, i.e., is there an autophagic flux threshold to breach?
The molecular distinction between ADCD and autosis
In order to evaluate the existence of an autophagic flux threshold, i.e., the level of autophagic activity that must be breached to induce cell death, the molecular hallmarks of ADCD and autosis must be considered. Given that autotic cells, but not cells displaying ADCD, can be rescued through treatment with Na+, K+-ATPase inhibitors, the possibility exists that autosis is not mediated entirely by the core autophagic machinery [12]. Although it has been elegantly revealed that knockdown of beclin1, ATG13 and ATG14 rescued cells from tat-Beclin 1 induced autosis, PI3K inhibition through 3-MA treatment only partially impaired this response and, most surprisingly, inhibition of lysosomal fusion through Bafilomycin-A1 treatment showed no rescue effect at all [12]. This is in stark contrast to scenarios of ADCD where death can be delayed by inhibition of both upstream and downstream regulators [44, 55]. Therefore, it is plausible that a highly, if not maximally enhanced autophagic flux is required for the upstream initiation of autotic death regulators and that this ‘lethal flux’ is higher than that associated with ADCD. Furthermore, autosis operates independently of the other cell death modalities, whereas true ADCD has primarily been shown to operate during developmental stages and appears to merely delay the onset of apoptosis or necrosis in other scenarios [56, 57]. The specificity and independent nature of autosis therefore makes it an attractive therapeutic target and an alternative to the control of ADCD.
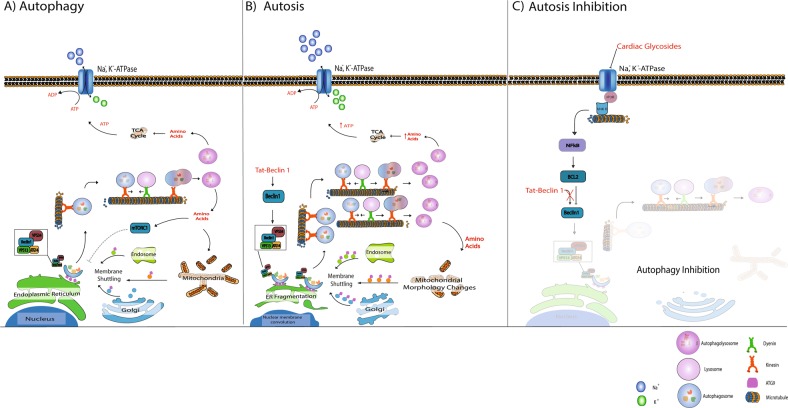
Further indicators of an excessively heightened autophagic flux that is beyond adaptive autophagic activity needed to promote autosis are the separation of the inner and outer nuclear membrane and the depletion of endoplasmic reticulum (ER) structures at the end stages of autosis (Fig. 3). The ER is a critical source and platform for autophagosomal biogenesis and its depletion may serve as a relative indication for autophagic flux [58]. Levine et al. have postulated that, as a result of ER depletion, the outer nuclear membrane is utilized as a ‘last resort’ to support autophagosome biogenesis, seeing as its use is prevented under basal autophagic flux conditions [58]. For a detailed outline of the differences between autosis and classical ADCD, the reader is referred to an excellent review by Lui & Levine [58].
Fig. 3.
Autosis onset and inhibition. a Under normal conditions, autophagosome production is activated in response to nutrient deprivation. Autophagosome formation is highly dependent on ATG9 mediated membrane shuttling between the autophagosome formation site -usually close to the endoplasmic reticulum (ER)- and membrane sources such as the Golgi, mitochondria and endosomes. Once autophagosomes are matured, they are transported along the microtubule system by molecular motors such as dynein and kinesin to merge with acidic lysosomes, giving rise to autolysosomes which release amino acids. b Beclin 1 over-stimulation (through targeted peptides such as tat-Beclin1) results in a high demand for membrane material to meet the requirements of excessively high autophagosome production. This is, at least in part, the cause of ER fragmentation and mitochondrial morphology changes observed during autosis. It is plausible that ATP generated from excessively high autophagic activity is utilised by the Na, K, ATPase to compensate for the high ATP production, depriving the cell of energy over time. c Inhibiting the NKA with cardiac glycosides decreases both the demand for ATP and autophagy associated ATP production through the NKA-IP3R microdomain signalling, thereby rescuing the cell from autosis
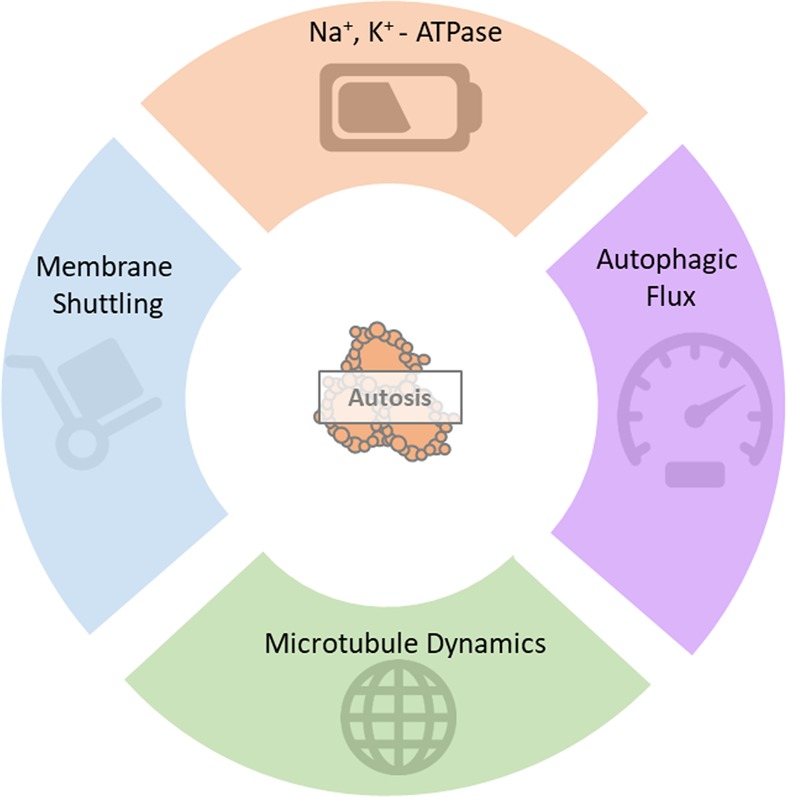
One of the few validated parameters crucial to autosis onset is the Na+, K+-ATPase pump (NKA-pump) activity [12, 59]. The exact mechanism by which the excessively high levels of autophagy together with Na+, K+-ATPase activity mediate cellular demise remains unclear. siRNA knockdown of the NKA-pump α1-subunit resulted in a pronounced decrease in starvation- and tat-Beclin1 induced autosis [12]. As of yet, no defined molecular mechanisms have been established between NKA-pump activity and autophagy regulation, which contributes to the lack of evidence for the exact mode though which the NKA-pump mediates autosis onset. However, existing literature about its involvement in various other cellular processes provides valuable insights into possible mechanisms. Firstly, the NKA interacts with the inositol 1,4,5,-trisphosphate (IP3)3 receptor (IP3R) when inhibited by the cardiac glycoside ouabain, forming a signalling microdomain capable of activating the NF-kB transcription factor [60]. Considering that NF-kB enables the activation of BCl2 which in turn inhibits Beclin1, it makes clear that digoxin, a same class cardiac glycoside as ouabain, was found to protect cells from autosis [12]. However, under nutrient-rich conditions, the same cardiac glycosides induce autophagy through the conventional mTOR signalling pathway [61].
Further conflicting evidence exists regarding the interaction of NKA with the cytoskeleton. For example, the formation of the NKA-IP3R microdomain is dependent on NKA binding to the Ankyrin B protein on the cytoskeleton [62–64], but acetylation of tubulin also inhibits NKA activity [65] even though it is an important requirement for NKA binding to the cytoskeleton [66]. What convolutes matters further is the fact that autophagosomal transport and lysosomal fusion is heavily dependent on acetylation of the same tubulin Lys40 residue required for NKA anchorage [67–71]. Therefore, the potential of cardiac glycosides to serve as autosis inhibitor seems highly context specific, with the IP3R-NFkB microdomain playing a role in autosis inhibition only under tat-Beclin1 inducing conditions (Fig. 3). If an increase in autophagic flux is capable to contribute to an enhanced ATP production, then the activation of the NKA could be viewed as a compensatory mechanism to utilise excessive ATP, depriving the cell of ATP, which, in time, results in cell death induction. Inhibiting the NKA counteracts this process, thereby ensuring decreased ATP demand and production through the NKA-IP3R microdomain signalling. An assessment of the ATP-consuming processes and their hierarchy [72] is required to further unravel the role of NKA activation and metabolic perturbation.
Another possible role player in autosis regulation is the only known transmembrane protein in the ATG-family, known as mammalian ATG9 (mATG9), the orthologue of Atg9 in yeast. mATG9 has been found to shuttle from the trans-Golgi network (TGN), ER, mitochondria and endosomes to the phagophore initiation site to provide lipids for phagophore formation (Fig. 3) [73, 74]. This shuttling is especially prominent during starvation-induced autophagy, where increased co-localization of ATG9 with LC3-II and defined endosome markers have been observed. Recent evidence from live cell imaging studies suggests that the association of ATG9 with the phagophore assembly site (PAS) is rather transient in nature, and that only a small number of ATG9 vesicles is recruited to the PAS [75]. This transient association is an important indication of the highly dynamic nature of autophagosomal assembly, and suggests that the shuttling can adapt rapidly to an increased autophagosomal demand. Therefore, the possibility exists that increased membrane shuttling by mATG9 under autophagy induction could contribute to the cause of ER fragmentation observed in early autosis. Moreover, mATG9 may be considered as a viable target for autosis induction, as it would allow for enhanced autophagosomal size and production frequency at the PAS level and warrants further investigation.
Although there are various possible molecular role players to be investigated as candidates for autosis induction, an important determining feature is the autophagic activity or autophagosomal degradation rate. Yet, the precise autophagic activity required to promote autosis remains to be elucidated. This is somewhat surprising, given that excessive autophagosomal degradation is routinely indicated as an integral parameter of autosis and yet it might be a key defining feature that sets autosis apart from ADCD. Future work is required that dissects the molecular relationship between efficient, adaptive and lethal autophagic responses in conditions of ADCD and autosis induction, where not only autophagic flux and ATG9 shuttling, but also ATP consumption and ER fragmentation is collectively and quantitatively determined.
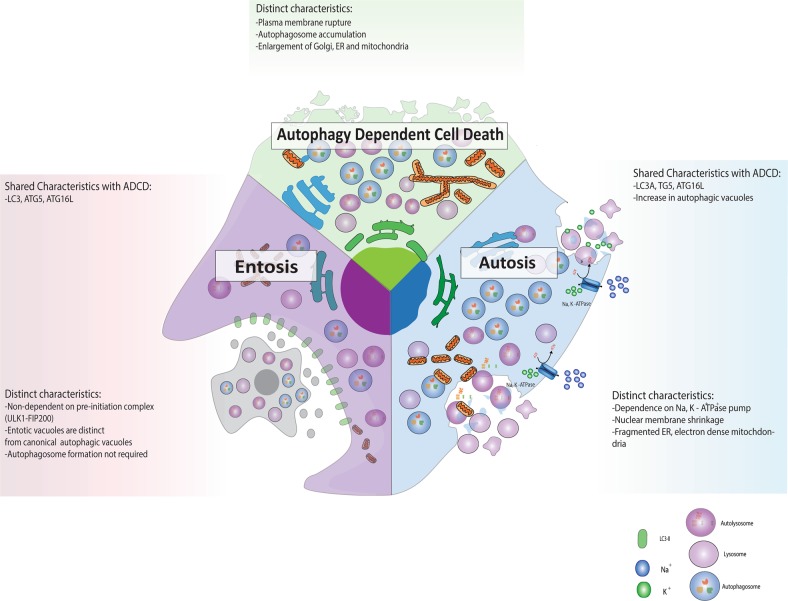
Finally, molecular overlap exists between ADCD, autosis and a third lethal autophagic response: entosis, the engulfment of a neighbouring cell (Fig. 5). The most notable shared trait is its dependence on LC3. During entosis LC3 associated phagocytosis (LAP) is critical for whole cell engulfment, although autophagosome formation does not take place due to a lack of phosphatidylserine expression, which results in immediate fusion of entotic vesicles with lysosomes. Furthermore, entotically engulfed cells exhibit features associated with ADCD, such as autophagosome accumulation, although autophagy is thought to be protective in this context as its inhibition induces apoptosis of the internalised cells [76]. Consequently, proteins involved in LC3 lipidation such as Vps34, ATG5 and ATG7 are important mediators of LAP and entosis. In contrast to autosis and ADCD, knockdown of proteins specific to autophagosome synthesis such as FIP200 and ATG13 have no effect on entotic activity, yet it protects from ADCD and autosis [12, 76]. Importantly, in the context of anti-cancer treatment, the engulfment of neighbouring cells through entosis can also enhance tumour formation [77], which highlights the possibility of exploiting the inherent upregulation of LC3 to enhance self-engulfment through autosis (by using autophagy inducers) as opposed to pro-tumourigenic entosis. Due to the lack of evidence pertaining to specific cellular fate, and the possibility for cells to escape entotic engulfment, it has now been proposed that entosis be referred to as a mode of internalisation only, and not a complete mode of cell death. In contrast, Perfettini et al. have recently suggested that entotic cell death be classified as type IV non-cell autonomous cell death [78]. It becomes clear that future work is required that enables precise relation of autophagic activity with the diversity of lethal autophagic responses.
Fig. 5.
Comparison of autophagy-dependent cell death (ADCD), autosis and entosis. All three modes of cell death rely on the expression and lipidation of LC3, yet, entosis does not require LC3 for autophagosome formation and entotic vacuoles fuse with lysosomes immediately. A most distinct feature of autosis is enhanced activity of the Na+, K+ -ATPase pump, ER fragmentation and electron dense mitochondria, whereas ADCD displays enlarged Golgi, ER and mitochondria
The autophagic flux threshold: concept or reality?
If the distinction between ADCD and autosis can be attributed to the variance in the rate at which lysosomes degrade autophagy substrates, is there evidence for the existence of an autophagic flux threshold that defines that rate? Given that a defined threshold of caspase activation and cytochrome release can lead to a ‘point of no return’ (PONR) that defines apoptosis induction and execution [23, 28, 79], a similar mechanism may govern the relationship between autophagic flux and autosis onset. An example supporting the concept of an autophagic ‘flux threshold’ can be found in studies that make use of mathematical modelling to predict cellular decision making [50, 80]. The negative feedback that exists between Beclin1 and caspases makes for robust predictive models in terms of apoptosis onset. When simulating the effect of increased stress levels on autophagic and apoptotic signalling, Kapuy et al. showed that initial autophagy activity during cellular stress is sufficient to inhibit apoptosis, however, with increased stress intensity caspase-mediated cleavage of Beclin1 ultimately results in apoptosis induction with decreased autophagy [37]. Before this cellular outcome manifests, there is indeed a point at which stress-induced autophagy reaches a maximum level and a new steady-state. Further modelling by Tavassoly et al. predicted that, after reaching a maximal level of autophagy, a decrease in autophagy is inevitable due to caspase signalling as a result of increased stress [80]. Although these examples are limiting in that they portray the behaviour of healthy cells under optimal conditions, they highlight the dynamic relationship that underpins autophagic activities and apoptosis onset. Intriguing experimental data has been gathered in the context of myeloma cancer cell death, where inhibition of caspase 10, which normally degrades BCL2 associated transcription factor1 (BCLAF1), results in such a high degree of uncontrolled autophagy activity that cell death ensues, without features of apoptosis [50]. Taken together, these data support the existence of a flux threshold, suggesting that future work that engages with the construction of minimal network models requires an inclusion of ADCD and autosis onset as additional expected outcome parameters.
In addition to previous modelling approaches, metabolic control analysis (MCA) [81] seems here highly applicable for the characterization of the autophagic system, as it is suitable for describing dynamic processes based on pathway intermediates and fluxes in response various metabolic cues. MCA allows not only to describe the steady-state flux in terms of its regulation and control, but also to calculate minimal or maximal conversion yields, based on energetic requirements. When applied to the autophagic pathway, it becomes clear that its process necessitates the completion of multiple steps while facilitating the flow of metabolites along the pathway to meet metabolic demands. Each step operates at defined rate, such as the rate at which lysosomes fuse with autophagosomes or the rate of autophagy substrate degradation by lysosomes [16, 82]. This rate may hence either be represented by the number of autophagosomes produced or consumed per time unit, or the number of autolysosomes produced per time unit [16]. For the autophagy system to be in steady state, the rate of each step in the pathway must be equal, making it possible to quantify autophagosome flux experimentally by inhibiting one of these steps [16]. This approach does not only allow for the precise quantification of the rate at which autophagosomes are produced in real time, but also for describing the transition time needed to clear the entire autophagosomal pool. Therefore, predicting the rate of degradation required for cell survival (non-lethal autophagic flux) and cellular demise (lethal autophagic flux) may be possible in the near future. It is furthermore plausible that this threshold is governed by the same molecular regulators involved in autosis control.
Therefore, evidence points towards the existence of an autophagic flux threshold suggesting that parameters such as NKA α1-subunit expression levels, autophagosomal degradation rate and autophagosome vesicle transport dynamics as well as ATG9 shuttling should be considered when assessing autophagic flux in the context of ADCD and autosis.
Autosis induction as treatment modality
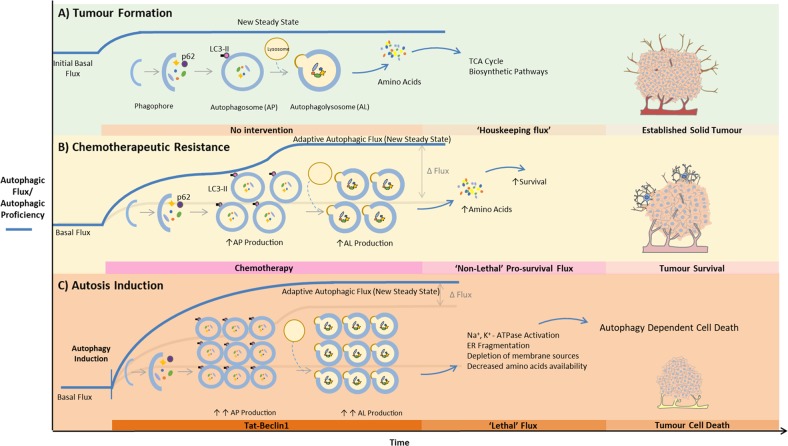
The concept of an autophagic flux threshold becomes highly apparent when applied to tumour biology (Fig. 4). During the process of tumour growth, a certain basal, yet usually already heightened autophagic flux is required to maintain its increasing metabolic demands. Under conditions of cytotoxic stress, enhanced autophagy has been implicated in chemotherapeutic resistance, which will require a non-lethal pro-survival flux well above that observed under basal conditions. Therefore, when autophagy is induced through a targeted intervention, the flux response necessary to elicit cell death should be significantly higher than both basal and pro-survival fluxes (Fig. 4).
Fig. 4.
Dependence of autosis on the autophagic flux threshold for cancer treatment. a In the course of tumour progression, a defined, usually heightened basal autophagic flux is required to maintain metabolic demands. b Under conditions of cytotoxic stress, enhanced autophagy has been implicated in chemotherapeutic resistance, which will require a non-lethal pro-survival flux well above that observed under basal conditions. c When autophagy is induced through a targeted intervention, the flux response necessary to elicit cell death must be significantly higher than both basal and pro-survival fluxes in order to induce autosis
Although limited evidence exists for autosis occurring in pathologies such as cancer and heart disease, reassessment of previous studies attributing cell death solely to ADCD could assist in strengthening the case for its existence outside a controlled environment. A key example is the reassessment of liver damage in patients with anorexia nervosa where autophagic markers were observed in the absence and presence of hepatocyte cell death [83, 84]. Although it was initially concluded that starvation-induced autophagy could have contributed to cell death, findings were reassessed after the first evidence for autosis came to light [83]. Scanning electron microscopy (SEM) evidence for distinct features of autosis could be found in 4 patient samples, as the cells displayed the same ER condensation and nuclear fragmentation [85]. Autosis has also recently been identified in cardiomyocytes in response to ischemia/reperfusion injury [86]. These examples highlight the possibility of autosis also occurring in cancer models displaying features of ADCD. Autophagy modulators (both inducers and inhibitors) have proven to sensitize several cancer types to chemotherapy [87, 88]. Yet, although therapeutic potential is enormous, clinical trials have remained largely unsuccessful due to the dose-limiting toxicity of autophagy modulators in combination with chemotherapeutics [89–91] as well as pharmacological, technical or experimental challenges associated with autophagy control. However, over-expression of autophagy inducing microRNAs (miRNAs) such as mir-15a and mir-16 have yielded promising sensitization effect [92]. When considering autophagy inhibition in therapy, an important contributing factor is its critical role in immune system function, and that global autophagy inhibition may result in immune-suppression, resulting in a pro-tumourigenic environment [93]. Furthermore, cancer ADCD has been observed in response to both global and targeted autophagy induction in the absence of chemotherapy [94–96]. In such cases the re-examination of these data for autosis markers is critical [97]. Moreover, when modulating autophagy as an anti-cancer measure, little attention is given to the non-cell- autonomous regulation associated with autophagy. Conventional cancer therapies have recently been shown to eradicate cancer cells through induction of non-cell autonomous death modalities such as entosis [98]. Hence, given the fact that autophagy and entosis share aspects of the molecular machinery (Fig. 5), careful attention must be given to the downstream effects of excessive autophagy modulation when targeting shared regulators such as ATG5 or LC3.
Although targeting autosis may present a plausible avenue for cell death control in cancer treatment, an accurate determination of autophagic flux will be crucial for successful clinical translation. This would require comprehensive assessment of the basal autophagosomal turnover rate in the established tumour itself. Growing tumour organoids from patient biopsy samples has shown immense improvement in curated treatment protocols which aim to pre-determine the exact drug regimen required to efficiently induce cell death [99]. A similar approach can be imagined for an autophagic flux assessment, so as to determine the precise level of induction required to promote autosis onset. Cancer cells have been found to display enhanced autophagic activity relative to the healthy surrounding tissue, in accordance with previous findings that different tissue types exhibit distinct levels of autophagic activity [100–103]. The same is likely the case for different tumour samples, each with a distinct autophagic flux signature. The basally enhanced levels of autophagy further support the validity of autosis induction as a potential therapeutic alternative seeing as the metabolic system is already susceptible to reaching a lethal threshold [104].
A major challenge remains to accurately target autophagy inducing compounds and peptides to the established tumour region. A major benefit of targeting autophagy lies in its inherent activity and function in all cells and tissues and would not necessarily require the utilisation of specialised drug delivery systems. For example, tat-Beclin treatment has been observed to enhance immunogenicity in vivo [105]. Given that immunotherapy is already being established as a viable clinical alternative to chemotherapy, the combination of precision controlled autophagy induction with targeted immunotherapy certainly warrants consideration [6].
Conclusion and future outlook
The addition of autosis to the growing library of specialised cell death modalities sheds much-needed light on the controversies surrounding ADCD. Its molecular definition and signature that differs from autophagy regulation finally makes it possible to discern between lethal and non-lethal autophagy. However, the degree to which autophagic flux must be enhanced to breach this threshold remains poorly defined. Furthermore, its dependence on cellular physiological functions such as the NKA-pump and ATP consuming processes is not fully understood. This review has outlined the conceptual differences between the role of autophagy in supporting RCD and its ability to operate as an independent cell death modality. We suggest the existence of an autophagic flux threshold that may play a critical role in defining and governing these cellular fate outcomes. Determining this threshold will require a more dynamic and systems-level understanding of both autophagic flux and RCD all together. These factors will prove crucial in order to accelerate both autophagy and autosis control in the clinical setting, so as to harness the therapeutic potential of autophagy modulation.
Edgar Allan Poe once declared; “The boundaries which divide Life from Death are at best shadowy and vague. Who shall say where the one ends, and the other begins?”. The answer, though powerful, may be much less poetic and depend on the activity of autophagosomes.
Acknowledgements
The authors wish to acknowledge financial support from the South African National Research Foundation (NRF), the South African Medical Research Council (SAMRC) and the Cancer Association of South Africa (CANSA).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by: M. Piacentini.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kumar-Sinha C, Chinnaiyan AM. Precision oncology in the age of integrative genomics. Nat Biotechnol. 2018;36:46–60. doi: 10.1038/nbt.4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schwartzberg L, Kim ES, Liu D, Schrag D. Precision Oncology: Who, How, What, When, and When Not? Am Soc Clin Oncol Educ Book Am Soc Clin Oncol Meet. 2017;37:160–9. doi: 10.1200/EDBK_174176. [DOI] [PubMed] [Google Scholar]
- 3.Tan P, He L, Han G, Zhou Y. Optogenetic immunomodulation: shedding light on antitumor immunity. Trends Biotechnol. 2017;35:215–26. doi: 10.1016/j.tibtech.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ford JM. Precision oncology: A new forum for an emerging field. JCO Precis Oncol 2017;1–2. [DOI] [PubMed]
- 5.Chiramel AI, Brady NR, Bartenschlager R. Divergent roles of autophagy in virus infection. Cells. 2013;2:83–104. doi: 10.3390/cells2010083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tanaka K, Matsuda N. Proteostasis and neurodegeneration: The roles of proteasomal degradation and autophagy. Biochim Biophys Acta - Mol Cell Res. 2014;1843:197–204. doi: 10.1016/j.bbamcr.2013.03.012. [DOI] [PubMed] [Google Scholar]
- 7.Goldsmith J, Levine B, Debnath J. Autophagy and Cancer Metabolism. Elsevier, 2014, pp 25–57. [DOI] [PMC free article] [PubMed]
- 8.Deter RL, Baudhuin P, de Duve C. Participation of lysosomes in cellular autophagy induced in rat liver by glucagon. J Cell Biol. 1967;35:C11–C16. doi: 10.1083/jcb.35.2.c11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nikoletopoulou V, Markaki M, Palikaras K, Tavernarakis N. Crosstalk between apoptosis, necrosis and autophagy. Biochim Biophys Acta - Mol Cell Res. 2013;1833:3448–59. doi: 10.1016/j.bbamcr.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 10.Galluzzi L, Vitale I, Aaronson SA, Abrams JM, Adam D, Agostinis P, et al. Molecular mechanisms of cell death: recommendations of the nomenclature committee on cell death 2018. Cell Death Differ. 2018;25:486–541. doi: 10.1038/s41418-017-0012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galluzzi L, Baehrecke EH, Ballabio A, Boya P, Bravo‐San Pedro JM, Cecconi F, et al. Molecular definitions of autophagy and related processes. EMBO J. 2017;36:1811–36. doi: 10.15252/embj.201796697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Y, Shoji-Kawata S, Sumpter RM, Wei Y, Ginet V, Zhang L, et al. Autosis is a Na+ ,K+ -ATPase-regulated form of cell death triggered by autophagy-inducing peptides, starvation, and hypoxia-ischemia. Proc Natl Acad Sci USA. 2013;110:20364–71. doi: 10.1073/pnas.1319661110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rybstein MD, Bravo-San Pedro JM, Kroemer G, Galluzzi L. The autophagic network and cancer. Nat Cell Biol. 2018;20:243–51. doi: 10.1038/s41556-018-0042-2. [DOI] [PubMed] [Google Scholar]
- 14.Galluzzi L, Bravo-San Pedro JM, Levine B, Green DR, Kroemer G. Pharmacological modulation of autophagy: therapeutic potential and persisting obstacles. Nat Rev Drug Discov. 2017;16:487. doi: 10.1038/nrd.2017.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaizuka T, Morishita H, Hama Y, Tsukamoto S, Matsui T, Toyota Y, et al. An Autophagic flux probe that releases an internal control. Mol Cell. 2016;64:835–49. doi: 10.1016/j.molcel.2016.09.037. [DOI] [PubMed] [Google Scholar]
- 16.Loos B, Toit A, du, Hofmeyr JHS. Defining and measuring autophagosome flux—concept and reality. Autophagy. 2014;10:2087–96. doi: 10.4161/15548627.2014.973338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition) Autophagy. 2016;12:1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lumkwana D, du Toit A, Kinnear C, Loos B. Autophagic flux control in neurodegeneration: Progress and precision targeting—Where do we stand? Prog Neurobiol. 2017;153:64–85. doi: 10.1016/j.pneurobio.2017.03.006. [DOI] [PubMed] [Google Scholar]
- 19.Wang SY, Yu QJ, Zhang RD, Liu B. Core signaling pathways of survival/death in autophagy-related cancer networks. Int J Biochem Cell Biol. 2011;43:1263–6. doi: 10.1016/j.biocel.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 20.Blum R, Kloog Y. Metabolism addiction in pancreatic cancer. Cell Death Dis. 2014;5:e1065. doi: 10.1038/cddis.2014.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Green DR, Galluzzi L, Kroemer G. Metabolic control of cell death. Science (80-) 2014;345:1250256–1250256. doi: 10.1126/science.1250256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsujimoto Y. Apoptosis and necrosis: intracellular ATP level as a determinant for cell death modes. Cell Death Differ. 1997;4:429–34. doi: 10.1038/sj.cdd.4400262. [DOI] [PubMed] [Google Scholar]
- 23.Loos B, Engelbrecht AM. Cell death: a dynamic response concept. Autophagy. 2009;5:590–603. doi: 10.4161/auto.5.5.8479. [DOI] [PubMed] [Google Scholar]
- 24.Dunlop EA, Tee AR. MTOR and autophagy: A dynamic relationship governed by nutrients and energy. Semin Cell Dev Biol. 2014;36:121–9. doi: 10.1016/j.semcdb.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 25.Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22:124–31. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanada T, Noda NN, Satomi Y, Ichimura Y, Fujioka Y, Takao T, et al. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem. 2007;282:37298–302. doi: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- 28.Loos B, Engelbrecht AM, Lockshin RA, Klionsky DJ, Zakeri Z. The variability of autophagy and cell death susceptibility: Unanswered questions. Autophagy. 2013;9:1270–85. doi: 10.4161/auto.25560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tasdemir E, Maiuri MC, Galluzzi L, Vitale I, Djavaheri-Mergny M, D’Amelio M, et al. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol. 2008;10:676–87. doi: 10.1038/ncb1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morselli E, Shen S, Ruckenstuhl C, Bauer MA, Marino G, Galluzzi L, et al. p53 inhibits autophagy by interacting with the human ortholog of yeast Atg17, RB1CC1/FIP200. Cell Cycle. 2011;10:2763–9. doi: 10.4161/cc.10.16.16868. [DOI] [PubMed] [Google Scholar]
- 31.Budanov AV, Karin M. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell. 2008;134:451–60. doi: 10.1016/j.cell.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maiuri MC, Malik SA, Morselli E, Kepp O, Criollo A, Mouchel PL, et al. Stimulation of autophagy by the p53 target gene Sestrin2. Cell Cycle. 2009;8:1571–6. doi: 10.4161/cc.8.10.8498. [DOI] [PubMed] [Google Scholar]
- 33.Vaseva AV, Marchenko ND, Ji K, Tsirka SE, Holzmann S, Moll UM. p53 opens the mitochondrial permeability transition pore to trigger necrosis. Cell. 2012;149:1536–48. doi: 10.1016/j.cell.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Narendra D, Tanaka A, Suen DFDF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2010;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Malik SA, Orhon I, Morselli E, Criollo A, Shen S, Marino G, et al. BH3 mimetics activate multiple pro-autophagic pathways. Oncogene. 2011;30:3918–29. doi: 10.1038/onc.2011.104. [DOI] [PubMed] [Google Scholar]
- 37.Kapuy O, Vinod PK, Mandl J, Bánhegyi G. A cellular stress-directed bistable switch controls the crosstalk between autophagy and apoptosis. Mol Biosyst. 2013;9:296–306. doi: 10.1039/c2mb25261a. [DOI] [PubMed] [Google Scholar]
- 38.Zalckvar E, Berissi H, Mizrachy L, Idelchuk Y, Koren I, Eisenstein M, et al. DAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-XL and induction of autophagy. EMBO Rep. 2009;10:285–92. doi: 10.1038/embor.2008.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wei Y, Pattingre S, Sinha S, Bassik M, Levine B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell. 2008;30:678–88. doi: 10.1016/j.molcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Russell RC, Tian Y, Yuan H, Park HW, Chang YY, Kim J, et al. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol. 2013;15:741–50. doi: 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Denton D, Nicolson S, Kumar S. Cell death by autophagy: facts and apparent artefacts. Cell Death Differ. 2012;19:87–95. doi: 10.1038/cdd.2011.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Galluzzi L, Morselli E, Vicencio JM, Kepp O, Joza N, Tajeddine N, et al. Life, death and burial: multifaceted impact of autophagy. Biochem Soc Trans. 2008;36:786–90. doi: 10.1042/BST0360786. [DOI] [PubMed] [Google Scholar]
- 43.Anding AL, Baehrecke EH. Autophagy in Cell Life and Cell Death. Curr Top Dev Biol. 2015;114:67–91. doi: 10.1016/bs.ctdb.2015.07.012. [DOI] [PubMed] [Google Scholar]
- 44.Shimizu S, Kanaseki T, Mizushima N, Mizuta T, Arakawa-Kobayashi S, Thompson CB, et al. Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nat Cell Biol. 2004;6:1221–8. doi: 10.1038/ncb1192. [DOI] [PubMed] [Google Scholar]
- 45.Fazi B, Bursch W, Fimia GM, Nardacci R, Piacentini M, Di Sano F, et al. Fenretinide induces autophagic cell death in caspase-defective breast cancer cells. Autophagy. 2008;4:435–41. doi: 10.4161/auto.5669. [DOI] [PubMed] [Google Scholar]
- 46.Karch J, Schips TG, Maliken BD, Brody MJ, Sargent MA, Kanisicak O et al. Autophagic cell death is dependent on lysosomal membrane permeability through Bax and Bak. Elife 2017; 6. 10.7554/eLife.30543. [DOI] [PMC free article] [PubMed]
- 47.Berry DL, Baehrecke EH. Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell. 2007;131:1137–48. doi: 10.1016/j.cell.2007.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Denton D, Shravage B, Simin R, Mills K, Berry DL, Baehrecke EH, et al. Autophagy, Not Apoptosis, Is Essential for Midgut Cell Death in Drosophila. Curr Biol. 2009;19:1741–6. doi: 10.1016/j.cub.2009.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Denton D, Shravage B, Simin R, Baehrecke EH, Kumar S. Larval midgut destruction in Drosophila: Not dependent on caspases but suppressed by the loss of autophagy. Autophagy. 2010;6:163. doi: 10.4161/auto.6.1.10601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lamy L, Ngo VN, Emre NCT, Shaffer AL, Yang Y, Tian E, et al. Control of autophagic cell death by caspase-10 in multiple myeloma. Cancer Cell. 2013;23:435–49. doi: 10.1016/j.ccr.2013.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marino G, Niso-Santano M, Baehrecke EH, Kroemer G. Self-consumption: the interplay of autophagy and apoptosis. Nat Rev Mol Cell Biol. 2014;15:81–94. doi: 10.1038/nrm3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Munoz-Pinedo C, Martin SJ. Autosis: a new addition to the cell death Tower of Babel. Cell Death Dis. 2014;5:e1319. doi: 10.1038/cddis.2014.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang X, Li W, Wang C, Leng X, Lian S, Feng J, et al. Inhibition of autophagy enhances apoptosis induced by proteasome inhibitor bortezomib in human glioblastoma U87 and U251 cells. Mol Cell Biochem. 2014;385:265–75. doi: 10.1007/s11010-013-1835-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Han K, Kim J, Choi M. Quantitative indices of autophagy activity from minimal models. Theor Biol Med Model. 2014;11:31. doi: 10.1186/1742-4682-11-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu L, Wan F, Dutta S, Welsh S, Liu Z, Freundt E, et al. Autophagic programmed cell death by selective catalase degradation. Proc Natl Acad Sci USA. 2006;103:4952 LP–4957. doi: 10.1073/pnas.0511288103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boya P, González-Polo RA, Casares N, Perfettini JL, Dessen P, Larochette N, et al. Inhibition of macroautophagy triggers apoptosis. Mol Cell Biol. 2005;25:1025–40. doi: 10.1128/MCB.25.3.1025-1040.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007;8:741. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- 58.Liu Y, Levine B. Autosis and autophagic cell death: the dark side of autophagy. Cell Death Differ. 2015;22:367–76. doi: 10.1038/cdd.2014.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shoji-Kawata S. Identification of a candidate therapeutic autophagy-inducing peptide. Seikagaku. 2015;87:481–4. [PubMed] [Google Scholar]
- 60.Miyakawa-Naito A, Uhlén P, Lal M, Aizman O, Mikoshiba K, Brismar H, et al. Cell signaling microdomain with Na,K-ATPase and Inositol 1,4,5-trisphosphate receptor generates calcium oscillations. J Biol Chem. 2003;278:50355–61. doi: 10.1074/jbc.M305378200. [DOI] [PubMed] [Google Scholar]
- 61.Wang Y, Qiu Q, Shen JJ, Li DD, Jiang XJ, Si SY, et al. Cardiac glycosides induce autophagy in human non-small cell lung cancer cells through regulation of dual signaling pathways. Int J Biochem Cell Biol. 2012;44:1813–24. doi: 10.1016/j.biocel.2012.06.028. [DOI] [PubMed] [Google Scholar]
- 62.Nelson WJ, Veshnock PJ. Ankyrin binding to (Na++ K+ )ATPase and implications for the organization of membrane domains in polarized cells. Nature. 1987;328:533. doi: 10.1038/328533a0. [DOI] [PubMed] [Google Scholar]
- 63.Stabach PR, Devarajan P, Stankewich MC, Bannykh S, Morrow JS. Ankyrin facilitates intracellular trafficking of α(1)-Na(+)-K(+)-ATPase in polarized cells. Am J Physiol - Cell Physiol. 2008;295:C1202–C1214. doi: 10.1152/ajpcell.00273.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang SH, Wang KL, Yang WK, Lee TH, Lo WY, Lee JD. Expression and potential roles of sodium-potassium ATPase and E-cadherin in human gastric adenocarcinoma. PLoS One. 2017;12:1–10. doi: 10.1371/journal.pone.0183692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Casale CH, Alonso A, del C, Barra HS. Brain plasma membrane Na+ , K+ -ATPase is inhibited by acetylated tubulin. Mol Cell Biochem. 2001;216:85–92. doi: 10.1023/a:1011029125228. [DOI] [PubMed] [Google Scholar]
- 66.Zampar G, Chesta M, Carbajal A, Chanaday N, et al. Acetylated tubulin associates with the fifth cytoplasmic domain of Na+ /K+ -ATPase: possible anchorage site of microtubules to the plasma membrane. Biochem J. 2009;422:129–37. doi: 10.1042/BJ20082410. [DOI] [PubMed] [Google Scholar]
- 67.Kast DJ, Dominguez R. The cytoskeleton–autophagy connection. Curr Biol. 2017;27:R318–R326. doi: 10.1016/j.cub.2017.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Köchl R, Hu XW, Chan EYW, Tooze SA. Microtubules facilitate autophagosome formation and fusion of autophagosomes with endosomes. Traffic. 2006;7:129–45. doi: 10.1111/j.1600-0854.2005.00368.x. [DOI] [PubMed] [Google Scholar]
- 69.Xie Rui, Nguyen Susan, McKeehan Wallace L, Liu Leyuan. Acetylated microtubules are required for fusion of autophagosomes with lysosomes. BMC Cell Biology. 2010;11(1):89. doi: 10.1186/1471-2121-11-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Geeraert C, Ratier A, Pfisterer SG, Perdiz D, Cantaloube I, Rouault A, et al. Starvation-induced hyperacetylation of tubulin is required for the stimulation of autophagy by nutrient deprivation. J Biol Chem. 2010;285:24184–94. doi: 10.1074/jbc.M109.091553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Santander VS, Bisig CG, Purro SA, Casale CH, Arce CA, Barra HS. Tubulin must be acetylated in order to form a complex with membrane Na+ ,K+ -ATPase and to inhibit its enzyme activity. Mol Cell Biochem. 2006;291:167–74. doi: 10.1007/s11010-006-9212-9. [DOI] [PubMed] [Google Scholar]
- 72.Buttgereit F, Brand MD. A hierarchy of ATP-consuming processes in mammalian cells. Biochem J. 1995;312:163–7. doi: 10.1042/bj3120163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mack HID, Zheng B, Asara JM, Thomas SM. AMPK-dependent phosphorylation of ULK1 regulates ATG9 localization. Autophagy. 2012;8:1197–214. doi: 10.4161/auto.20586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Feng Y, Backues SK, Baba M, Heo JM, Harper JW, Klionsky DJ. Phosphorylation of Atg9 regulates movement to the phagophore assembly site and the rate of autophagosome formation. Autophagy. 2016;12:648–58. doi: 10.1080/15548627.2016.1157237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Koyama-Honda I, Itakura E, Fujiwara TK, Mizushima N. Temporal analysis of recruitment of mammalian ATG proteins to the autophagosome formation site. Autophagy. 2013;9:1491–9. doi: 10.4161/auto.25529. [DOI] [PubMed] [Google Scholar]
- 76.Florey Oliver, Kim Sung Eun, Sandoval Cynthia P., Haynes Cole M., Overholtzer Michael. Autophagy machinery mediates macroendocytic processing and entotic cell death by targeting single membranes. Nature Cell Biology. 2011;13(11):1335–1343. doi: 10.1038/ncb2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Krajcovic M, Overholtzer M. Mechanisms of ploidy increase in human cancers: a new role for cell cannibalism. Cancer Res. 2012;72:1596–601. doi: 10.1158/0008-5472.CAN-11-3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Martins I, Raza SQ, Voisin L, Dakhli H, Law F, De Jong D, et al. Entosis: The emerging face of non-cell-autonomous type IV programmed death. Biomed J. 2017;40:133–40. doi: 10.1016/j.bj.2017.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.MAJNO G, LA GATTUTAM, THOMPSON TE. Cellular death and necrosis: chemical, physical and morphologic changes in rat liver. Virchows Arch Pathol Anat Physiol Klin Med. 1960;333:421–65. doi: 10.1007/BF00955327. [DOI] [PubMed] [Google Scholar]
- 80.Tavassoly I, Parmar J, Shajahan-Haq A, Clarke R, Baumann W, Tyson J. Dynamic modeling of the interaction between autophagy and apoptosis in mammalian cells. CPT Pharmacomet Syst Pharmacol. 2015;4:263–72. doi: 10.1002/psp4.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kacser H, Burns JA, Kacser H, Fell DA. The control of flux. Biochem Soc Trans. 1995;23:341–66. doi: 10.1042/bst0230341. [DOI] [PubMed] [Google Scholar]
- 82.du Toit A, Hofmeyr JS, Gniadek TJ, Loos B. Measuring autophagosome flux. Autophagy 2018;14:1060–71. [DOI] [PMC free article] [PubMed]
- 83.Rautou P, Cazals–Hatem D, Moreau R, Francoz C, Feldmann G, Lebrec D, et al. Acute liver cell damage in patients with anorexia nervosa: a possible role of starvation-induced hepatocyte autophagy. Gastroenterology. 2008;135:840–848.e3. doi: 10.1053/j.gastro.2008.05.055. [DOI] [PubMed] [Google Scholar]
- 84.van Niekerk G, Loos B, Nell T, Engelbrecht AM. Autophagy—A free meal in sickness-associated anorexia. Autophagy. 2016;12:727–34. doi: 10.1080/15548627.2016.1147672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kheloufi M, Boulanger CM, Codogno P, Rautou PE. Autosis occurs in the liver of patients with severe anorexia nervosa. Hepatology. 2015;62:657–8. doi: 10.1002/hep.27597. [DOI] [PubMed] [Google Scholar]
- 86.Nah J, Zhai P, Levine B, Sadoshima J. Autosis is Triggered by Ischemia/reperfusion in the Heart. Circulation. 2017;136:A18532 LP–A18532. [Google Scholar]
- 87.Bhat P, Kriel J, Priya BS, Basappa, Shivananju NS, Loos B. Modulating autophagy in cancer therapy: Advancements and challenges for cancer cell death sensitization. Biochem Pharmacol. 2018;147:170–82. doi: 10.1016/j.bcp.2017.11.021. [DOI] [PubMed] [Google Scholar]
- 88.Kriel J, Müller-Nedebock K, Maarman G, Mbizana S, Ojuka E, Klumperman B et al. Coordinated autophagy modulation overcomes glioblastoma chemoresistance through disruption of mitochondrial bioenergetics. Sci Rep. 2018;8:10348. [DOI] [PMC free article] [PubMed]
- 89.Rangwala R, Leone R, Chang YC, Fecher LA, Schuchter LM, Kramer A, et al. Phase I trial of hydroxychloroquine with dose-intense temozolomide in patients with advanced solid tumors and melanoma. Autophagy. 2014;10:1369–79. doi: 10.4161/auto.29118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rangwala R, Chang YC, Hu J, Algazy KM, Evans TL, Fecher LA, et al. Combined MTOR and autophagy inhibition: Phase I trial of hydroxychloroquine and temsirolimus in patients with advanced solid tumors and melanoma. Autophagy. 2014;10:1391–402. doi: 10.4161/auto.29119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rosenfeld MR, Ye X, Supko JG, Desideri S, Grossman SA, Brem S, et al. A phase I/II trial of hydroxychloroquine in conjunction with radiation therapy and concurrent and adjuvant temozolomide in patients with newly diagnosed glioblastoma multiforme. Autophagy. 2014;10:1359–68. doi: 10.4161/auto.28984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Huang N, Wu J, Qiu W, Lyu Q, He J, Xie W, et al. MiR-15a and miR-16 induce autophagy and enhance chemosensitivity of Camptothecin. Cancer Biol Ther. 2015;16:941–8. doi: 10.1080/15384047.2015.1040963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gewirtz DA, Tyutyunyk-Massey L, Landry JW. The potentially conflicting cell autonomous and cell non-autonomous functions of autophagy in mediating tumor response to cancer therapy. Biochem Pharmacol. 2018;153:46–50. doi: 10.1016/j.bcp.2018.01.048. [DOI] [PubMed] [Google Scholar]
- 94.Lamy Laurence, Ngo Vu N., Emre N.C. Tolga, Shaffer Arthur L., Yang Yandan, Tian Erming, Nair Vinod, Kruhlak Michael J., Zingone Adriana, Landgren Ola, Staudt Louis M. Control of Autophagic Cell Death by Caspase-10 in Multiple Myeloma. Cancer Cell. 2013;23(4):435–449. doi: 10.1016/j.ccr.2013.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yuen Kwan Law B, Gordillo-Martnez F, Qing Qu Y, Zhang N, Wei Xu S, Saul Coghi P, et al. Thalidezine, a novel AMPK activator, eliminates apoptosis-resistant cancer cells through energy-mediated autophagic cell death. Oncotarget. 2017;8:30077–91. doi: 10.18632/oncotarget.15616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Elgendy M, Sheridan C, Brumatti G, Martin SJ. Oncogenic Ras-induced expression of Noxa and Beclin-1 promotes autophagic cell death and limits clonogenic survival. Mol Cell. 2011;42:23–35. doi: 10.1016/j.molcel.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 97.Begley CG, Ellis LM. Raise standards for preclinical cancer research. Nature. 2012;483:531. doi: 10.1038/483531a. [DOI] [PubMed] [Google Scholar]
- 98.Martins I, Raza SQ, Voisin L, Dakhli H, Allouch A, Law F et al. Anticancer chemotherapy and radiotherapy trigger both non-cell-autonomous and cell-autonomous death article. Cell Death Dis 2018; 9. 10.1038/s41419-018-0747-y. [DOI] [PMC free article] [PubMed]
- 99.Weeber F, Ooft SN, Dijkstra KK, Voest EE. Tumor organoids as a pre-clinical cancer model for drug discovery. Cell Chem Biol. 2017;24:1092–1100. doi: 10.1016/j.chembiol.2017.06.012. [DOI] [PubMed] [Google Scholar]
- 100.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–11. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H, et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011;25:717–29. doi: 10.1101/gad.2016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lock R, Kenific CM, Leidal AM, Salas E, Debnath J. Autophagy-dependent production of secreted factors facilitates oncogenic RAS-driven invasion. Cancer Discov. 2014;4:466–79. doi: 10.1158/2159-8290.CD-13-0841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Galluzzi Lorenzo, Pietrocola Federico, Bravo‐San Pedro José Manuel, Amaravadi Ravi K, Baehrecke Eric H, Cecconi Francesco, Codogno Patrice, Debnath Jayanta, Gewirtz David A, Karantza Vassiliki, Kimmelman Alec, Kumar Sharad, Levine Beth, Maiuri Maria Chiara, Martin Seamus J, Penninger Josef, Piacentini Mauro, Rubinsztein David C, Simon Hans‐Uwe, Simonsen Anne, Thorburn Andrew M, Velasco Guillermo, Ryan Kevin M, Kroemer Guido. Autophagy in malignant transformation and cancer progression. The EMBO Journal. 2015;34(7):856–880. doi: 10.15252/embj.201490784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Shoji-Kawata S, Sumpter R, Leveno M, Campbell GR, Zou Z, Kinch L, et al. Identification of a candidate therapeutic autophagy-inducing peptide. Nature. 2013;494:201–6. doi: 10.1038/nature11866. [DOI] [PMC free article] [PubMed] [Google Scholar]