Dear Editor,
Type VI secretion system (T6SS) adopts a contact-dependent secretion mechanism to deliver various lethal effectors into prokaryotic or eukaryotic cells.1,2 Due to its antibacterial and anti-eukaryotic activity, T6SS provides a powerful weapon for bacteria to kill their competitors and attackers.2 T6SS has been shown to resemble a bacteriophage tail-like nanomachine of more than 13 components.3 The whole complex can be divided into four subassemblies: a membrane core complex of TssJLM (TssJ, TssL, and TssM), a baseplate of TssAEGFK (TssA, TssE, TssG, TssF, and TssK) with VgrG spike complex, a tail complex composed of the Hcp tube and TssBC (TssB and TssC) sheath, and a cytoplasmic ATPase ClpV.4 The membrane core complex serves as a docking station for the baseplate and also a channel for the passage of the Hcp tube after sheath contraction, and it is made up of the inner membrane proteins TssL and TssM, and the outer membrane lipoprotein TssJ.2,5 Partial structural information of the T6SS membrane core complex has been reported in recent years,2,6,7 including the crystal structures of TssJ, a part of TssL and TssM, and a low-resolution negative-staining electron microscopy (EM) structure of the whole complex. This structural information has revealed that the T6SS membrane core complex does not form a pore on the outer membrane as other secretion channels do, such as T2SS and T3SS.8,9 More precise structural information of the T6SS membrane core complex is necessary to understand the secretion through T6SS.
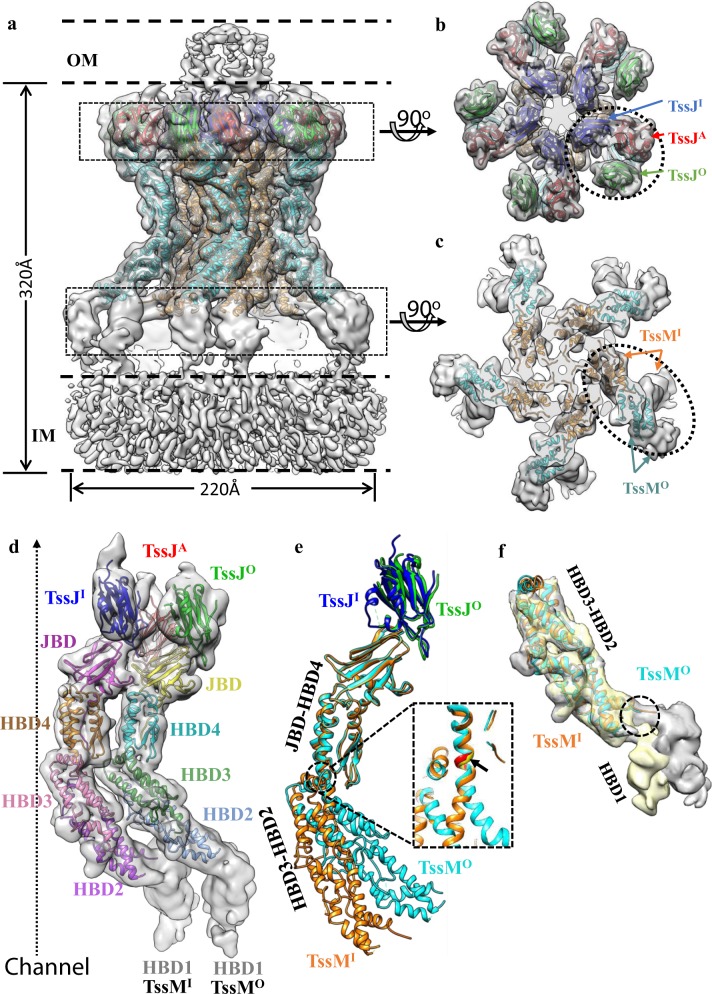
To obtain more structural details, we resolved the structure of a 1.7-MDa T6SS membrane core complex from enteroaggregative Escherichia coli (EAEC) by electron cryo-microscopy (cryo-EM) at an overall 4.0 Å resolution (Supplementary information, Figs. S1, S2 and Table S1). The T6SS membrane core complex was resolved as a rocket-shaped pentamer with a diameter of ~220 Å and a length of ~320 Å, spanning the outer and inner membranes (Fig. 1a), similar to the previously reported negative-staining EM structure.2 Local resolution analysis of the complex indicated varying resolutions (Supplementary information, Fig. S1f). The tip complex, which attaches to the outer membrane and consists of TssJ and part of TssM,2 had the highest resolution of 3.8 Å, enabling unambiguous model building (Supplementary information, Figs. S1, S3). The domains near the inner membrane seem to be flexible and consist of TssL and part of TssM (Fig. 1a). TssL was co-purified in the complex, (Supplementary information, Fig. S1a) but was missing in the 3D reconstruction, probably owing to its flexibility in the complex.
Fig. 1.
Overall structure of the T6SS membrane core complex and comparison of the TssJ-TssM subcomplexes in the inner and outer layer. a Side view of the T6SS membrane core complex. The model is superimposed with a density map low-pass filtered to 6 Å. TssMI, TssMO, TssJI, TssJO, and TssJA are colored in orange, cyan, blue, green, and red, respectively. The positions of the inner membrane (IM) and the outer membrane (OM) are labeled by dashed lines. b, c Top views of the regions boxed out from the density map in a. Three TssJ subunits in b and two TssM subunits in c labeled by the dotted ellipsoids belong to one asymmetric unit. d The asymmetric unit. The model is superimposed with a density map low-pass filtered to 6 Å. The TssM domains, JBD and HBD1–4, and three TssJ subunits are drawn with different colors. e Structural alignment of the JBD-HBD4 from the inner and outer TssM subunits (RMSD is 0.768 Å). The flexible hinge is enlarged in the dashed section. f Structural alignment of HBD2-HBD3 from the inner and outer TssM subunits (RMSD is 0.747 Å). Density maps with HBD1 densities are superimposed with the aligned model, and different relative orientations of HBD1 in inner (gray) and outer (yellow) TssM subunits are shown. The hinge region is labeled by the dashed circle
TssM is the major part of the complex. This 1129-residue protein contains an N-terminal cytoplasmic domain (residues 1–386) with three transmembrane helices and a large periplasmic region (residues 387–1129).7 The former is located at the inner membrane, and is missing in the reconstruction with TssL. According to our cryo-EM structure and the secondary structure prediction,7 the periplasmic region of TssM can be structurally divided into five domains: the TssJ binding domain (JBD), and four helix-bundle domains (HBD1–4) (Fig. 1d). JBD and HBD2–4 belong to the tip complex, which were resolved at near-atomic resolution. HBD1 binds to the inner membrane, and was resolved at nanometer resolution (Fig. 1d). Ten TssM subunits constitute a double-layer channel with five TssM pillars in each layer (Fig. 1c), similar to previous observations by negative-staining EM.2 The resolved regions of the membrane core complex have a C5 symmetry, and each asymmetric unit consists of two TssM subunits (TssMO in the outer layer and TssMI in the inner layer) and three TssJ subunits (TssJO bound to TssMO, TssJI bound to TssMI, and an additional TssJA) (Fig. 1a–c).
Unexpectedly, we found that an additional TssJA subunit was attached to two pairs of TssJ-TssM complex in an asymmetric unit (Fig. 1b), leading to a 3:2 stoichiometric ratio of TssJ and TssM in one asymmetrical unit. This differs from the 2:2 ratio observed in the X-ray heterodimeric structure of TssJ-TssMCt and proposed in the negative-staining EM map of a fully assembled T6SS membrane core complex.2 However, our model with 3:2 stoichiometric ratio is consistent with the previously reported negative-staining EM map,2 in which an extra density fits the additional TssJA (Supplementary information, Fig. S4). The 3:2 ratio between the lipoproteins and its binding proteins is unique and differs from the 1:1 ratio among those resolved in other bacterial secretion systems, such as AspS-GspD in type II secretion system and VirB7-VirB9/10 in the type IV secretion system.10,11 TssJA forms interactions with adjacent TssJI and TssMO (Supplementary information, Fig. S5). The interaction area between TssJA and TssJI is ~552 Å2 and that between TssJA and TssMO is ~580 Å2 (Supplementary information, Fig. S5d). The presence of TssJA enhances the interaction and the pairing relationship between two adjacent TssJ-TssM pairs in one asymmetric unit.
Other than the additional TssJA, the high-resolution structure also enabled comparisons between subunits and domains in the large complex. Structural comparison between TssMI and TssMO reveal obvious conformational differences (Fig. 1e). Both TssMI and TssMO have a curved-in shape, where TssMO has slightly larger inward curvature than TssMI (Fig. 1e). Structural alignment between TssMI and TssMO reveals that the five TssM domains, JBD and HBD1–4, can be divided into three rigid groups: JBD-HBD4, HBD3-HBD2, and HBD1 (Fig. 1e, f). The relative orientation between JBD-HBD4 and HBD3-HBD2 and that between HBD3-HBD2 and HBD1 differ in the inner and outer TssM subunits (Fig. 1e, f). HBD4 and HBD3 are connected by a long α-helix (residue 851–891). By comparing the conformation in TssMI and TssMO, this helix is bent in the middle, near a region of residues P870/A871/A872. Therefore, this region might be able to serve as a hinge to allow the relative rotation between JBD-HBD4 and HBD2-HBD3 (Fig. 1e). Another hinge can be found between the HBD1 and HBD2 domains (Fig. 1f). By aligning the HBD2-HBD3 regions of TssMI and TssMO, it is apparent that the relative orientation between HBD2-HBD3 and HBD1 differs (Fig. 1f). These differences between TssMI and TssMO suggest a possible conformational change in TssM, which might be useful to form an open channel.
Both our cryo-EM structure and the previous negative-staining EM structure solved the T6SS membrane core complex at the closed state.6 Inner TssM subunits define a very narrow channel, limiting the passage of the Hcp tube.2 Based on the C10 symmetrized TssJ/TssM model for Hcp tube secretion,6 the five inner TssM subunits must re-localize to the outer section of the complex to form a bigger ring along with the five outer TssM subunits. However, by our measurement for the inner diameter of the channel in a model of C10 symmetrized TssMs (Supplementary information, Fig. S6), the channel at the hinge region between JBD-HBD4 and HBD2-HBD3 is still too narrow to allow the Hcp passage. This implied that conformational changes of TssMs are necessary. These flexible hinges provide a possibility for TssMs to undergo outward conformational changes during Hcp secretion.
In summary, our study reveals the architecture of TssJLM membrane complex. Based on the high-resolution information above, our structure highlights two features that may be involved in the channel opening. One is an additional TssJA subunit attached to two TssJ-TssM subcomplexes, resulting in a 3:2 stoichiometric ratio of TssJ and TssM in one asymmetrical unit. The other is the conformational differences between TssMI and TssMO, exhibiting two flexible hinges in TssM. While the actual conformational changes or subunit rearrangements during the secretion are still unknown, these flexible hinges may provide a clue to understand the possible conformational changes of TssMs during the TssJLM channel opening.
Supplementary information
Acknowledgements
This work was supported by funds from the National Natural Science Foundation of China (31722015 and 31570730), the National Key Research and Development Program (2016YFA0501102 and 2016YFA0501902), Advanced Innovation Center for Structural Biology, Tsinghua-Peking Joint Center for Life Sciences, and One Thousand Talent Program by the State Council of China. We acknowledge Tsinghua University Branch of China National Center for Protein Sciences Beijing for providing facility support for cryo-EM and computation.
Author contributions
M.Y., Z.Y., and X.L. designed experiments. M.Y. and Z.Y. performed the cryo-EM and biochemistry experiments. All authors contributed to the data analysis and manuscript preparation.
Competing interests
The authors declare no competing interests.
Footnotes
Accession codes
The density map of the T6SS membrane core complex is available in EMDB with entry code: EMD-9747. Model of the T6SS membrane core complex is deposited in the Protein Data Bank with entry code: 6IXH.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Meng Yin and Zhaofeng Yan.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41422-018-0130-7.
References
- 1.Russell AB, Peterson SB, Mougous JD. Nat. Rev. Microbiol. 2014;12:137–148. doi: 10.1038/nrmicro3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Durand E, et al. Nature. 2015;523:555–560. doi: 10.1038/nature14667. [DOI] [PubMed] [Google Scholar]
- 3.Cascales E. EMBO Rep. 2008;9:735–741. doi: 10.1038/embor.2008.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basler Marek. Type VI secretion system: secretion by a contractile nanomachine. Philosophical Transactions of the Royal Society B: Biological Sciences. 2015;370(1679):20150021. doi: 10.1098/rstb.2015.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aschtgen MS, Gavioli M, Dessen A, Lloubes R, Cascales E. Mol. Microbiol. 2010;75:886–899. doi: 10.1111/j.1365-2958.2009.07028.x. [DOI] [PubMed] [Google Scholar]
- 6.Durand E, et al. J. Biol. Chem. 2012;287:14157–14168. doi: 10.1074/jbc.M111.338731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Felisberto-Rodrigues C, et al. PLoS. Pathog. 2011;7:e1002386. doi: 10.1371/journal.ppat.1002386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan Z, Yin M, Xu D, Zhu Y, Li X. Nat. Struct. Mol. Biol. 2017;24:177–183. doi: 10.1038/nsmb.3350. [DOI] [PubMed] [Google Scholar]
- 9.Worrall LJ, et al. Nature. 2016;540:597–601. doi: 10.1038/nature20576. [DOI] [PubMed] [Google Scholar]
- 10.Yin M, Yan Z, Li X. Nat. Microbiol. 2018;3:581–587. doi: 10.1038/s41564-018-0148-0. [DOI] [PubMed] [Google Scholar]
- 11.Chandran V, et al. Nature. 2009;462:1011–5. doi: 10.1038/nature08588. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.