The NLRP3 inflammasome detects a variety of pathogen-derived and host-derived signals, but its mechanism of activation has long remained uncharacterized. In a recent study in Nature, Chen et al. demonstrate that NLRP3 activators induce the disassembly of the trans-Golgi network (TGN) and that NLRP3 is targeted for activation at the dispersed TGN (dTGN) by ionic bonding between its conserved polybasic region and phosphatidylinositol-4-phosphates on the dTGN.
NLRP3 is a cytosolic pattern recognition receptor that assembles a multiprotein signaling complex, the NLRP3 inflammasome, upon detecting pathogens or cellular stress. This complex recruits and activates the protease caspase-1(Casp1), which cleaves interleukin (IL)-1 family proteins, pro-IL-1β and pro-IL-18 to their mature bioactive forms; and processes the pore-forming protein Gasdermin D (GSDMD) to induce a form of lytic cell death known as pyroptosis.1 While many studies show an important role of NLRP3 in host defense, it also plays a deleterious role in chronic inflammatory disorders and gain-of-function mutations in NLRP3 are associated with numerous hereditary inflammatory diseases (e.g., CAPS, neonatal onset multisystem inflammatory disorder).2 Therefore, there is an urgent need to better understand the activation mechanisms of NLRP3.
NLRP3 has a unique ability to sense a wide variety of structurally unrelated molecules ranging from whole pathogens, insoluble particulates and endogenous danger signals, indicating that these molecules likely elicit a common stress signal that is sensed by NLRP3. Indeed, potassium efflux, mitochondrial damage, reactive oxygen species, lysosomal rupture and calcium mobilization have all been proposed to be the common event sensed by NLRP3. However, despite more than 10 years of intensive research, the field has yet to reach a consensus opinion. In a recent study published in Nature, the group of Zhijian ‘James’ Chen (referred to as Chen et al.3) reports an unexpected finding that dispersion of the trans-golgi network (TGN) is the early and common stress event that is required for NLRP3 activation in response to diverse agonists.
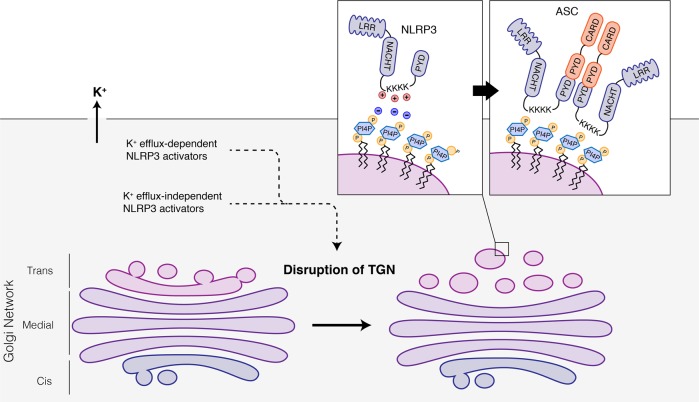
To investigate the subcellular location of NLRP3 assembly, Chen et al. established an elegant in vitro assay in which they permeabilized the plasma membrane of HEK-293T cells expressing the inflammasome adaptor ASC and caspase-1 with perfringolysin O (PFO), before treating them with fractionated extracts of stimulated NLRP3-positive but ASC-negative cells. Surprisingly, NLRP3 co-migrated strongly with markers of the TGN in the fractions which elicited robust Casp1 processing in PFO-permeabilized ASC-Casp1-expressing HEK-293T cells. To investigate this further, the authors examined the changes in morphology and location of subcellular organelles and the NLRP3 protein via microscopy following inflammasome stimulation. Remarkably, all five structurally unrelated NLRP3 agonists: nigericin, ATP, gramicidin, imiquimod and CL097 triggered disassembly of the TGN onto which NLRP3 formed distinct puncta. Disassembled TGN (dTGN)-localized NLRP3 also initiated ASC polymerization, consistent with the idea that TGN disassembly is the common stress event sensed by NLRP3. (Fig. 1)
Fig. 1.
Diverse NLRP3 agonists trigger disassembly of the trans-Golgi network (TGN)exposing PI4P microdomains to recruit NLRP3 through its ionic bonding between its polybasic KKKK motif and the negatively charged PtdIns4Ps. NLRP3 aggregates on disassembled TGN and nucleates polymerization of the adaptor protein ASC
To further understand the molecular requirements for NLRP3-dTGN interaction, the authors generated truncated and point mutants of NLRP3 and found that deletion of four consecutive lysine residues between the PYD domain and the NACHT domain of NLRP3 abrogated its recruitment to the dTGN. They further demonstrated that this polybasic sequence, which is evolutionarily highly conserved, mediates binding to phosphatidylinotisol 4-phosphate (PtdIns4P) microdomain present on the dTGN. This was a surprising finding, since the leucine-rich repeat (LRR) domain of NLRP3 was often believed to mediate ligand sensing,2 much like the LRR of other innate immune sensors such as Toll-like receptor.4 This is, however, consistent with a recent report demonstrating that LRR of NLRP3 is not required for ASC oligomerization and Casp1 processing upon stimulation with classical NLRP3 agonists.4 Nevertheless, although recruitment of NLRP3 to the dTGN is an essential step for activation, it by itself is not sufficient, since induced targeting of NLRP3 to the TGN using the PH domain of OSBP did not yet lead to NLRP3 activation in the absence of NLRP3 agonist.
The discovery that TGN disassembly is a common stress event required for NLRP3 inflammasome assembly has undoubtedly expanded our current knowledge on the activation mechanisms of this inflammasome, and has important implications to the field. How canonical NLRP3 activators initiate the dispersion of the TGN is unclear and awaits further clarification. NLRP3 activity is tightly regulated by post-translational modifications including phosphorylation and ubiquitination. It is tempting to speculate that such modifications may alter the net charge of NLRP3, or sterically inhibit the interactions between the KKKK motif and PtdIns4P on dTGN. Furthermore, GSDMD and MLKL, two death effectors downstream of the non-canonical inflammasome and necrosome respectively, were shown to disrupt internal membranes including those of the Golgi.5–7 It will be interesting to investigate whether GSDMD or MLKL promotes NLRP3 activation by directly inducing dispersion of the TGN.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Kaiwen W. Chen, Dave Boucher
References
- 1.Broz P, Dixit VM. Nat. Rev. Immunol. 2016;16:407–420. doi: 10.1038/nri.2016.58. [DOI] [PubMed] [Google Scholar]
- 2.Schroder K, Tschopp J. Cell. 2010;140:821–832. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- 3.Chen J, Chen ZJ. Nature. 2018;564:71–76. doi: 10.1038/s41586-018-0761-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hafner-Bratkovic I, et al. Nat. Commun. 2018;9:5182. doi: 10.1038/s41467-018-07573-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ding J, et al. Nature. 2016;535:111–116. doi: 10.1038/nature18590. [DOI] [PubMed] [Google Scholar]
- 6.Liu X, et al. Nature. 2016;535:153–158. doi: 10.1038/nature18629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oberst A. FEBS J. 2016;283:2616–2625. doi: 10.1111/febs.13520. [DOI] [PMC free article] [PubMed] [Google Scholar]