In a recent paper published in Cell Discovery , Sun et al. report a novel chemical approach to rapidly and reversibly deplete endogenous proteins globally in both small and large animals via the PROTAC (PROteolysis TArgeting Chimera) approach. This strategy not only provides a powerful tool for protein function studies in vivo, but also highlights the potential of using PROTAC in future human cancer therapies.
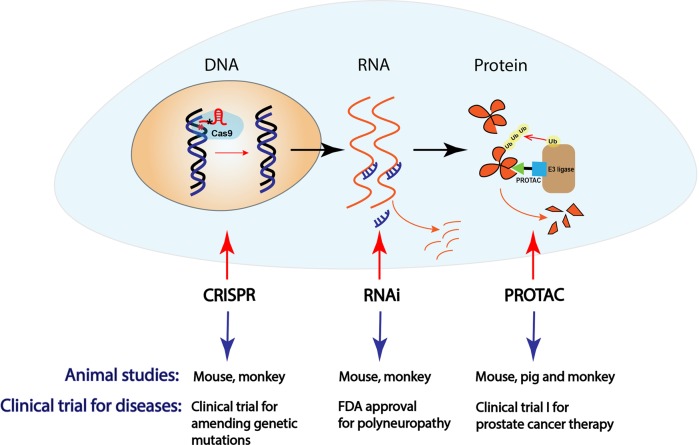
In cancer therapies, targeting tumor-addicted oncoproteins is a promising strategy. To this end, small molecules targeting protein catalytic activities have been successfully developed and approved for cancer therapies in clinic, including but not limited to the small-molecule inhibitors targeting kinases, DNA and histone methyltransferases and deacetylases. However, a large amount of oncoproteins, such as transcriptional factors, chromatin modulators and small GTPases, are hard to be directly targeted pharmaceutically and termed undruggable targets.1 As illustrated in Fig. 1, there are three potential approaches to inactivate an oncogene or disease-causing genes, at the DNA, RNA or protein level with CRISPR, RNAi (RNA interference) or PROTAC (PROteolysis TArgeting Chimera), respectively. The CRISPR technique has been predicted with huge potential to amend defective genomic DNA,2 especially DNA mutations, to potentially cure these diseases. The first clinical trial for CRISPR (CTX001, CRISPR Therapeutics and Vertex Pharmaceuticals) has been recently issued for treating β-thalassemia (NCT03655678) and sickle cell (NCT03745287) in both Europe and U.S., although the off-target side effect might limit the future usage of this approach in other human disorders. Meantime, RNAi technique has been designed for targeted therapies via destabilizing specific disease-causing RNA species,3 and the first RNAi drug Onpattro (also known as Patisiran, Alnylam Pharmaceuticals) has been released in 2017 for the treatment of polyneuropathy caused by hereditary transthyretin-mediated amyloidosis (hATTR amyloidosis). However, off-target toxicity and inefficient drug delivery is still the major limitation for this approach. Given the limits for CRISPR and RNAi, as well as other small-molecule inhibitors, the PROTAC technology has recently attracted a lot of attention, due to its unique feature. PROTAC is designed to target protein of interest (usually oncoprotein) for degradation by hijacking the endogenous E3 ligase and ubiquitin proteasome system (UPS).4
Fig. 1.
The schematic illustration of various strategies to inactivate an oncogene or other disease-causing genes at the DNA, RNA or protein level by CRISPR, RNAi or PROTAC, respectively
Like bi-specific antibodies, PROTACs act as bridging molecules for E3 ligase and protein of interest (Fig. 1). The first proof-of-concept PROTAC, PROTAC-1 is a peptide-based PROTAC containing phosphopeptide, which was initially designed to bind β-TRCP to degrade methionine aminopeptidase 2 (MetAP2);5 other peptide-based PROTACs contain hydroxyl peptide that binds pVHL to subsequently target FKBP12 or Androgen Receptor (AR) for degradation.6 The first generation of PROTACs mainly depends on peptide, while small molecule-based PROTACs recently emerge due to the discovery of specific ligand for E3 ligase, such as thalidomide and its derivatives that bind CRL4CRBN to target IKZF1/3 for degradation.7 Various PROTACs have been reported to effectively target and destabilize proteins of interest such as BRD4 and BTK in either CRL2pVHL-based or CRL4CRBN-based manner.8 The first oral PROTAC drug (ARV-110, targeting AR for degradation) has recently been approved by FDA for phase I clinical trial in treating patients with metastatic castration resistant prostate cancer (mCRPC) in 2019. These findings shed light on the impressive approach to directly target proteins of interest. However, the systematical evaluation and preclinical studies of PROTACs in animals, especially in large animals, have not been well investigated yet.
In a recent paper in Cell Discovery, Sun et al. have systematically investigated the potential usage of PROTACs in mice and large animals, such as pigs and rhesus monkeys. They observed that the PROTAC approach could markedly deplete the targeted proteins such as FKBP12 and BTK in vivo.9 Moreover, to provide a preclinical evidence that PROTAC is a potentially powerful tool to combat human diseases, Sun et al. investigated the efficacy of PROTACs in large and small animal models, and revealed that PROTAC-mediated depletion of FKBP12 by oral administration occurred in most organs or tissues (except brain, which is accessible by intracerebroventricular injection), with a constant effect for about one week after a single treatment. Furthermore, they also observed that PROTAC-mediated degradation of targeted protein has specific biological consequences corresponding to the physiological function of the target protein. For instance, ablation of FKBP12 by PROTACs could disturb cardiac function in mouse and rhesus monkeys, which phenocopies Fkbp12 conditional knockout in mice.10 More importantly, this disturbance is reversible and the effect on cardiac function disappears 22 days after withdrawal of the FKBP12-PROTAC.9 All these novel findings suggest the efficiency and reversible potential of PROTAC approach in animals, and provide a robust basis for future clinical trials of cancer therapies in human patients (Fig. 1).
While the elegant studies by Sun et al. provide a promising perspective for the PROTAC approach with rapid and reversible efficacy and less cost in animals, it is still a big challenge for designing on-target PROTACs. Identification of the appropriate binding ligands for E3 ligases and targeted proteins to reduce off-target toxicity will be the focus of further studies.
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Dang CV, Reddy EP, Shokat KM, Soucek L. Nat. Rev. Cancer. 2017;17:502–508. doi: 10.1038/nrc.2017.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cong L, et al. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bumcrot D, Manoharan M, Koteliansky V, Sah DW. Nat. Chem. Biol. 2006;2:711–719. doi: 10.1038/nchembio839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sakamoto KM. Pediatr. Res. 2010;67:505–508. doi: 10.1203/PDR.0b013e3181d35017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sakamoto KM, et al. Proc. Natl Acad. Sci. USA. 2001;98:8554–8559. doi: 10.1073/pnas.141230798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schneekloth JS, Jr., et al. J. Am. Chem. Soc. 2004;126:3748–3754. doi: 10.1021/ja039025z. [DOI] [PubMed] [Google Scholar]
- 7.Fischer ES, et al. Nature. 2014;512:49–53. doi: 10.1038/nature13527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Winter GE, et al. Science. 2015;348:1376–1381. doi: 10.1126/science.aab1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun, X. et al. Cell Dis.10.1038/s41421-018-0079-1 (2019).
- 10.Xin HB, et al. Nature. 2002;416:334–338. doi: 10.1038/416334a. [DOI] [PubMed] [Google Scholar]