Abstract
Nonribosomal peptide synthetases (NRPSs) are involved in the biosynthesis of numerous peptide and peptide-like natural products that have been exploited in medicine, agriculture, and biotechnology, among other fields. As a consequence, there have been considerable efforts aimed at understanding how NRPSs orchestrate the assembly of these natural products. This review highlights several recent examples that continue to expand upon the fundamental knowledge of NRPS mechanism and includes (1) the discovery of new NRPS substrates and the mechanism by which these sometimes structurally complex substrates are made, (2) the characterization of new NRPS activities and domains that function during the process of peptide assembly, and (3) the various catalytic strategies that are utilized to release the NRPS product. These findings continue to strengthen the predictive power for connecting genes-to-products, thereby facilitating natural product discovery and development in the Genomics Era.
Keywords: NRPS, Natural Products, Bioinformatics, Discovery, Domain, Mechanism
Introduction
The field of natural products has undergone a rebirth since the turn of the millennium owing to the realization that the established natural product repertoire of a given organism appears to be greatly under-representative of what is predictable from genomic information [2,6,18]. There are now several examples in the literature highlighting the power of harnessing this genomic information into the isolation and characterization of novel natural products, a process that is often referred to as genome mining. The predictive power that is guiding genome mining has been particularly fruitful when applied to the class of secondary metabolites known as nonribosomal peptides (NRPs), which are typically assembled from megaenzyme complexes called nonribosomal peptide synthetases (NRPSs). Among other notable pioneers [49,50,68–70], Christopher T. Walsh and coworkers have made several breakthrough contributions that have been integral for establishing a functional and mechanistic paradigm for NRPS-guided biosynthesis of natural products [exemplified in [5,54,66,92,97,104,89]]. In this review, we highlight several NRPS systems that have been characterized within the past decade that expand upon and refine the mechanistic foundation that was initially established by Walsh and others. These new variations of the established paradigm, which often introduce unusual chemical features that extend well beyond the chemistry of proteinogenic amino acids, now offer new genetic fingerprints that will enable future efforts to take advantage of genome mining for NRP discovery.
The NRPS Paradigm
The fundamental enzymology as well as recent advances in the structure and function of NRPS has been reviewed elsewhere in several noteworthy publications and in this special issue [69,24,111,15,28,78,38,52,71]. To summarize briefly, NRPS are generally multimodular proteins, wherein each module is responsible for the incorporation of a single amino acid into an NRP. A minimal module consists of three domains: a condensation (C) domain, an adenylation (A) domain, and a carrier protein [CP] domain, which is often termed a thiolation (T) or peptidyl-carrier protein (PCP) when specifically discussing NRPS. A notable variation to the minimal module is the loading or initiation module, which often consists of simply an A and CP domain. The catalytic cycle of an NRPS is preceded by the posttranslational modification of a Ser residue of the CP domain with a 4-phosphopantetheinyl (Ppant) group. This reaction, catalyzed by a phosphopantetheinyltransferase (PPTase) that is encoded by a discrete gene, generates the reactive sulfhydryl functionality that is essential for the ensuing thiotemplated chemistry. In this review, it is assumed that the CP, if present, is already posttranslationally modified with the Ppant group. Following the generation of the holo CP domain, the A domain selects a carboxylic acid-containing substrate, activates the acid via ATP-dependent adenylation, and transfers the acid to the partnered CP domain via thioesterification to form an acyl-S-CP. Although the monomeric building blocks are often proteinogenic α-amino acids, this is not always the case as many non-proteinogenic α-amino acids, β-keto acids, β-amino acids, among others, are known to be activated and loaded by certain A domains. After thioesterification the C domain then catalyzes amide or ester bond formation between upstream and downstream CP-thioesterified substrates. The growing chain is passed from module to module in an assembly-line fashion as (amino) acid units are added until the peptide product is released from the terminal CP domain. Although variations are known, this final off-loading reaction is typically catalyzed by a thioesterase (TE) domain embedded just downstream of the terminal CP domain. The TE-catalyzed reaction generally proceeds by hydrolysis or intramolecular cyclization leading to a free acid or lactone/lactam, respectively.
Shortly after the discovery and characterization of the first NRPS, a co-linearity rule was proposed that stated that the ordering of modules within an NRPS from N- to C-terminus directs the linear sequence of amino acids in the resulting NRP. This rule has held true for many NRPSs, which has enabled bioinformatic approaches for predicting the identity of an NRP from genome sequence and architectural organization. Likewise, the co-linearity rule has spurred significant efforts aimed at defining the substrate specificity of the catalytic A and C domains, resulting not only in predictive algorithms such as the NRPS code for A domains [87,81,19,96] but also in defining different sub-types of C domains [21,80,86]. Structural studies and the utilization of chemical probes for a few model NRPS have added yet another spatial dimension, which has rapidly progressed from an easy-to-imagine assembly line of linearly connected domains to one of a series of stacked channels with highly mobile elements [26,103,113,44,51,48,25]. These efforts have afforded a logical, working mechanism that describes how domains of a given module or domains between modules interact to process the substrates and intermediates.
Despite the significant advances toward establishing an NRPS biosynthetic paradigm, several biochemical studies have now revealed what whole genome sequencing first uncovered: that microorganisms harbor many NRPS biosynthetic gene clusters that appear architecturally unique when compared to the relatively well-studied model systems that have yielded the co-linearity rule and NRPS code. For example, it is now apparent that domains or didomains can be encoded as single proteins, or modules can exist with unusual organization of the individual domains, each of which serves to complicate the bioinformatic predictions, particularly in the cases without a correlated NRP product [110]. In addition to the potential of having an unusual modular or domain architecture, NRPS can harbor additional catalytic domains—herein referred to as auxiliary domains—that significantly expand upon the chemistry of a substrate by performing oxidations, cyclizations, epimerizations, and halogenations, to name a few. NRPS-catalyzed processes can also incorporate several discrete-acting enzymes, the timing of which can occur at all stages, i.e., before or after thioesterification of the CP domain, following C domain-catalyzed amide bond formation, or after product release. Finally, yet another level of diversity among NRPS has been realized evolutionarily through mutations to any of the essential or auxiliary domains or the discrete-acting enzymes. These additional “dimensions”, although adding a layer of complexity that needs to be considered in defining an NRPS mechanism, significantly enhance the structural diversity that can be achieved from the products of NRPS. Discussed herein are select examples from the recent literature that highlight this point.
Beyond the Chosen Twenty
The A domain initiates peptide synthesis by specifically selecting the carboxylic acid-containing substrate for incorporation. Due to this role, the A domain is considered the gatekeeper of an NRPS assembly line. As previously noted, the importance of the A domain in substrate selectivity has resulted in efforts from multiple groups toward generating an NRPS code, which utilizes structurally homology models and active site mapping of an A domain to predict the most likely substrate. The NRPS code continues to be refined as biochemical studies have revealed the large breath of substrates that can be activated, which now extends well beyond the twenty proteinogenic amino acids. Highlighted below are four relatively recent examples of biochemical studies that have revealed a new substrate selectivity for A domains, thus continuing to add to the already large repertoire of carboxylic acid-containing precursors that are incorporated into NRPs.
S-adenosylmethionine joins the list.
Colibactin is a genotoxin produced by several enterobacteria including uropathogenic and probiotic strains of Escherichia coli. A genomic island encoding minimally nineteen proteins (annotated here as ClbA-S as defined in [43]), including several NRPS modules and polyketide synthases (PKSs), was initially linked to the biosynthesis of colibactin in 2006 [75,43]. With the exception of the self-resistance hydrolase ClbS, all of the gene products were determined to be essential for genotoxicity. Bioinformatic analysis subsequently enabled the prediction of the colibactin structure as an unusual hybrid peptide-polyketide, yet the final metabolic product of this gene cluster remained uncertain [8,14,57,105,108].
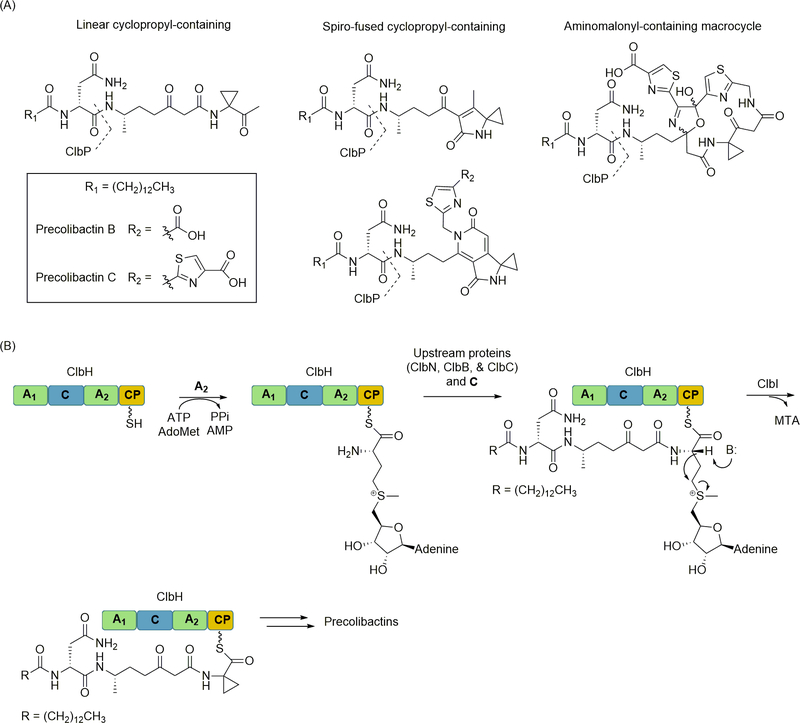
ClbP was one of the first gene products biochemically characterized, revealing this protein functions as a peptidase that cleaves an N-terminal acylated D-Asn as the likely final biosynthetic step to yield the bioactive natural product [7,13,22]. The discovery of this “prodrug” production strategy inspired several groups to independently prepare ΔclbP strains and examine the metabolome of these mutants. As a result of these efforts, a number of precolibactins were identified that represent shunt or advanced products from nearly every stage of the scaffold extension process (Figure 1A). These precolibactins can be categorized into three structural types (in order of early-to-late stage intermediates): a linear cyclopropyl-containing precolibactin, spiro-fused cyclopropyl-containing precolibactins, or an aminomalonyl-containing macrocyclic precolibactin. The last of this group, which is the largest precolibactin that has been identified to date [56], also contains a cyclopropyl group embedded within the macrocycle. Importantly, the cyclopropyl group appears to be the defining functionality that causes DNA interstrand crosslinking for the mature colibactin, as a spirocyclopropyl-containing-precolibactin has been demonstrated to cause alkylation and crosslinking in vivo and in vitro [117,11].
Figure 1.
NRPS-catalyzed cyclopropyl group formation during colibactin biosynthesis. (A) Structures of representative precolibactins isolated from ΔclbP mutant strains. The site of ClbP-catalyzed hydrolysis is indicated. (B) The role of the NRPS ClbH in cyclopropyl formation. The A2 domain of ClbH selectively activates and loads S-adenosyl-l-methionine to the CP domain. ClbI, a multidomain polyketide synthase, presumably utilizes general base catalysis to initiate cyclopropyl formation with concomitant release of 5′-methylthioadenosine. AdoMet, S-adenosyl-l-methionine; MTA, 5′-methylthioadenosine.
The cyclopropyl functionality is found in other microbial and plant natural products, and for the most part, the biochemical details for its biogenesis have been established. For colibactin biosynthesis, a distinct mechanism involving an NRPS and a PKS was uncovered [106,121]. The first evidence that an NRPS was involved came from gene inactivation experiments, wherein a ΔclbH mutant strain was unable to produce cyclopropane-containing precolibactins [8,57]. The ClbH protein consists of sequential A1, C, A2, and CP domains, with A1 having a predicted specificity toward l-Ser while A2 specificity could not be predicted (Figure 1B). Therefore, the bioinformatics analysis did not provide significant insight into how ClbH facilitated cyclopropylation or whether other proteins were involved. Comparative HRMS, following feeding of the producing strain with potential 13C-labeled precursors, revealed incorporation of L-[U-13C]-Met into the cyclopropyl group, suggesting that ClbH incorporates l-Met or an l-Met-derived metabolite [107]. Using a combination of in vitro and in vivo experiments [121], the A2 domain of ClbH was demonstrated to selectively activate the nonproteinogenic amino acid S-adenosyl-l-methionine (AdoMet) and incorporate this amino acid building block into the growing precolibactin chain (Figure 1B). Following A2-catalyzed thioesterification to generate AdoMet-S-CP, the C domain of ClbH catalyzes amide bond formation with the upstream intermediate. This reaction is followed by cyclopropylation, which is dependent upon the inclusion of the protein ClbI, a PKS that consists of sequential ketosynthase, acyltransferase, and acyl carrier protein domains. Utilizing site-directed mutagenesis, the ketosynthase domain of ClbI was deemed essential for cyclopropyl formation. Although the biochemical details of how ClbI catalyzes cyclopropyl formation remains unclear, the ability of ClbH to incorporate AdoMet expands upon the catalog of NRPS-incorporated α-l-amino acids and affords a new specificity code embedded within A2.
d-amino acid precursors.
In addition to novel l-amino acids exemplified with AdoMet, the A domain specificity code continues to be refined to include d-amino acids. As one interesting example, NRPS-mediated incorporation of d-Ala into the ansatrienins was recently delineated. The ansatrienins are members of the ansamycin-type antibiotics that are isolated from several different strains of Streptomyces [100]. As a family, the ansamycins are structurally characterized by an aromatic component bridged by a PKS-derived macrocyclic lactam, which together has been termed the ansa ring (Figure 2). The ansatrienins contain a cyclohexanoyl-d-Ala side chain that is attached to the ansa ring, thereby differentiating themselves from other ansamycins [101]. Initial studies probing the biosynthetic origin of the cyclohexanoyl-d-Ala side chain revealed that the d-Ala moiety likely originates from free d-Ala as opposed to l-Ala [115]. Bioinformatic analysis of the ansatrienin gene cluster from Streptomyces sp. XZQH13 revealed a single NRPS protein named AstC that was predicted to be involved in the direct incorporation of this d-Ala [93]. Typically, d-amino acid-containing metabolites derived from NRPS are first loaded to the CP domain as the l-isomer and epimerized by a specialized epimerization or duel condensation-epimerization domain that is embedded in the NRPS. However, AstC does not contain such epimerization domains, but instead consists of a tri-domain structure of A, CP, and TE domains, which is an overall unusual domain architecture for NRPS. Similar to the A domain of ClbH that incorporated AdoMet during colibactin biosynthesis, the specificity of the A domain of AstC could not be predicted using available bioinformatic tools. Thus it was unclear from bioinformatics if and how AstC orchestrates d-Ala incorporation.
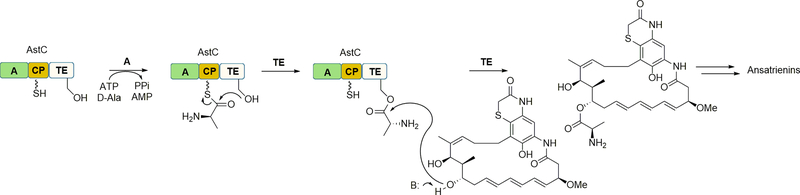
Figure 2.
The role of the NRPS AstC in d-alanylation incorporation during the biosynthesis of ansatrienins. The A domain of AstC selectively activates and loads d-Ala to the CP domain. The TE domain then catalyzes an intramolecular acyl group transfer to a conserved Ser to form an acyl-enzyme intermediate. This reaction is followed by a TE-catalyzed intermolecular esterification to incorporate d-Ala into the ansatrienin scaffold.
To assign the function of AstC, the encoding gene was inactivated and the resulting ΔastC mutant strain was shown to produce several ansatrienin precursors lacking the cyclohexanoyl-d-Ala side chain [55,93], demonstrating an essential role for AstC in d-Ala incorporation. One of these new, ansatrienin precursors was then used as a substrate to test the activity of recombinant AstC, revealing that this NRPS catalyzed an ATP-dependent d-alanylation at the expected hydroxyl group. Furthermore, the formation of the ester bond was dependent upon the CP domain, as a point mutation of the Ser residue revealed the Ppant modification was essential. Additional biochemical characterization revealed that other d-amino acids including d-Ser, d-Thr, and d-Val and other small amino acids including β-alanine and glycine were incorporated, enabling the semisynthesis of several unnatural ansatrienins. In total the results were consistent with a mechanism wherein the A domain first activates and loads d-Ala to the CP domain. Subsequently, the TE domain catalyzes two sequential acyl transfer reactions: the first an intramolecular transfer of d-Ala to the conserved Ser residue of the TE domain, followed by an intermolecular esterification to the ansa ring, the latter not unlike intramolecular esterification catalyzed by several NRPS TE domains.
In addition to the expansion of the A domain specificity to include d-Ala, a couple of interesting discoveries were made upon the characterization of AtsC. Prior to these studies, the handful of known NRPSs consisting of the A-CP-TE tridomain architecture were found to catalyze homodimerizations of aromatic α-keto acids via formation of carbon-carbon bonds [3,109,125]. For example, the tridomain NRPS EhcA involved in echoside biosynthesis was shown to convert two molecules of phenylpyruvic acid to polyporic acid via carbon-carbon bond-forming events [125]. In the case of AstC, the A-CP-TE tridomain architecture is utilized instead to catalyze a distinct chemistry involving C-O bond formation. Secondly, although not very common to date, ester bond-forming NRPSs that utilize a thioesterified acceptor and free hydroxyl-containing donor were previously ascribed to C domains [58,119]. AstC, however, utilizes a TE domain to catalyze the comparable intermolecular esterification.
Amino-group carrier proteins for assembling amino acid precursors.
While a single-step epimerization is needed to convert l-Ala to the nonproteinogenic isomer used by AstC, many NRPS-incorporated nonproteinogenic amino acids require multiple enzymatic steps to yield highly unusual amino acids. One such example is found in the biosynthesis of vazabitide A (Figure 3A), which is a dipeptide that was discovered by searching for so-called amino group carrier proteins (AmCP) that are homologous to the protein LysW utilized in the biosynthesis of l-Lys and l-Arg in certain thermophilic microorganisms [41]. During the biosynthesis of l-Lys and l-Arg in these thermophiles, the γ-carboxylate of a C-terminal Glu residue of LysW is covalently modified with l-2-aminoadipic acid or l-Glu, reactions that are catalyzed by the ATP-dependent ligases LysX or ArgX, respectively. Several complementary enzymatic steps ensue, culminating in the release of l-Lys or l-ornithine, respectively, the latter of which is the precursor of l-Arg. The genus Streptomyces and most bacteria, on the other hand, utilize LysW-independent pathways—the diaminopimelate pathway and N-acetyl glutamate pathway—for l-Lys and l-Arg biosynthesis, respectively. Nonetheless, the whole genome analysis of hundreds of Streptomyces strains has now revealed that several encode LysW-like proteins in addition to the proteins for the canonical l-Lys and l-Arg biosynthetic pathways. Intriguingly, Streptomyces sp. SANK 60404 encodes a LysW-like protein and other carrier protein-dependent l-Lys/l-Arg biosynthetic enzymes in a region clustered with two proteins with sequence similarity to NRPS. As a result, it was hypothesized that the LysW homologs, which were called amino-group carrier proteins (AmCP), were instead utilized to generate potentially new nonproteinogenic amino acids that served as a substrates for NRPS-mediated peptide biosynthesis.
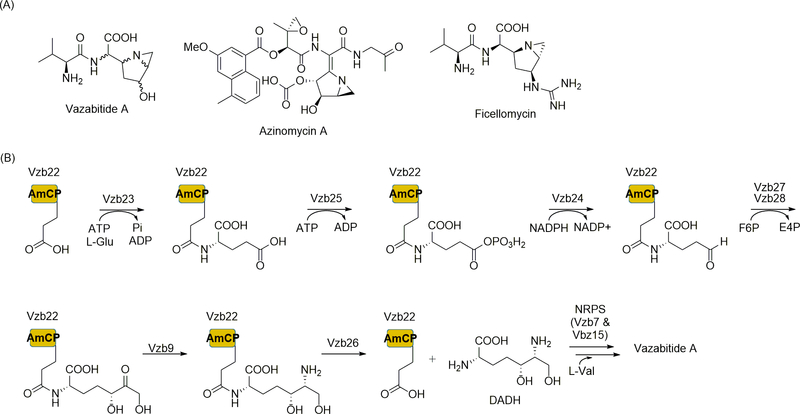
Figure 3.
NRPS-catalyzed incorporation of unusual nonproteinogenic amino acids that require multiple biosynthetic steps. (A) Structures of natural products containing a 1-azabicyclo[3.1.0]hexane moiety. (B) Biosynthesis of the probable NRPS substrate (2S,5R,6R)-2,6-diamino-5,7-dihydroxy-heptanoic acid (DADH). The biosynthetic pathway uses the LysW-like protein Vzb22 as an amino-group carrier protein (AmCP). F6P, fructose-6-phosphate; E4P, erythrose-4-phosphate.
To determine the amino acid product of this LysW-utilizing pathway, a combination of gene inactivation experiments and in vitro characterization of the recombinant proteins was employed to monitor the covalent modification of the LysW-like protein, Vzb22, by HR-MS [41]. The results revealed that the pathway is initiated by Vzb23, an ATP-dependent l-Glu ligase that generates an isopeptide bond at the C-terminus of Vbz22 (Figure 3B). Once covalently attached to the Vbz22, Vbz25 catalyzes phosphorylation of the δ-carboxylic acid followed by Vbz24-catalyzed reduction of the phosphate-carboxylate mixed anhydride to an aldehyde. Vbz27 and Vbz28 subsequently utilize this aldehyde as an aldol acceptor to catalyze the addition of a glycoaldehyde fragment from the breakdown of fructose-6-phosphate in a TPP-dependent transketolase reaction. Vbz9, which has sequence homology to the known l-Lys biosynthesis protein LysJ, catalyzes transamination followed by Vzb26-catalyzed hydrolysis to yield the unusual amino acid (2S,5R,6R)-2,6-diamino-5,7-dihydroxyheptanoic acid (DADH). The metabolite that incorporates DADH was ultimately identified as vazabitide A, a dipeptide consisting of l-Val and a 1-azabicyclo[3.1.0]hexane-containing amino acid, the latter of which is derived from DADH via uncharacterized steps. As expected, the formation of the final dipeptide was shown to require vzb7, which encodes one of the two NRPSs within the gene cluster. Vzb7 is predicted to consist of a C, A, CP, and TE domain, and the A domain specificity for Vzb7 could not be readily predicted by sequence, consistent with a role in activating the newly discovered amino acid DADH. The remaining NRPS Vzb15 was predicted to consist of a Val-specific A and CP domain, which is likewise consistent with the structure of vazabitide A. The ability of these two NRPS to generate the dipeptide between a proteinogenic and nonproteinogenic amino acid, the latter originating from the action of minimally eight specialized proteins, now awaits biochemical characterization. Nevertheless, defining DADH biosynthesis that is highlighted by AmCP-dependent steps has already led to the discovery of other novel natural products via genome mining [46].
To date, azinomycin and ficellomycin are the only other known natural products to contain a 1-azabicyclo[3.1.0]hexane moiety (Figure 3A). Preliminary characterization of the biosynthetic pathway leading to the azabicycle-containing amino acid of azinomycin suggested that the pathway begins with l-Glu, yet the three enzymes (AziC2, AziC3, and AziC4) that initiate this pathway appear to resemble the canonical bacterial pathway for l-ornithine biosynthesis, i.e., independent of an AmCP [41]. For ficellomycin, the azabicycle-containing amino acid was proposed to be biosynthesized with l-Glu attached to a peptidyl CP via the traditional thioester linkage [61]. Intriguingly, however, a LysW homolog Fic22 was annotated in the ficellomycin gene cluster, yet a role for the putative AmCP was not incorporated into the proposed biosynthetic pathway. Contrastingly, the initial annotation of the azinomycin gene cluster did not include a LysW homolog [123]; however, our re-analysis of the deposited sequence suggests a small open reading frame encoding a putative LysW-like protein is located between aziC7 and aziC4. Fic22 and this previously missed ORF, which we annotate as AziC12 to be consistent with the past annotation, not only have an overall high sequence identity (55%) to Vbz22 but also have the same 13 residues at the C-terminus including the terminal Glu residue that undergoes posttranslational modification, suggesting that the azabicycle for all three pathways employs a comparable AmCP-dependent biosynthetic strategy.
α-keto acids and ester synthase modules.
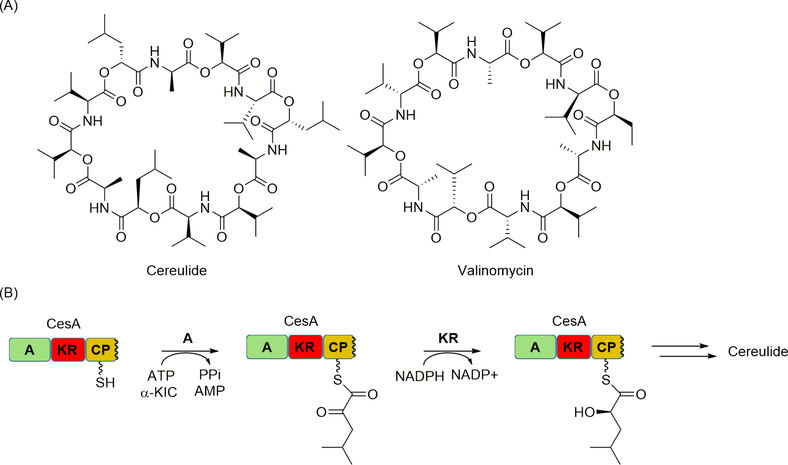
Precursors incorporated by NRPSs have expanded beyond amino acids. Cereulide and valinomycin are unusual peptides each composed of a macrocyclic core with alternating amide and ester bonds in their structures (Figure 4A) [67]. Using isotopic enrichment experiments with the cereulide producer, the α-hydroxy acids (l-hydroxyisovalerate and d-hydroxyisocapreate) were first demonstrated to originate from their amino acid precursors (l-Val and l-Leu, respectively). However, it was unclear how the α-hydroxy acids were incorporated in what appeared to be a natural product derived from an NRPS [53]. The biosynthetic gene clusters for both cereulide and valinomycin were subsequently cloned and sequenced, and bioinformatic analysis revealed two homologous NRPS (annotated as CesA and CesB for cereulide) for a total of four modules. Interestingly, the N-terminal of both CesA and CesB contained A domains whose substrate specificity could not be predicted through bioinformatic analysis. Additionally, the cognate downstream CP domain was preceded by a predicted dehydrogenase domain that is similar to the beta-ketoacyl reductase (KR) domains found in PKSs. The A-KR-CP of CesA and CesB were subsequently produced as tridomain-containing proteins and characterized in vitro, and both A domains were found to selectively activate and load α-keto acids over their corresponding α-hydroxy acids or α-amino acids (Figure 4B). Furthermore, the downstream KR domains were demonstrated to stereoselectively catalyze ketoreduction to yield the α-hydroxy thioester, thus affording the hydroxyl group for downstream esterification.
Figure 4.
NRPS-catalyzed mechanism of α-hydroxy acid incorporation. (A) Structures of α-hydroxy acid-containing NRPs cereulide and valinomycin. (B) Role of CesA in the incorporation of a α-hydroxy acid through A domain-catalyzed activation and loading of the corresponding α-keto acid and KR domain-catalyzed reduction. α-KIC, α-ketoisocaproic acid.
The characterization of the cereulide NRPS coincided with several noteworthy discoveries. First and foremost was the discovery that the A domains activate α-keto acids, thus establishing yet another specificity code for A domains. Secondly, the results uncovered a new NRPS domain, the KR domain, that contributes an additional functionality not previously realized with NRPSs. Interestingly, the insertion of the KR domain was proposed to occur between a flexible loop between A8 and A9 of the A domain as discussed in the next section with embedded methyltransferase (MT) domains, thus providing insight into how Nature has engineered NRPSs to incorporate functional domains. Finally, based on the structure of the final product, the C domain at the C-terminus of CesA is predicted to catalyze ester bond formation, thus adding to the growing list of esterifying C domains.
More than the Minimum
The prior examples highlight the diversity of carboxylate-containing substrates that can be incorporated based on the selectivity of the A domain. However, chemical modifications can also occur following the A domain selection and thioesterification to the CP. Often these modifications are a consequence of harboring auxiliary domains that are embedded within the NRPS minimal module. These additional domains—exemplified by the aforementioned KR domain—can be sandwiched between two domains within a minimal module or embedded directly within the A domain. Furthermore, they can be catalytic domains that modify the thioesterified intermediate or noncatalytic domains that serve as scaffolds to recruit catalytic proteins that function in trans. The following section provides additional examples of recently characterized auxiliary domains that expand upon the chemistry that can be achieved with NRPS.
F is for Formylation.
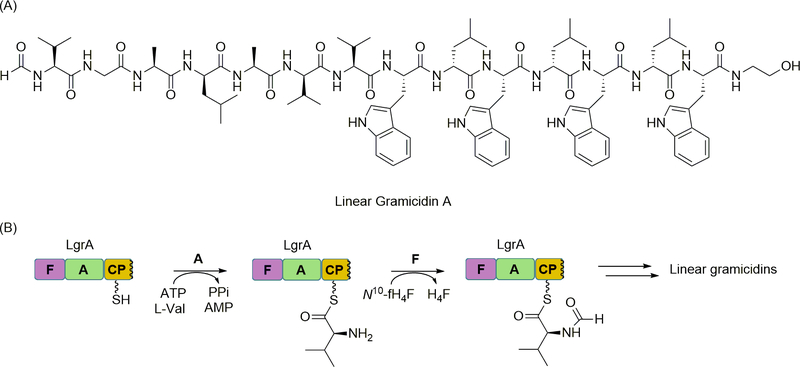
N-formylation of methionyl-tRNA is a critical step for initiating ribosomal protein biosynthesis in bacteria. Despite the stark contrast in the biosynthetic mechanism, the N-formyl group is also observed in the final product of some NRP including linear gramicidin (Figure 5A). As previously noted, a typical NRPS assembly line begins with an initiation module consisting of A and CP domains [112]. However, formylation (F) domains have been found at the beginning of some NRPS, creating an apparent F-A-CP initiation module [90]. These F domains appear to have originated through horizontal gene transfer from sugar formyltransferase genes despite functioning on markedly different substrates. Biochemical characterization of the F domain found in the NRPS for linear gramicidin biosynthesis has now revealed how this domain functions in the context of the NRPS assembly line.
Figure 5.
NRPS-catalyzed formylation. (A) Structure of the N-formyl-containing NRP, linear gramicidin A. (B) Role of LgrA in the incorporation of an N-formyl group through A domain-catalyzed activation and loading of l-Val and F domain-catalyzed formylation. N10-fH4F, N10-formyltetrahydrofolate; fH4F, formyltetrahydrofolate.
The N-terminus of linear gramicidin consists of either an N-formyl-l-Val or N-formyl-l-Ile [47]. The biochemical characterization of the A domain of the initiation module of the linear gramicidin NRPS (LgrA), which was produced in E. coli as a F-A-CP tridomain protein (Figure 5B), revealed a specificity toward small hydrophobic amino acids, notably with highest activity toward the expected amino acids of l-Val and l-Ile. Contrastingly, N-formyl-l-Val was not accepted as a substrate by the A domain, providing initial evidence that N-formylation occurs following thioesterification. Activity of the F domain was next directly examined using N10-formyltetrahydrofolate (N10-fH4F) as the formyl group donor. Following the loading of the CP domain with l-Val, LC-MS of the base-hydrolyzed product revealed complete conversion of l-Val to N-formyl-l-Val. In contrast no reaction was observed with free l-Val or synthetic l-Val-AMP as substrates, thus revealing that formylation only occurs when the amino acid is loaded onto the CP domain. To demonstrate that the CP-linked N-formyl-l-Val is processed by the downstream C domain, elongation assays were performed using a dimodular recombinant LgrA (F-A1-CP-C-A2-CP, where A1 and A2 activate l-Val and Gly, respectively). HPLC-MS analysis showed that the expected dipeptide N-formyl-l-Val-Gly was indeed produced. Dipeptide formation was not observed in reactions without the N10-fH4F cofactor. This result implies that the C domain linking A1 and A2 will only process N-formyl-l-Val as the acyl donor, which is consistent with the observation that only N-formylated gramicidins are produced from the strain [114].
At the time of the functional assignment of the F domain, very little was known about the structure other than its modest sequence similarity to previously discovered sugar formyltransferases. Since this time, several crystal structures of the first module of LgrA have been solved, each capturing distinct conformations representing the different catalytic steps involved in initiating the NRPS assembly line [84,83]. These structures illustrate that the F domain is connected to the rest of the initiation module through an interface with the A domain that is distinct from any other A domain interface such as the C-A domain interface found in elongation and termination modules [103]. The connection between the F-A domain is rigid, while the C-terminal portion of the A domain (often described as the Asub domain) and the A-CP connection appears quite dynamic and undergo significant conformational changes during catalysis. These rigid and mobile elements will likely be important criteria to consider for efforts aimed at engineering novel NRPS.
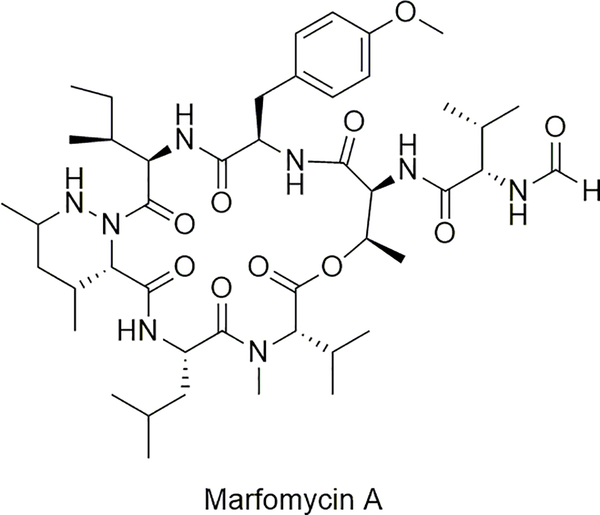
A potential alternative mechanism for introducing formyl groups into NRPS-derived natural products has recently been described [59]. A series of cycloheptadepsipeptides represented by marformycin A (Figure 6) was isolated from deep sea-derived Streptomyces drozdowiczii SCSIO 10141, and all contain either an N-formyl-l-allo-Ile or N-formyl-l-Val residue [124]. Bioinformatic analysis of the marformycin gene cluster revealed two notable differences when compared with the linear gramicidin gene cluster: the putative N-formyltransferase is a discrete protein (MfnA) as opposed to an NRPS domain, and the initiation module of the NRPS (MfnC) consists of an unusual C-A-CP domain architecture as opposed to the F-A-CP module of LgrA. Inactivation of mfnA eliminated production of all of the marformycins, and inactivation of the gene mfnK encoding the A domain of the fifth NRPS module yielded a strain producing a shunt peptide product that was still N-formylated [59]. Thus, similar to linear gramicidins, the formylation occurs early in the biosynthesis. However, whether N-formylation occurs prior to or following amino acid loading, and whether the C domain plays a role in this chemistry, remains to be determined.
Figure 6.
Structure of the NRPS-derived marformycin A that uses a distinct mechanism for N-formylation relative to the linear gramicidins.
M is for Methylation.
Shortly after linking sequence with function, A domains were described as containing ten core motifs (a1-a10) with highly conserved sequence [29,71,96,19]. Structural analysis revealed that nearly all of the ten motifs are positioned around the active site, with the a1-a7 motifs forming a larger N-terminal sub-domain and the a8-a10 motifs forming a small C-terminal sub-domain. Snapshots from multiple NRPS systems at various stages of the catalytic cycle have shown that the region between the two sub-domains functions like a hinge to enable large conformational changes to reorient the C-terminal sub-domain [38]. Interestingly, several auxiliary domains—instead of being found between two distinct domains—have now been discovered that are embedded within the A domain itself, primarily localized between the a8 and a9 motifs of the mobile C-terminal sub-domain but also appearing between the a2 and a3 motifs. These embedded domains, which are commonly domains with predicted methyltransferase (M) activity, have been the subject of several recent studies aimed at defining how they work in concert with their so-called interrupted A domains.
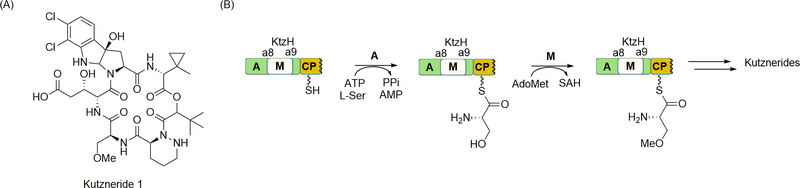
Kutznerides are cyclic hexadepsipeptides that consist entirely of nonproteinogenic amino acids, the majority of which require multiple enzymes for their biosynthesis (Figure 7A) [12,79]. One of the relatively simple residues in kutznerides is O-methyl-l-Ser. The A domain of the second module of the NRPS KtzH was predicted through bioinformatics analysis to activate and load l-Ser to the downstream CP [32], which was confirmed in vitro using the recombinant protein (Figure 7B) [126]. Importantly, O-methyl-l-Ser was not recognized, suggesting that l-Ser is modified following the attachment to the CP. Inspection of the A domain sequence uncovered a putative M domain embedded between the a8 and a9 motifs, and by using radiolabeled AdoMet as a the methyl donor, it was shown that the M domain was functional and able to methylate l-seryl-S-CP. Although the regiochemistry of methylation (in other words, O-methylation versus hypothetically possible N-methylation) was not confirmed at this time, the results were the first example of reconstituting the activity of an embedded M domain, revealing that methylation follows thioesterification.
Figure 7.
NRPS-catalyzed O-methylation. (A) Structure of a representative of the cyclic NRP kutznerides. (B) Role of KtzH in the incorporation of methylated amino acids through A domain-catalyzed activation and loading of l-Ser followed by M domain-catalyzed O-methylation. AdoMet, S-adenosyl-l-methionine; SAH, S-adenosyl-l-homocysteine.
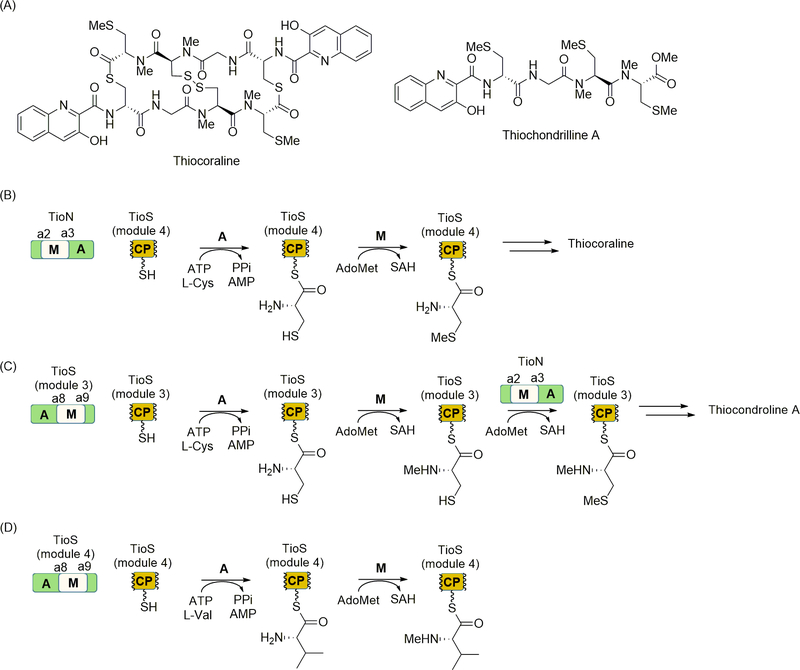
In addition to O-methylation, both N-methylation of the amide backbone and S-methylation of l-Cys residues are occasionally observed in NRPs. The DNA bisintercalator thiocoraline is an example of such an NRP containing both modifications [85]. The structure of thiocoraline consists of a symmetrical dimer of two pentapeptides, each with an N-methyl-l-Cys at the fourth residue of the peptide (inserted by module 3) and an N,S,-dimethyl-l-Cys as the fifth residue (inserted by module 4) (Figure 8A) [62]. Modules 3 and 4 make up a two module-containing NRPS, TioS. Similar to the A domain of KtzH, both of the A domains in TioS are embedded with M domains between the a8 and a9 motif, suggesting that one or both are involved in the incorporation of N-methyl-l-Cys [72]. A discrete and apparently extra A domain protein called TioN is also encoded within the gene cluster [1]. The specificity of the TioN A domain could not be bioinformatically predicted, and interestingly, this A domain contains an embedded M domain inserted between the a2 and a3 motif. The domain architecture of TioN, along with closer sequence analysis of the embedded M domain, suggested that this M domain is the missing S-methyltransferase. It was unclear, however, how the three A domains, each embedded with a unique MT domain, worked together to incorporate the two methylated cysteines.
Figure 8.
NRPS-catalyzed N- and S-methylation. (A) Structure of thiocoraline and thiochondrilline A. (B) Role of TioN in the incorporation of methylated amino acids through A domain-catalyzed activation and loading of l-Cys to the CP domain of TioS module 3 followed by M domain-catalyzed S-methylation. (C) Role of TioS module 3 for the incorporation of methylated amino acids through A domain-catalyzed activation and loading of l-Cys to the CP domain of TioS module 3, M domain-catalyzed N-methylation, and TioN M domain-catalyzed S-methylation to generate the dimethylated intermediate. (D) Activity of TioS module 4 including A domain-catalyzed activation and loading of l-Val to the CP domain of TioS module 4 followed by M domain-catalyzed N-methylation. The role of TioS module 4 in thiocoraline and thiochondrilline A biosynthesis remains unknown. AdoMet, S-adenosyl-l-methionine; SAH, S-adenosyl-l-homocysteine.
To establish the biosynthetic mechanism for NRPS-directed N- and S-methylation, the function of the TioN A domain was initially established (Figure 8B) [1]. Somewhat unexpectedly, the A domain of TioN was demonstrated to preferentially activate and load l-Cys to the module 4 CP domain, which was produced as a single domain-containing protein. Subsequently, TioN was shown to catalyze methylation of l-Cys-S-CP, and although it was believed to be S-methylation, the regiochemistry of methylation was not determined. Following the functional assignment of TioN, the role of TioS was investigated in closer detail [72]. The thiocoraline gene cluster also produces thiochondrilline A, which is a pentapeptide that is, in essence, half of the thiocoraline dimer with a few structural variations (Figure 8A) [116]. One of the differences is found in the fourth residue, wherein thiochondrilline A contains an N,S,-dimethyl-l-Cys in place of N-methyl-l-Cys in the thiocoraline monomeric unit (the latter is oxidized to a disulfide to connect the monomeric units in the final natural product). To establish how the N,S,-dimethyl-l-Cys is incorporated, the module 3 A-M didomain and the module 3 CP domain were independently produced for characterization of the recombinant proteins (Figure 8C). Unlike the A domain of TioN, the module 3 A domain was predicted to activate l-Cys based on the NRPS specificity code and, as expected, selectively loaded l-Cys to the CP. Furthermore, the M domain of TioS module 3 was active, catalyzing N-methylation. In a series of experiments that took advantage of the relaxed specificity of the TioS and TioN A domains, the TioS M domain was determined to first catalyze N-methylation followed by the TioN M domain-catalyzed S-methylation, thus generating the dimethylated product.
The results with TioN and module 3 of TioS raised several interesting questions including (1) why both M-embedded A domains of TioS and TioN are able to load l-Cys to the CP domain of module 3, (2) whether the functional redundancy occurs with the native, full-length NRPS in vivo, and (3) what is the function, if any, of the A domain of module 4 of TioS. In an attempt to answer the last question, biochemical and structural characterization of the A-M didomain of module 4 of TioS was very recently reported (Figure 8D) [73]. This A domain was predicted to activate l-Val based on the NRPS specificity code, although no such residue is found in thiocoraline or thiochondrilline A. Nonetheless, the results were consistent with this prediction, as the A domain preferentially recognized and loaded l-Val to the module 4 CP domain, which was produced as a single-domain protein in this study. The embedded M domain was likewise functional and catalyzed N-methylation of l-Val-S-CP. The biological relevance of l-Val activation, loading, and methylation, however, still remains unclear, although it was predicted to be a nonfunctional, evolutionary remnant of the thiocoraline/thiochondrilline A gene cluster.
The discovery and characterization of A domains with embedded M domains has exposed an unexpected architectural layout that Nature employs to introduce chemistry into NRPs, which could conceivably be exploited for NRPS engineering. Indeed, modifying A domains to remove or incorporate embedded M domains has already shown some promise. The M domain of KtzH, for example, can be excised from the A domain without compromising adenylation activity [65]. Perhaps more exciting is the result toward artificially embedding an unmodified A domain with one or more M domains to make unnatural A-M domain hybrids. In this instance, the A domain of module 1 of Ecm6, which preferentially activates and loads l-Ser during the biosynthesis of the DNA bisintercalator echinomycin (a structural relative of thiocoraline), was successfully engineered by inserting the M domain of either KtzH or TioS between the a8 and a9 motifs [65]. Both of these unnatural A-M domain hybrids retained their preferential adenylation of l-Ser with minimal effect on the catalytic efficiency. Furthermore, both were still able to load l-Ser to the cognate CP domain, and both were able to methylate l-Ser-S-CP. Although several questions remain—for example, whether downstream NRPS elements can process the artificially methylated amino acids—these proof-of-concept studies illustrate the potential for generating engineered A domains.
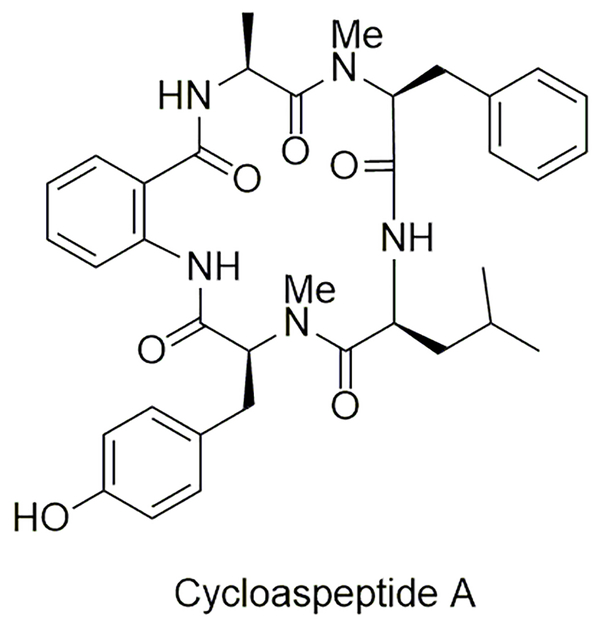
Fungal NRPSs are known to have some unique functional and mechanistic features relative to the bacterial counterparts. One of these differences appears to be the mechanism of incorporation of methylated amino acids into NRPs. The biosynthetic gene cluster for cycloaspeptides, N-methylated cyclic pentapeptides from Penicillium spp., was recently reported (Figure 9) [23]. As expected, a five module NRPS was uncovered, but instead of having embedded M domains within the two A domains involved in the incorporation of the two N-methylated amino acids, a completely separate gene (pscyA) encoding a putative methyltransferase was found adjacent to the NRPS-encoding gene. Inactivation of pscyA yielded a mutant strain unable to biosynthesize any cycloaspeptides. However, feeding of the appropriate N-methylated amino acids (either N-methyl-l-Phe or N-methyl-l-Tyr) restored the production in the ΔpscyA mutant strain, suggesting that PscyA catalyzes methylation of the free amino acids prior to NRPS-mediated incorporation. Although it remains a possibility that the respective A domains have a relaxed substrate specificity such that the methylated amino acids can substitute for the nonmethylated version when fed to the production media, the results minimally have revealed a mechanism that requires a methyltransferase to function in trans, thereby differentiating from the in cis architecture found in bacterial NRPS.
Figure 9.
Structure of a representative fungal-derived cycloaspeptide that is biosynthesized by an NRPS that employs a distinct mechanism for N-methylation relative to kutnzerides and thiocoraline.
X is for Oxidation.
Many peptides derived from NRPS incorporate substrates that undergo oxidation during biosynthesis. Similar to the group transfer reactions described above, this chemistry is often performed by a catalytic domain that is inserted between two domains of the minimal module of an NRPS. Perhaps the best studied of these oxidation domains is the so-called Ox domain involved in thiazole formation. In vitro characterization of the Ox domains involved in the biosynthesis bleomycin and epothilone, among others, has established their activity as flavin-dependent oxidase that function in cis on a CP-bound thiazoline [27,89]. Redox chemistry during peptide assembly, however, has now expanded beyond reactions catalyzed by NRPS-embedded domains: recent studies have established alternative redox strategies with catalysts functioning in trans, highlighted below with the discovery of the X domain.
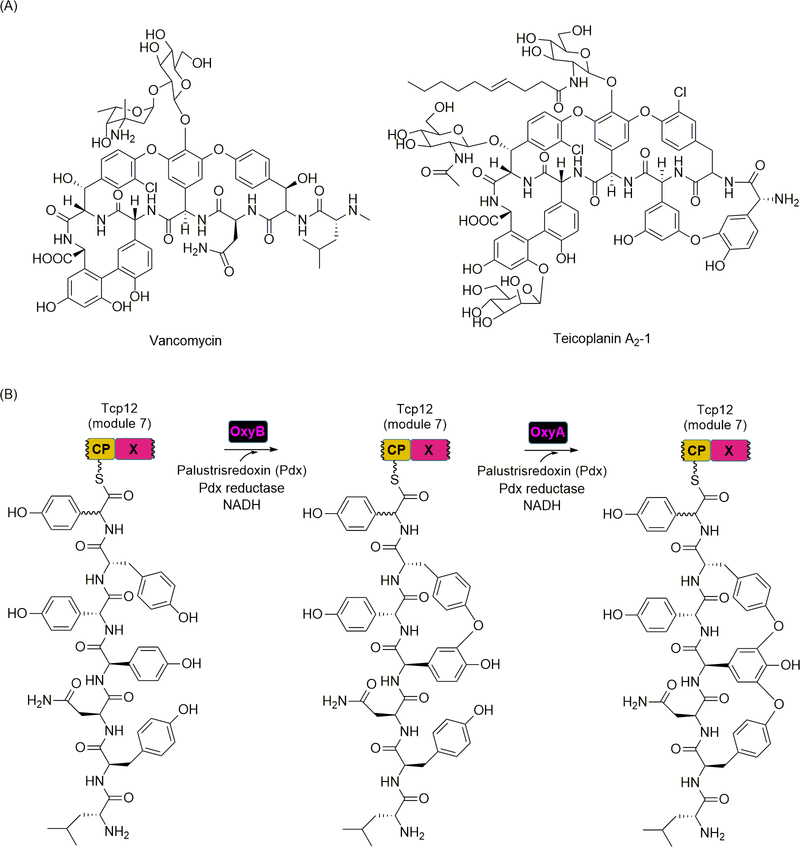
During the biosynthesis of glycopeptide antibiotics (GPAs), which includes the clinically used antibiotics vancomycin and teicoplanin, several phenolic and aryl bridges are made between nonproteinogenic aromatic amino acids to form a highly cross-linked, rigid structure (Figure 10A) [118]. Four P450 oxygenase-like proteins annotated as OxyA, B, C, and E are encoded in the gene cluster, and in vivo experiments initially indicated that the oxidation reactions catalyzed by these four enzymes occur while the peptide substrate is covalently bound to the NRPS assembly line [120,9,35]. Furthermore, gene inactivations also indicated a specific sequence of oxidations for creating these phenolic and aryl bridges, with OxyB catalyzing the first reaction [102,10,99,39]. However, the detailed catalytic strategy for inserting the phenolic and aryl bridges remained mostly unknown until an uncharacterized domain in the final NRPS module for GPA biosynthesis was interrogated [98]. Phylogenic analysis indicated that this uncharacterized domain was most closely related to C domains, but it was believed to be inactive as an amide bond-forming catalyst due to mutations in the highly conserved HHxxxDG motif that is essential for condensation activity [123]. This domain was named the X domain and hypothesized to be important for the oxidation process.
Figure 10.
NRPS recruitment of oxidative tailoring enzymes. (A) Structures of representative glycopeptide antibiotics. (B) Role of the auxiliary X domain of Tcp12 in phenolic bridging by binding of OxyB and OxyA for oxidation in trans. Palustrisredoxin (Pdx) is a [2Fe-2S]ferrodoxin from Rhodopseudomonas palustris that is able to act as an electron relay, accepting electrons from the NADH and flavin-dependent Pdx reductase and transferring them to the P450 enzymes OxyB and OxyA.
Using recombinant proteins from the teicoplanin biosynthetic pathway, the X domain, produced as a single-domain protein, was shown to interact with OxyA, OxyB, and OxyC by using native polyacrylamide gel electrophoresis and gel filtration [42]. Only OxyE, which catalyzes the phenolic coupling of residues 1 and 3 at the N-terminus of the final heptapeptide, did not display any binding interactions. Although there was variability in the affinity, similar results were obtained with the Oxy enzymes from other GPA biosynthetic pathways, suggesting this phenomenon is not unique to the teicoplanin NRPS. Crystal structures of the X domain alone and in complex with OxyBtei were solved, leading to the identification of residues that are important for protein-protein interactions, which primarily occur through hydrogen bonds and salt bridges. Mutation of select residues in the X domain proposed to be important for these interactions abolished the binding to OxyBtei, supporting the structural assignment. Finally, the catalytic activity of OxyBtei and homologs from other GPA biosynthetic pathways was reconstituted with a simplified heptapeptide precursor that was thioesterified to the CP, produced as a CP-X didomain protein, of the last NRPS module (Figure 10B). The results revealed the X domain was essential or significantly enhanced oxidative phenolic coupling. Additionally, the aforementioned site-directed mutants of the X domain were unable to support OxyB-catalyzed oxidation, further establishing the importance of the X domain. Interestingly, OxyB activity was not observed when substituting the substrate with a hexapeptide attached to the CP domain of the penultimate NRPS module (also produced as a single-domain protein), suggesting that—in addition to recognizing and binding the X domain—OxyB only recognizes a heptapeptide-S-CP. Finally, OxyA was included in the reaction with OxyB, and a product with the two expected phenolic bridges was detected (Figure 10B). These and other results led to the conclusion that the X domain functions to recruit and bind three of the four aryl-aryl or phenolic coupling enzymes, thus establishing a conceptually new strategy for the mediation of redox chemistry during NRP biosynthesis.
Product Release
The final aminoacyl-S-CP is typically released from the NRPS in a reaction catalyzed by a TE domain, which is generally located adjacent to and downstream of the terminal CP domain. TE domains have structural similarity to proteins of the α/β hydrolase superfamily, which contain a catalytic triad consisting of an essential Ser residue that forms an acyl-enzyme intermediate with the substrate during catalysis [31,103,16]. Biochemical studies with TE domains are consistent with this well-established catalytic strategy, wherein the peptide is first transferred from the aminoacyl-S-CP to the hydroxyl group of Ser to form a covalent acyl-enzyme intermediate. This intermediate is subsequently hydrolyzed or undergoes intramolecular lactamization or lactonization [17,36,37,45,94]. Alternatively, some NRPSs can have reductase (R) domains in place of the terminal TE. In contrast to TEs, the R domain has sequence similarity to short-chain dehydrogenases, and biochemical studies have shown that R domains catalyze an NADPH-dependent, 2- or 4-electron reduction of the aminoacyl-S-CP to release the peptide product as an aldehyde or alcohol, respectively. Examples of this mechanism are found during the biosynthesis of linear gramicidin and lyngbatoxin [82,91], among others, and have been nicely reviewed elsewhere [28]. Since the original discovery and characterization of both TE and R domains, new or interesting variations of the established off-loading mechanisms have been described, examples of which are now provided.
TE-catalyzed Dieckmann condensation.
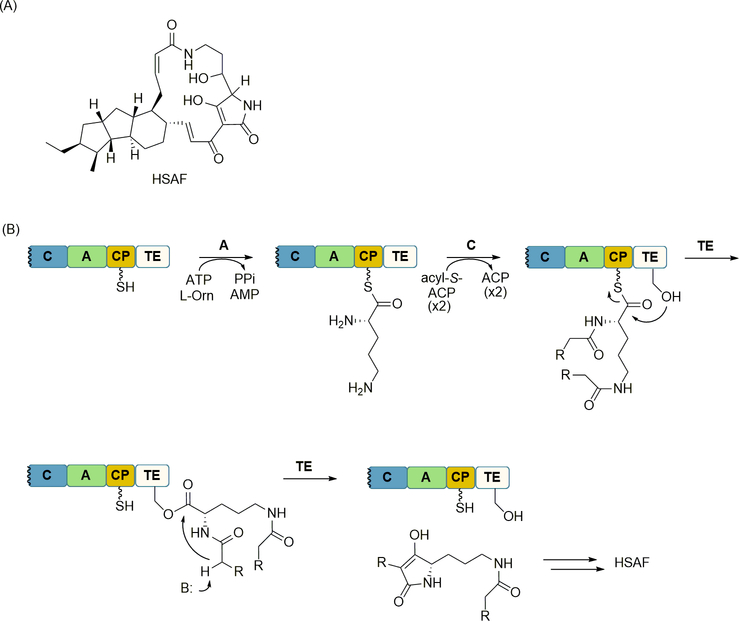
Tetramic acids are found in several natural products, including polyketides and fungal NRPs such as equisetin, cyclopropiazonic acid, and pretenillin, among others. Based on sequence analysis, the NRPS required for the biosynthesis of these fungal natural products is terminated by what initially appeared to be an R domain. However, this R domain, which was designated as an R* domain, was shown to catalyze product release by a non-reductive Dieckmann condensation of the aminoacyl-S-CP, yielding a peptide product containing a tetramic acid [95]. Heat-Stable Antifungal Factor (HSAF) is a tetramic acid-containing natural product that, in contrast to the aforementioned natural products, is isolated from a bacterial strain (Figure 11A). The biosynthetic gene cluster of HSAF was found to include a gene encoding a single PKS/NRPS hybrid, with the PKS harboring one module for polyketide assembly and the NRPS harboring a minimal C-A-CP module [30,64]. Instead of ending with the predicted R* domain, however, the C-terminus of the NRPS contained a TE domain with sequence similarity to the traditional TE of the α/β hydrolase superfamily (Figure 11B). Using a combination of in vivo and in vitro experiments, the PKS/NRPS protein was shown to be sufficient for generating a tetramic acid-containing product, and the A domain of the NRPS module was shown to selectively activate and load l-ornithine to the CP domain [63]. These and other results were consistent with a mechanism wherein, after l-ornithine-S-CP formation, the C domain catalyzes amide bond formation to both the α- and δ-amines using a CP-linked polyketide as the acyl donor, and the TE catalyzes release of the tetramic acid-containing product via a Dieckmann-type condensation. In total, the results support a second mechanism of Dieckmann-type condensation for NRPS off-loading. Also noteworthy is the discovery of a TE that catalyzes C-C bond formation, a process that typically yields a new C-O bond (lactonization or hydrolysis) or C-N bond (lactamization).
Figure 11.
NRPS-catalyzed Dieckmann-type condensation for product release and tetramic acid formation. (A) Structure of the bacterial natural product Heat Stable Activating Factor (HSAF). (B) Role of the lone NRPS in tetramic acid formation including A domain-catalyzed activation and loading of l-Orn, C domain-catalyzed amide bond formation, and TE domain-catalyzed Dieckmann-type condensation to release the product from the NRPS.
TE-catalyzed β-lactone/lactam formation.
As previously mentioned, TE-catalyzed intramolecular cyclization is a common mechanism for peptide release from an NRPS. In this case the formation of an acyl-enzyme intermediate is followed by intramolecular nucleophile attack of a side-chain hydroxyl or amine group to generate lactones and lactams, respectively. The side-chain nucleophile is typically several amino acids removed from the acyl functionality, leading to multi-residue macrocycles exemplified by the 10-residue macrocycles found in tyrocidine A and daptomycin and by the 6-residue macrocycles of marfomycin A (Figure 6) and kutzneride 1 (Figure 7A). Contrastingly, some NRPS-derived natural products contain C-terminal β-lactones or β-lactams, and the biochemical details for their respective biosynthesis have recently been deciphered.
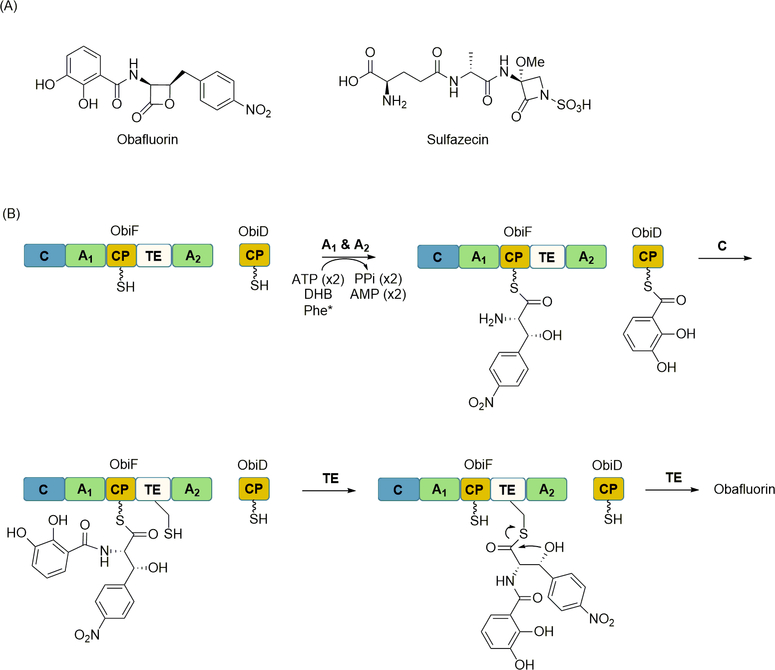
The β-lactone-containing obafluorin contains a single amide bond linking a 2,3-dihydroxybenzoic acid and a lactone-modified β-hydroxy-p-nitro-homophenylalanine (Figure 12A). Identification of the biosynthetic gene cluster revealed it encodes two proteins with similarity to NRPSs: ObiD, which consists of a single CP domain, and ObiF, which consists of sequential C-A1-CP-TE-A2 domains [88]. As expected from inspection of the obafluorin structure, the A domains were shown to selectively activate 2,3-dihydroxybenzoic acid and β-hydroxy-p-nitro-homophenylalanine. Reconstitution of the system in vitro led to the synthesis of the unstable β-lactone product along with the hydrolyzed variant, suggesting that the TE releases the product via β-lactonization. Interestingly, in contrast to the typical TE catalytic triad, the ObiF TE contains a Cys in place of the typical Ser. By “reverse” engineering to a Ser-containing TE, ObiF was shown to produce only the free acid. This and additional biochemical data supported the role of the TE as the β-lactone forming enzyme, first catalyzing transacylation of the dipeptide precursor from the CP domain to the Cys of the TE domain, then catalyzing intramolecular attack to generate a strained β-lactone ring (Figure 12B). Shortly after the discovery of TE-catalyzed β-lactone formation, the mechanistic details regarding the biosynthesis of a comparable β-lactam ring of the tripeptide sulfazecin were reported (Figure 12A) [76]. The same Ser-to-Cys substitution was also found within the TE domain of the terminal module of the NRPS SulM. In vitro characterization of the SulM TE revealed it indeed catalyzes β-lactam formation. Similar to the mutational analysis of the ObiF TE, “reverse” engineering of the SulM TE abolished β-lactamization activity and led to the sole production of the hydrolyzed tripeptide. In total these results suggest that a Ser-to-Cys substitution within the catalytic triad of the otherwise canonical TE could potentially serve as a sequence fingerprint for identifying or engineering new β-lactamization/β-lactonization catalysts.
Figure 12.
NRPS-catalyzed formation of β-lactones and β-lactams. (A) Structure of representative β-lactone and β-lactam-containing NRPs (B) Role of ObiF and ObiD in lactone synthesis including A domain-catalyzed activation and loading of DHB and Phe*, C domain-catalyzed amide bond formation to generate the pseudo dipeptide, and TE domain-catalyzed lactonization to release the product from the NRPS. A similar strategy is utilized to generate the β-lactam in sulfacezin. DHB, 2,3-dihydroxybenzoic acid; Phe*, β-hydroxy-p-nitro-homophenylalanine.
C domain-catalyzed product release.
Several cyclic NRPs from fungi are generated by an NRPS that terminates with a C domain in place of the TE domain that would typically be found in a bacterial NRPS. Biochemical studies of these NRPS systems have demonstrated that the C domains, which have been termed CT domains, catalyze intramolecular cyclization of the final aminoacyl-S-CP [40,20,77,34]. Structural investigation of the NRPSs involved in the biosynthesis of the indole alkaloid fumiquinazoline F have revealed that CT domains have an overall fold that is similar to the canonical C domains of bacterial NRPSs with a few key differences that help explain the unique intramolecular cyclization reaction [122]. For example, while a typical C domain contains a binding pocket for the acylated acceptor CP domain, access to this pocket appears to be blocked within the CT structure. Additionally, the extensive contacts within certain peripheral helices and loops greatly limit the access of water or other competing nucleophiles. These and other noted structural features of the CT domain suggest that, unlike bacterial TE domains, CT catalysis does not rely on the formation of an acyl-enzyme intermediate, thus establishing another mechanism for macrocyclization for NRPS product release.
Nonenzymatic Product Release.
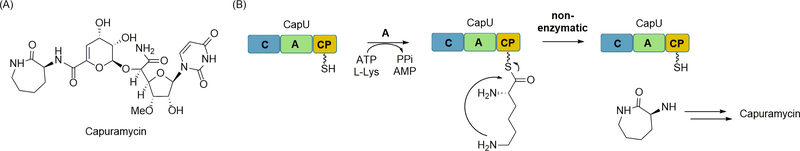
Some peptides appear to be generated by NRPSs lacking an obvious catalytic domain for product release. One example is found during the biosynthesis of capuramycin, which is structurally highlighted by a disaccharide core bonded to an l-α-amino-ε-caprolactam that originates from l-Lys (Figure 13A) [74]. The biosynthetic gene cluster encodes two NRPSs: CapU containing a minimal module (a C, A, and CP domain) and CapV, a single C domain-containing protein [33]. Since no TE was present, it was originally hypothesized that the C domain of CapV catalyzed intramolecular lactamization, not unlike the previously noted CT domain of fungal NRPS. As predicted from the bioinformatics analysis, biochemical characterization of the A domain of CapU revealed a preference for activation and loading l-Lys to the CP domain (Figure 13B). Unexpectedly, however, neither C domain was required nor accelerated lactamization [60]. Using synthetic l-Lys thioesters as controls, the lactamization was demonstrated to occur spontaneously in aqueous conditions at rates comparable to reactions with CapU. Therefore, it appears that both C domains are non-functional leading to nonenzymatic product release from the NRPS. A comparable phenomenon has been suggested for the biosynthesis of gliotoxin, a dipeptide-derived diketopiperazine from Aspergillus fumigatus [4]. The results also raise an intriguing question into how other NRPS involved in the biosynthesis of NRPs containing noncyclized l-Lys (or other amino acids with nucleophilic side-chains such at the l-ornithine activated by the A domain of the HSAF NRPS) avoid lactamization during the peptide chain extension process.
Figure 13.
Nonenzymatic product release from NRPS. (A) Structure of the aminocaprolactam-containing natural product capuramycin. (B) Role of CapU in aminocaprolactam biosynthesis including A domain-catalyzed activation and loading of l-Lys followed by nonenzymatic lactamization to release the product from the NRPS.
Summary and Outlook
We have highlighted several discoveries that have expanded upon the already remarkable function and mechanism of NRPSs. These discoveries include (1) the characterization of A domains with novel substrate specificity, thereby expanding upon the known and already diverse pool of substrates that are incorporated by NRPS; (2) the functional assignment of new domains that are considered an extension of the minimal NRPS module; and (3) new mechanisms for releasing products from an NRPS. This information, along with the knowledge garnered from the pioneering efforts of Christopher T. Walsh and others, continue to enhance the accuracy in the predictive power for connecting genomes to natural products. Continued efforts toward defining the function and mechanism of novel NRPS domains will undeniably improve upon and accelerate the discovery and development of natural products guided solely by the genetic code, a guiding principle for many research groups in the Genomic Era.
Acknowledgements
We appreciate the topical and intellectual discussions with Dr. Sylvie Garneau-Tsodikova and Taylor Lundy (Department of Pharmaceutical Sciences, University of Kentucky). Natural product-inspired research in the Van Lanen laboratory is supported in part by National Institutes of Health Grants AI128862, AI087849, and CA217255.
References
- 1.Al-Mestarihi AH, Villamizar G, Fernandez J, Zolova OE, Lombo F, Garneau-Tsodikova S (2014) Adenylation and S-methylation of cysteine by the bifunctional enzyme TioN in thiocoraline biosynthesis. J Am Chem Soc 136:17350–17354 [DOI] [PubMed] [Google Scholar]
- 2.Bachmann BO, Van Lanen SG, Baltz RH (2014) Microbial genome mining for accelerated natural products discovery: is a renaissance in the making? J Ind Microbiol Biotechnol 41:175–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balibar CJ, Howard-Jones AR, Walsh CT (2007) Terrequinone A biosynthesis through L-tryptophan oxidation, dimerization and bisprenylation. Nat Chem Biol 3:584–592 [DOI] [PubMed] [Google Scholar]
- 4.Balibar CJ, Walsh CT (2006) GliP, a multimodular nonribosomal peptide synthetase in Aspergillus fumigatus, makes the diketopiperazine scaffold of gliotoxin. Biochemistry 45:15029–15038 [DOI] [PubMed] [Google Scholar]
- 5.Belshaw PJ, Walsh CT, Stachelhaus T (1999) Aminoacyl-CoAs as probes of condensation domain selectivity in nonribosomal peptide synthesis. Science 284:486–489 [DOI] [PubMed] [Google Scholar]
- 6.Bentley SD, Chater KF, Cerdeno-Tarraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, Bateman A, Brown S, Chandra G, Chen CW, Collins M, Cronin A, Fraser A, Goble A, Hidalgo J, Hornsby T, Howarth S, Huang CH, Kieser T, Larke L, Murphy L, Oliver K, O’Neil S, Rabbinowitsch E, Rajandream MA, Rutherford K, Rutter S, Seeger K, Saunders D, Sharp S, Squares R, Squares S, Taylor K, Warren T, Wietzorrek A, Woodward J, Barrell BG, Parkhill J, Hopwood DA (2002) Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141–147 [DOI] [PubMed] [Google Scholar]
- 7.Bian X, Fu J, Plaza A, Herrmann J, Pistorius D, Stewart AF, Zhang Y, Muller R (2013) In vivo evidence for a prodrug activation mechanism during colibactin maturation. Chembiochem 14:1194–1197 [DOI] [PubMed] [Google Scholar]
- 8.Bian X, Plaza A, Zhang Y, Muller R (2015) Two more pieces of the colibactin genotoxin puzzle from Escherichia coli show incorporation of an unusual 1-aminocyclopropanecarboxylic acid moiety. Chem Sci 6:3154–3160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bischoff D, Bister B, Bertazzo M, Pfeifer V, Stegmann E, Nicholson GJ, Keller S, Pelzer S, Wohlleben W, Sussmuth RD (2005) The biosynthesis of vancomycin-type glycopeptide antibiotics--a model for oxidative side-chain cross-linking by oxygenases coupled to the action of peptide synthetases. Chembiochem 6:267–272 [DOI] [PubMed] [Google Scholar]
- 10.Bischoff D, Pelzer S, Holtzel A, Nicholson GJ, Stockert S, Wohlleben W, Jung G, Sussmuth RD (2001) The biosynthesis of vancomycin-type glycopeptide antibiotics-new insights into the cyclization steps. Angew Chem Int Ed Engl 40:1693–1696 [DOI] [PubMed] [Google Scholar]
- 11.Bossuet-Greif N, Vignard J, Taieb F, Mirey G, Dubois D, Petit C, Oswald E, Nougayrede JP (2018) The colibactin genotoxin generates DNA interstrand cross-links in infected cells. MBio 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Broberg A, Menkis A, Vasiliauskas R (2006) Kutznerides 1–4, depsipeptides from the actinomycete Kutzneria sp. 744 inhabiting mycorrhizal roots of Picea abies seedlings. J Nat Prod 69:97–102 [DOI] [PubMed] [Google Scholar]
- 13.Brotherton CA, Balskus EP (2013) A prodrug resistance mechanism is involved in colibactin biosynthesis and cytotoxicity. J Am Chem Soc 135:3359–3362 [DOI] [PubMed] [Google Scholar]
- 14.Brotherton CA, Wilson M, Byrd G, Balskus EP (2015) Isolation of a metabolite from the pks island provides insights into colibactin biosynthesis and activity. Org Lett 17:1545–1548 [DOI] [PubMed] [Google Scholar]
- 15.Condurso HL, Bruner SD (2012) Structure and noncanonical chemistry of nonribosomal peptide biosynthetic machinery. Nat Prod Rep 29:1099–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bruner SD, Weber T, Kohli RM, Schwarzer D, Marahiel MA, Walsh CT, Stubbs MT (2002) Structural basis for the cyclization of the lipopeptide antibiotic surfactin by the thioesterase domain SrfTE. Structure 10:301–310 [DOI] [PubMed] [Google Scholar]
- 17.Cao S, Yang Y, Ng NL, Guo Z (2005) Macrolactonization catalyzed by the terminal thioesterase domain of the nonribosomal peptide synthetase responsible for lichenysin biosynthesis. Bioorg Med Chem Lett 15:2595–2599 [DOI] [PubMed] [Google Scholar]
- 18.Challis GL (2008) Genome mining for novel natural product discovery. J Med Chem 51:2618–2628 [DOI] [PubMed] [Google Scholar]
- 19.Challis GL, Ravel J, Townsend CA (2000) Predictive, structure-based model of amino acid recognition by nonribosomal peptide synthetase adenylation domains. Chem Biol 7:211–224 [DOI] [PubMed] [Google Scholar]
- 20.Clevenger KD, Ye R, Bok JW, Thomas PM, Islam MN, Miley GP, Robey MT, Chen C, Yang K, Swyers M, Wu E, Gao P, Wu CC, Keller NP, Kelleher NL (2018) Interrogation of benzomalvin biosynthesis using fungal artificial chromosomes with metabolomic scoring (FAC-MS): Discovery of a benzodiazepine synthase activity. Biochemistry 57:3237–3243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clugston SL, Sieber SA, Marahiel MA, Walsh CT (2003) Chirality of peptide bond-forming condensation domains in nonribosomal peptide synthetases: the C5 domain of tyrocidine synthetase is a (D)C(L) catalyst. Biochemistry 42:12095–12104 [DOI] [PubMed] [Google Scholar]
- 22.Cougnoux A, Gibold L, Robin F, Dubois D, Pradel N, Darfeuille-Michaud A, Dalmasso G, Delmas J, Bonnet R (2012) Analysis of structure-function relationships in the colibactin-maturating enzyme ClbP. J Mol Biol 424:203–214 [DOI] [PubMed] [Google Scholar]
- 23.de Mattos-Shipley KMJ, Greco C, Heard DM, Hough G, Mulholland NP, Vincent JL, Micklefield J, Simpson TJ, Willis CL, Cox RJ, Bailey AM (2018) The cycloaspeptides: uncovering a new model for methylated nonribosomal peptide biosynthesis. Chem Sci 9:4109–4117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwarzer D, Finking R, Marahiel MA (2003) Nonribosomal peptides: From genes to products. Nat Prod Rep 20:275–287 [DOI] [PubMed] [Google Scholar]
- 25.Drake EJ, Miller BR, Shi C, Tarrasch JT, Sundlov JA, Allen CL, Skiniotis G, Aldrich CC, Gulick AM (2016) Structures of two distinct conformations of holo-non-ribosomal peptide synthetases. Nature 529:235–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Drake EJ, Nicolai DA, Gulick AM (2006) Structure of the EntB multidomain nonribosomal peptide synthetase and functional analysis of its interaction with the EntE adenylation domain. Chem Biol 13:409–419 [DOI] [PubMed] [Google Scholar]
- 27.Du L, Chen M, Sanchez C, Shen B (2000) An oxidation domain in the BlmIII non-ribosomal peptide synthetase probably catalyzing thiazole formation in the biosynthesis of the anti-tumor drug bleomycin in Streptomyces verticillus ATCC15003. FEMS Microbiol Lett 189:171–175 [DOI] [PubMed] [Google Scholar]
- 28.Du L, Lou L (2010) PKS and NRPS release mechanisms. Nat Prod Rep 27:255–278. doi: 10.1039/b912037h [DOI] [PubMed] [Google Scholar]
- 29.Conti E, Stachelhaus T, Marahiel MA, Brick P (1997) Structural basis for the activation of phenylalanine in the non-ribosomal biosynthesis of gramicidin S. EMBO J 16:4174–4183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu F, Zaleta-Rivera K, Zhu X., Huffman J, Millet JC, Harris SD, Yuen G, Li X-C, and Du L (2007) Structure and biosynthesis of Heat-Stable Antifungal Factor (HSAF), a broad-spectrum antimycotic with a novel mode of action. Antimicrob Agents and Chemother 51:64–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frueh DP, Arthanari H, Koglin A, Vosburg DA, Bennett AE, Walsh CT, Wagner G (2008) Dynamic thiolation-thioesterase structure of a non-ribosomal peptide synthetase. Nature 454:903–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fujimori DG, Hrvatin S, Neumann CS, Strieker M, Marahiel MA, Walsh CT (2007) Cloning and characterization of the biosynthetic gene cluster for kutznerides. Proc Natl Acad Sci U S A 104:16498–16503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Funabashi M, Yang Z, Nonaka K, Hosobuchi M, Fujita Y, Shibata T, Chi X, Van Lanen SG (2010) An ATP-independent strategy for amide bond formation in antibiotic biosynthesis. Nat Chem Biol 6:581–586 [DOI] [PubMed] [Google Scholar]
- 34.Gao X, Haynes SW, Ames BD, Wang P, Vien LP, Walsh CT, Tang Y (2012) Cyclization of fungal nonribosomal peptides by a terminal condensation-like domain. Nat Chem Biol 8:823–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Geib N, Woithe K, Zerbe K, Li DB, Robinson JA (2008) New insights into the first oxidative phenol coupling reaction during vancomycin biosynthesis. Bioorg Med Chem Lett 18:3081–3084 [DOI] [PubMed] [Google Scholar]
- 36.Grunewald J, Kopp F, Mahlert C, Linne U, Sieber SA, Marahiel MA (2005) Fluorescence resonance energy transfer as a probe of peptide cyclization catalyzed by nonribosomal thioesterase domains. Chem Biol 12:873–881 [DOI] [PubMed] [Google Scholar]
- 37.Grunewald J, Sieber SA, Marahiel MA (2004) Chemo- and regioselective peptide cyclization triggered by the N-terminal fatty acid chain length: the recombinant cyclase of the calcium-dependent antibiotic from Streptomyces coelicolor. Biochemistry 43:2915–2925 [DOI] [PubMed] [Google Scholar]
- 38.Gulick AM (2009) Conformational dynamics in the Acyl-CoA synthetases, adenylation domains of non-ribosomal peptide synthetases, and firefly luciferase. ACS Chem Biol 4:811–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hadatsch B, Butz D, Schmiederer T, Steudle J, Wohlleben W, Sussmuth R, Stegmann E (2007) The biosynthesis of teicoplanin-type glycopeptide antibiotics: assignment of P450 mono-oxygenases to side chain cyclizations of glycopeptide A47934. Chem Biol 14:1078–1089 [DOI] [PubMed] [Google Scholar]
- 40.Hai Y, Tang Y (2018) Biosynthesis of long-chain N-acyl amide by a truncated polyketide synthase-nonribosomal peptide synthetase hybrid megasynthase in fungi. J Am Chem Soc 140:1271–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hasebe F, Matsuda K, Shiraishi T, Futamura Y, Nakano T, Tomita T, Ishigami K, Taka H, Mineki R, Fujimura T, Osada H, Kuzuyama T, Nishiyama M (2016) Amino-group carrier-protein-mediated secondary metabolite biosynthesis in Streptomyces. Nat Chem Biol 12:967–972 [DOI] [PubMed] [Google Scholar]
- 42.Haslinger K, Peschke M, Brieke C, Maximowitsch E, Cryle MJ (2015) X-domain of peptide synthetases recruits oxygenases crucial for glycopeptide biosynthesis. Nature 521:105–109 [DOI] [PubMed] [Google Scholar]
- 43.Homburg S, Oswald E, Hacker J, Dobrindt U (2007) Expression analysis of the colibactin gene cluster coding for a novel polyketide in Escherichia coli. FEMS Microbiol Lett 275:255–262 [DOI] [PubMed] [Google Scholar]
- 44.Hur GH, Meier JL, Baskin J, Codelli JA, Bertozzi CR, Marahiel MA, Burkart MD (2009) Crosslinking studies of protein-protein interactions in nonribosomal peptide biosynthesis. Chem Biol 16:372–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Keating TA, Ehmann DE, Kohli RM, Marshall CG, Trauger JW, Walsh CT (2001) Chain termination steps in nonribosomal peptide synthetase assembly lines: directed acyl-S-enzyme breakdown in antibiotic and siderophore biosynthesis. Chembiochem 2:99–107 [DOI] [PubMed] [Google Scholar]
- 46.Matsuda K, Hasebe F, Shiwa Y, Kanesaki Y, Tomita T, Yoshikawa H, Shin-ya K, Kuzuyama T, Nishiyama M (2017) Genome mining of amino group carrier protein-mediated machinery: Discovery and biosynthetic characterization of a natural product with unique hydrazone unit. ACS Chem Bio 12:124–131 [DOI] [PubMed] [Google Scholar]
- 47.Kessler N, Schuhmann H, Morneweg S, Linne U, Marahiel MA (2004) The linear pentadecapeptide gramicidin is assembled by four multimodular nonribosomal peptide synthetases that comprise 16 modules with 56 catalytic domains. J Biol Chem 279:7413–7419 [DOI] [PubMed] [Google Scholar]
- 48.Kim WE, Patel A, Hur GH, Tufar P, Wuo MG, McCammon JA, Burkart MD (2018) Mechanistic probes for the epimerization domain of nonribosomal peptide synthetases. Chembiochem. doi: 10.1002/cbic.201800439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kleinkauf H, Gevers W (1969) Nonribosomal polypeptide synthesis: the biosynthesis of a cyclic peptide antibiotic, gramicidin S. Cold Spring Harb Symp Quant Biol 34:805–813 [DOI] [PubMed] [Google Scholar]
- 50.Kleinkauf H, von Dohren H (1990) Nonribosomal biosynthesis of peptide antibiotics. Eur J Biochem 192:1–15 [DOI] [PubMed] [Google Scholar]
- 51.Konno S, Ishikawa F, Suzuki T, Dohmae N, Burkart MD, Kakeya H (2015) Active site-directed proteomic probes for adenylation domains in nonribosomal peptide synthetases. Chem Commun (Camb) 51:2262–2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kudo F, Miyanaga A, Eguchi T (2019) Structural basis of the nonribosomal codes for nonproteinogenic amino acid selective adenylation enzymes in the biosynthesis of natural products. J Ind Microbiol Biotechnol, in press [DOI] [PubMed] [Google Scholar]
- 53.Kuse M, Franz T, Koga K, Suwan S, Isobe M, Agata N, Ohta M (2000) High incorporation of l-amino acids to cereulide, an emetic toxin from Bacillus cereus. Bioorg Med Chem Lett 10:735–739 [DOI] [PubMed] [Google Scholar]
- 54.Lambalot RH, Walsh CT (1995) Cloning, overproduction, and characterization of the Escherichia coli holo-acyl carrier protein synthase. J Biol Chem 270:24658–24661 [DOI] [PubMed] [Google Scholar]
- 55.Li X, Zhu J, Shi G, Sun M, Guo Z, Wang H, Lu C, Shen Y (2016) Deletion of the side chain assembly reveals diverse post-PKS modifications in the biosynthesis of ansatrienins. RSC Advances 6:88571–88579 [Google Scholar]
- 56.Li ZR, Li J, Gu JP, Lai JY, Duggan BM, Zhang WP, Li ZL, Li YX, Tong RB, Xu Y, Lin DH, Moore BS, Qian PY (2016) Divergent biosynthesis yields a cytotoxic aminomalonate-containing precolibactin. Nat Chem Biol 12:773–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li ZR, Li Y, Lai JY, Tang J, Wang B, Lu L, Zhu G, Wu X, Xu Y, Qian PY (2015) Critical intermediates reveal new biosynthetic events in the enigmatic colibactin pathway. Chembiochem 16:1715–1719 [DOI] [PubMed] [Google Scholar]
- 58.Lin S, Van Lanen SG, Shen B (2009) A free-standing condensation enzyme catalyzing ester bond formation in C-1027 biosynthesis. Proc Natl Acad Sci U S A 106:4183–4188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu J, Wang B, Li H, Xie Y, Li Q, Qin X, Zhang X, Ju J (2015) Biosynthesis of the anti-infective marformycins featuring pre-NRPS assembly line N-formylation and O-methylation and post-assembly line C-hydroxylation chemistries. Org Lett 17:1509–1512 [DOI] [PubMed] [Google Scholar]
- 60.Liu X, Jin Y, Cui Z, Nonaka K, Baba S, Funabashi M, Yang Z, Van Lanen SG (2016) The role of a nonribosomal peptide synthetase in l-lysine lactamization during capuramycin biosynthesis. Chembiochem 17:804–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu Y, Li M, Mu H, Song S, Zhang Y, Chen K, He X, Wang H, Dai Y, Lu F, Yan Z, Zhang H (2017) Identification and characterization of the ficellomycin biosynthesis gene cluster from Streptomyces ficellus. Appl Microbiol Biotechnol 101:7589–7602 [DOI] [PubMed] [Google Scholar]
- 62.Lombo F, Velasco A, Castro A, de la Calle F, Brana AF, Sanchez-Puelles JM, Mendez C, Salas JA (2006) Deciphering the biosynthesis pathway of the antitumor thiocoraline from a marine actinomycete and its expression in two Streptomyces species. Chembiochem 7:366–376 [DOI] [PubMed] [Google Scholar]
- 63.Lou L, Chen H, Cerny RL, Li Y, Shen Y, Du L (2012) Unusual activities of the thioesterase domain for the biosynthesis of the polycyclic tetramate macrolactam HSAF in Lysobacter enzymogenes C3. Biochemistry 51:4–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lou L, Qian G, Xie Y, Hang J, Chen H, Zaleta-Rivera K, Li Y, Shen Y, Dussault PH, Liu F, Du L (2011) Biosynthesis of HSAF, a tetramic acid-containing macrolactam from Lysobacter enzymogenes. J Am Chem Soc 133:643–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lundy TA, Mori S, Garneau-Tsodikova S (2018) Engineering bifunctional enzymes capable of adenylating and selectively methylating the side chain or core of amino acids. ACS Synth Biol 7:399–404 [DOI] [PubMed] [Google Scholar]
- 66.Luo L, Kohli RM, Onishi M, Linne U, Marahiel MA, Walsh CT (2002) Timing of epimerization and condensation reactions in nonribosomal peptide assembly lines: kinetic analysis of phenylalanine activating elongation modules of tyrocidine synthetase B. Biochemistry 41:9184–9196 [DOI] [PubMed] [Google Scholar]
- 67.Magarvey NA, Ehling-Schulz M, Walsh CT (2006) Characterization of the cereulide NRPS alpha-hydroxy acid specifying modules: activation of alpha-keto acids and chiral reduction on the assembly line. J Am Chem Soc 128:10698–10699 [DOI] [PubMed] [Google Scholar]
- 68.Marahiel MA (1992) Multidomain enzymes involved in peptide synthesis. FEBS Lett 307:40–43 [DOI] [PubMed] [Google Scholar]
- 69.Marahiel MA (1997) Protein templates for the biosynthesis of peptide antibiotics. Chem Biol 4:561–567 [DOI] [PubMed] [Google Scholar]
- 70.Marahiel MA, Krause M, Skarpeid HJ (1985) Cloning of the tyrocidine synthetase 1 gene from Bacillus brevis and its expression in Escherichia coli. Mol Gen Genet 201:231–236 [DOI] [PubMed] [Google Scholar]
- 71.Marahiel MA, Stachelhaus T, Mootz HD (1997) Modular peptide synthetases involved in nonribosomal peptide synthesis. Chem Rev 97:2651–2674 [DOI] [PubMed] [Google Scholar]
- 72.Mori S, Garzan A, Tsodikov OV, Garneau-Tsodikova S (2017) Deciphering nature’s intricate way of N,S-dimethylating l-cysteine: Sequential action of two bifunctional adenylation domains. Biochemistry 56:6087–6097 [DOI] [PubMed] [Google Scholar]
- 73.Mori S, Pang AH, Lundy TA, Garzan A, Tsodikov OV, Garneau-Tsodikova S (2018) Structural basis for backbone N-methylation by an interrupted adenylation domain. Nat Chem Biol 14:428–430 [DOI] [PubMed] [Google Scholar]
- 74.Muramatsu Y, Miyakoshi S, Ogawa Y, Ohnuki T, Ishii MM, Arai M, Takatsu T, Inukai M (2003) Studies on novel bacterial translocase I inhibitors, A-500359s. III. Deaminocaprolactam derivatives of capuramycin: A-500359 E, F, H; M-1 and M-2. J Antibiot (Tokyo) 56:259–267 [DOI] [PubMed] [Google Scholar]
- 75.Nougayrede JP, Homburg S, Taieb F, Boury M, Brzuszkiewicz E, Gottschalk G, Buchrieser C, Hacker J, Dobrindt U, Oswald E (2006) Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science 313:848–851 [DOI] [PubMed] [Google Scholar]
- 76.Oliver RA, Li R, Townsend CA (2018) Monobactam formation in sulfazecin by a nonribosomal peptide synthetase thioesterase. Nat Chem Biol 14:5–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Parkinson EI, Tryon JH, Goering AW, Ju KS, McClure RA, Kemball JD, Zhukovsky S, Labeda DP, Thomson RJ, Kelleher NL, Metcalf WW (2018) Discovery of the tyrobetaine natural products and their biosynthetic gene cluster via metabologenomics. ACS Chem Biol 13:1029–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Payne JA, Schoppet M, Hansen MH, Cryle MJ (2016) Diversity of nature’s assembly lines - recent discoveries in non-ribosomal peptide synthesis. Mol Biosyst 13:9–22 [DOI] [PubMed] [Google Scholar]
- 79.Pohanka A, Menkis A, Levenfors J, Broberg A (2006) Low-abundance kutznerides from Kutzneria sp. 744. J Nat Prod 69:1776–1781 [DOI] [PubMed] [Google Scholar]
- 80.Rausch C, Hoof I, Weber T, Wohlleben W, Huson DH (2007) Phylogenetic analysis of condensation domains in NRPS sheds light on their functional evolution. BMC Evol Biol 7:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rausch C, Weber T, Kohlbacher O, Wohlleben W, Huson DH (2005) Specificity prediction of adenylation domains in nonribosomal peptide synthetases (NRPS) using transductive support vector machines (TSVMs). Nucleic Acids Res 33:5799–5808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Read JA, Walsh CT (2007) The lyngbyatoxin biosynthetic assembly line: chain release by four-electron reduction of a dipeptidyl thioester to the corresponding alcohol. J Am Chem Soc 129:15762–15763 [DOI] [PubMed] [Google Scholar]
- 83.Reimer JM, Aloise MN, Harrison PM, Schmeing TM (2016) Synthetic cycle of the initiation module of a formylating nonribosomal peptide synthetase. Nature 529:239–242 [DOI] [PubMed] [Google Scholar]
- 84.Reimer JM, Harb I, Ovchinnikova OG, Jiang J, Whitfield C, Schmeing TM (2018) Structural insight into a novel formyltransferase and evolution to a nonribosomal peptide synthetase tailoring domain. ACS Chem Biol 13:3161–3172 [DOI] [PubMed] [Google Scholar]
- 85.Romero F, Espliego F, Perez Baz J, Garcia de Quesada T, Gravalos D, de la Calle F, Fernandez-Puentes JL (1997) Thiocoraline, a new depsipeptide with antitumor activity produced by a marine Micromonospora. I. Taxonomy, fermentation, isolation, and biological activities. J Antibiot (Tokyo) 50:734–737 [DOI] [PubMed] [Google Scholar]
- 86.Roongsawang N, Lim SP, Washio K, Takano K, Kanaya S, Morikawa M (2005) Phylogenetic analysis of condensation domains in the nonribosomal peptide synthetases. FEMS Microbiol Lett 252:143–151 [DOI] [PubMed] [Google Scholar]
- 87.Rottig M, Medema MH, Blin K, Weber T, Rausch C, Kohlbacher O (2011) NRPSpredictor2--a web server for predicting NRPS adenylation domain specificity. Nucleic Acids Res 39:W362–3673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Schaffer JE, Reck MR, Prasad NK, Wencewicz TA (2017) Beta-lactone formation during product release from a nonribosomal peptide synthetase. Nat Chem Biol 13:737–744 [DOI] [PubMed] [Google Scholar]
- 89.Schneider TL, Shen B, Walsh CT (2003) Oxidase domains in epothilone and bleomycin biosynthesis: thiazoline to thiazole oxidation during chain elongation. Biochemistry 42:9722–9730 [DOI] [PubMed] [Google Scholar]
- 90.Schoenafinger G, Schracke N, Linne U, Marahiel MA (2006) Formylation domain: an essential modifying enzyme for the nonribosomal biosynthesis of linear gramicidin. J Am Chem Soc 128:7406–7407 [DOI] [PubMed] [Google Scholar]
- 91.Schracke N, Linne U, Mahlert C, Marahiel MA (2005) Synthesis of linear gramicidin requires the cooperation of two independent reductases. Biochemistry 44:8507–8513 [DOI] [PubMed] [Google Scholar]
- 92.Shaw-Reid CA, Kelleher NL, Losey HC, Gehring AM, Berg C, Walsh CT (1999) Assembly line enzymology by multimodular nonribosomal peptide synthetases: the thioesterase domain of E. coli EntF catalyzes both elongation and cyclolactonization. Chem Biol 6:385–400 [DOI] [PubMed] [Google Scholar]
- 93.Shi G, Shi N, Li Y, Chen W, Deng J, Liu C, Zhu J, Wang H, Shen Y (2016) D-Alanylation in the assembly of ansatrienin side chain is catalyzed by a modular NRPS. ACS Chem Biol 11:876–881 [DOI] [PubMed] [Google Scholar]
- 94.Sieber SA, Tao J, Walsh CT, Marahiel MA (2004) Peptidyl thiophenols as substrates for nonribosomal peptide cyclases. Angew Chem Int Ed Engl 43:493–498 [DOI] [PubMed] [Google Scholar]
- 95.Sims JW, Schmidt EW (2008) Thioesterase-like role for fungal PKS-NRPS hybrid reductive domains. J Am Chem Soc 130:11149–11155 [DOI] [PubMed] [Google Scholar]
- 96.Stachelhaus T, Mootz HD, Marahiel MA (1999) The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem Biol 6:493–505 [DOI] [PubMed] [Google Scholar]
- 97.Stachelhaus T, Walsh CT (2000) Mutational analysis of the epimerization domain in the initiation module PheATE of gramicidin S synthetase. Biochemistry 39:5775–5787 [DOI] [PubMed] [Google Scholar]
- 98.Stegmann E, Frasch HJ, Wohlleben W (2010) Glycopeptide biosynthesis in the context of basic cellular functions. Curr Opin Microbiol 13:595–602 [DOI] [PubMed] [Google Scholar]
- 99.Stegmann E, Pelzer S, Bischoff D, Puk O, Stockert S, Butz D, Zerbe K, Robinson J, Sussmuth RD, Wohlleben W (2006) Genetic analysis of the balhimycin (vancomycin-type) oxygenase genes. J Biotechnol 124:640–653 [DOI] [PubMed] [Google Scholar]
- 100.Sugita M, Natori Y, Sasaki T, Furihata K, Shimazu A, Seto H, Otake N (1982) Studies on mycotrienin antibiotics, a novel class of ansamycins. I. Taxonomy, fermentation, isolation and properties of mycotrienins I and II. J Antibiot (Tokyo) 35:1460–1466 [DOI] [PubMed] [Google Scholar]
- 101.Sugita M, Sasaki T, Furihata K, Seto H, Otake N (1982) Studies on mycotrienin antibiotics, a novel class of ansamycins. II. Structure elucidation and biosynthesis of mycotrienins I and II. J Antibiot (Tokyo) 35:1467–1473 [DOI] [PubMed] [Google Scholar]
- 102.Süssmuth RD, Pelzer S, Nicholson G, Walk T, Wohlleben W, Jung G (1999) New advances in the biosynthesis of glycopeptide antibiotics of the vancomycin type from A mycolatopsis mediterranei. 38:1976–1979 [DOI] [PubMed] [Google Scholar]
- 103.Tanovic A, Samel SA, Essen LO, Marahiel MA (2008) Crystal structure of the termination module of a nonribosomal peptide synthetase. Science 321:659–663 [DOI] [PubMed] [Google Scholar]
- 104.Trauger JW, Kohli RM, Mootz HD, Marahiel MA, Walsh CT (2000) Peptide cyclization catalysed by the thioesterase domain of tyrocidine synthetase. Nature 407:215–218 [DOI] [PubMed] [Google Scholar]
- 105.Trautman EP, Healy AR, Shine EE, Herzon SB, Crawford JM (2017) Domain-targeted metabolomics delineates the heterocycle assembly steps of colibactin biosynthesis. J Am Chem Soc 139:4195–4201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Van Lanen SG (2017) Biosynthesis: SAM cycles up for colibactin. Nat Chem Biol 13:1059–1061 [DOI] [PubMed] [Google Scholar]
- 107.Vizcaino MI, Crawford JM (2015) The colibactin warhead crosslinks DNA. Nat Chem 7:411–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Vizcaino MI, Engel P, Trautman E, Crawford JM (2014) Comparative metabolomics and structural characterizations illuminate colibactin pathway-dependent small molecules. J Am Chem Soc 136:9244–9247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wackler B, Schneider P, Jacobs JM, Pauly J, Allen C, Nett M, Hoffmeister D (2011) Ralfuranone biosynthesis in Ralstonia solanacearum suggests functional divergence in the quinone synthetase family of enzymes. Chem Biol 18:354–360 [DOI] [PubMed] [Google Scholar]
- 110.Walsh CT (2017) Are highly morphed peptide frameworks lurking silently in microbial genomes valuable as next generation antibiotic scaffolds? Nat Prod Rep 34:687–693 [DOI] [PubMed] [Google Scholar]
- 111.Fischbach MA, Walsh CT (2006) Assembly-line enzymology for polyketide and nonribosomal peptide antibiotics: Logic, machinery, and mechanisms. Chem Rev 106:3468–3496 [DOI] [PubMed] [Google Scholar]
- 112.Weissman KJ (2015) The structural biology of biosynthetic megaenzymes. Nat Chem Biol 11:660–672 [DOI] [PubMed] [Google Scholar]
- 113.Weissman KJ, Muller R (2008) Crystal structure of a molecular assembly line. Angew Chem Int Ed Engl 47:8344–8346 [DOI] [PubMed] [Google Scholar]
- 114.Witkop R Sa B(1965) Gramicidin A. VI. The synthesis of valine-and isoleucine-gramicidin A. J Am Chem Soc 87:2020–2027 [DOI] [PubMed] [Google Scholar]
- 115.Wu TS, Duncan J, Tsao SW, Chang CJ, Keller PJ, Floss HG (1987) Biosynthesis of the ansamycin antibiotic ansatrienin (mycotrienin) by Streptomyces collinus. J Nat Prod 50:108–118 [DOI] [PubMed] [Google Scholar]
- 116.Wyche TP, Hou Y, Braun D, Cohen HC, Xiong MP, Bugni TS (2011) First natural analogs of the cytotoxic thiodepsipeptide thiocoraline A from a marine Verrucosispora sp. J Org Chem 76:6542–6547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Xue M, Shine E, Wang W, Crawford JM, Herzon SB (2018) Characterization of natural colibactin-nucleobase adducts by tandem mass spectrometry and isotopic labeling. Support for DNA alkylation by cyclopropane ring opening. Biochemistry 57:6391–6394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yim G, Thaker MN, Koteva K, Wright G (2014) Glycopeptide antibiotic biosynthesis. J Antibiot (Tokyo) 67:31–41 [DOI] [PubMed] [Google Scholar]
- 119.Zaleta-Rivera K, Xu C, Yu F, Butchko RA, Proctor RH, Hidalgo-Lara ME, Raza A, Dussault PH, Du L (2006) A bidomain nonribosomal peptide synthetase encoded by FUM14 catalyzes the formation of tricarballylic esters in the biosynthesis of fumonisins. Biochemistry 45:2561–2569 [DOI] [PubMed] [Google Scholar]
- 120.Zerbe K, Woithe K, Li DB, Vitali F, Bigler L, Robinson JA (2004) An oxidative phenol coupling reaction catalyzed by oxyB, a cytochrome P450 from the vancomycin-producing microorganism. Angew Chem Int Ed Engl 43:6709–6713 [DOI] [PubMed] [Google Scholar]
- 121.Zha L, Jiang Y, Henke MT, Wilson MR, Wang JX, Kelleher NL, Balskus EP (2017) Colibactin assembly line enzymes use S-adenosylmethionine to build a cyclopropane ring. Nat Chem Biol 13:1063–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhang J, Liu N, Cacho RA, Gong Z, Liu Z, Qin W, Tang C, Tang Y, Zhou J (2016) Structural basis of nonribosomal peptide macrocyclization in fungi. Nat Chem Biol 12:1001–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zhao Q, He Q, Ding W, Tang M, Kang Q, Yu Y, Deng W, Zhang Q, Fang J, Tang G, Liu W (2008) Characterization of the azinomycin B biosynthetic gene cluster revealing a different iterative type I polyketide synthase for naphthoate biosynthesis. Chem Biol 15:693–705 [DOI] [PubMed] [Google Scholar]
- 124.Zhou X, Huang H, Li J, Song Y, Jiang R, Liu J, Zhang S, Hua Y, Ju J (2014) New anti-infective cycloheptadepsipeptide congeners and absolute stereochemistry from the deep sea-derived Streptomyces drozdowiczii SCSIO 10141. Tetrahedron 70:7795–7801 [Google Scholar]
- 125.Zhu J, Chen W, Li YY, Deng JJ, Zhu DY, Duan J, Liu Y, Shi GY, Xie C, Wang HX, Shen YM (2014) Identification and catalytic characterization of a nonribosomal peptide synthetase-like (NRPS-like) enzyme involved in the biosynthesis of echosides from Streptomyces sp. LZ35. Gene 546:352–358 [DOI] [PubMed] [Google Scholar]
- 126.Zolova OE, Garneau-Tsodikova S (2014) KtzJ-dependent serine activation and O-methylation by KtzH for kutznerides biosynthesis. J Antibiot (Tokyo) 67:59–64 [DOI] [PubMed] [Google Scholar]