Abstract
Runx2 plays an essential role in embryonic disc tissue development in mice. However, the role of runt-related transcription factor 2 (Runx2) in postnatal disc tissue growth and development has not been defined. In the present studies, we generated Runx2 conditional knockout (KO) mice (Runx2Agc1ER), in which Runx2 was deleted in Aggrecan-expressing cells in disc tissue at postnatal 2-weeks of age. We then analyzed changes in disc tissue growth and development using histology and immunohistochemical methods in 3-month-old mice. We found that large vacuolated notochordal cells were accumulated in the nucleus pulposus (NP) in Runx2 KO mice. The growth plate cartilage tissue in the disc was thicker in Runx2 KO mice. We also found a significant upregulation of Indian hedgehog (Ihh) expression in the cells in NP cells and in annulus fibrosus cells of Runx2 KO mice. These results demonstrated that Runx2 may play an important role in postnatal disc tissue development through interacting with Ihh signaling.
Keywords: growth plate (GP) cartilage, Indian hedgehog (Ihh), intervertebral disc (IVD), notochordal cells, runt-related transcription factor 2 (Runx2)
1 |. INTRODUCTION
Intervertebral disc (IVD) is a specialized connective tissue which consists of three distinct interdependent tissues: the central viscous nucleus pulposus (NP), the outer fibrillary annulus fibrosus (AF) and the cartilage growth plate (GP) and endplate (EP), which anchor the disks to the adjacent vertebral body. The NP is composed of a proteoglycan and water gel held together loosely by an irregular network of type II collagen and elastin fibers. It contains two distinct cell types: large clusters of notochordal cells (M. McCann & Seguin, 2016) and smaller, more disperse chondrocyte-like cells (Roberts, 2000). There are large vacuoles inside the notochordal cells and these vacuoles increase the cell size of notochordal cells and occupy about 80% of the cell volume (Hunter, Bianchi, Cheng, & Muldrew, 2007). The presence of these large intracellular vacuoles makes notochordal cells directly contribute to the mechanical properties of the notochord (Hunter et al., 2007). Actually, these notochordal cell vacuoles are lysosome related organelles that function in axis and spine morphogenesis (Ellis, Bagwell, & Bagnat, 2013). In most vertebrates, including humans and mice, there is a progressive loss of large vacuolated notochord cells immediately after birth and the NP becomes populated by small chondrocyte-like NP cells (Hunter, Matyas, & Duncan, 2003; Roberts, 2000). Interestingly, the loss of large vacuolated notochordal cells is also associated with the onset of disc degeneration, suggesting that these cells are required for the maintenance of the NP (Aguiar, Johnson, & Oegema, 1999; Boos et al., 2002; Hunter et al., 2003).
Understanding signals which control the normal IVD growth and differentiation may provide potential therapies for the treatment of disc degenerative disease. The runt-related transcription factor 2 (Runx2) is an essential transcription factor regulating endochondral ossification through control of chondrocyte proliferation and differentiation (Chen et al., 2014; Komori 2008; Zaidi et al., 2002). Over the past two decades, major progress has been made in the understanding the critical role of Runx2 in regulating osteoblast differentiation. In addition, Runx2 has also been reported to regulate the chondrocyte differentiation and hypertrophy (Kamekura et al., 2006; Liao et al., 2017; Takarada et al., 2013). In mice, Runx2 expression has been detected in prehypertrophic and hypertrophic chondrocytes (Kamekura et al., 2006; Sato et al., 2008) and osteoblast lineage cells (Ducy, Zhang, Geoffroy, Ridall, & Karsenty, 1997). Although the role of Runx2 in long bone has been well documented, less is known about its role in disc tissue.
In 1997, Runx2 was found to be essential for embryonic skeletal development and vertebral bone formation in mice (Ducy et al., 1997). During embryonic development, Runx2 expression was detected to be restricted to cells of the ossification centers of vertebrae before they became mineralized (Ducy et al., 1997). Subsequently, another study demonstrated that, during mouse embryogenesis, Runx2, but not Runx1 or Runx3, was expressed in the IVD and Runx2 expression was significantly increased in the mouse model of IVD degeneration and in patients with IVD degeneration (Sato et al., 2008). These studies suggest that Runx2 is essential for the disc development during embryogenesis, and may play an important role in disc degeneration after birth. However, the effect of Runx2 on postnatal disc tissue development has not been fully defined.
Agc1-CreER transgenic mice are a mouse model which could be used to target cartilage cells at postnatal and adult stages (Henry et al., 2009). In the present studies, we generated Runx2Agc1ER conditional KO mice in which Runx2 is deleted in Aggrecan-expressing cells. We analyzed the disc phenotype of Runx2 knockout (KO) mice and found that the transition of notochordal cell to chondrocyte-like cells in the NP was inhibited in Runx2Agc1ER KO mice. Furthermore, Indian hedgehog (Ihh) expression was found to be significantly increased in NP and AF areas of Runx2 KO mice. Our findings demonstrate that Runx2 expression is required for postnatal disc tissue growth and development.
2 |. MATERIALS
2.1 |. Animals
Runx2Agc1ER mice were generated by breeding Runx2flox/flox mice (Liao et al., 2017; Takarada et al., 2013) with Agc1-CreER mice (Henry et al., 2009). Agc1-CreER transgenic mice were obtained from Jackson laboratories (Bar Harbor, ME). Runx2Agc1ER conditional KO mice were administered with tamoxifen (1 mg/10 g body weight, intraperitoneal injection [i.p.] injection, daily for 5 days) at age of 2-week-old. Mice were killed at age of 3-month-old for microcomputed tomography (μCT) and histologic analyses. Cre-negative littermates were used as controls, n = 5 per group. Animal protocol of this study has been approved by the IACUC of the Rush University Medical Center and all experimental methods and procedures were carried out in accordance with the approved guidelines.
2.2 |. Cre-recombination efficiency
To examine whether Agc1-CreER could efficiently target disc cells in postnatal mice, Agc1-CreER transgenic mice (Henry et al., 2009) were bred with ROSAmT/mG reporter mice to generate Agc1-CreER; ROSAmT/mG mice. Agc1-CreER; ROSAmT/mG mice express red fluorescence in all cell types before Cre-recombination and express green fluorescence following recombination after tamoxifen induction. Agc1-CreER; ROSAmT/mG mice are administrated tamoxifen at 2-week-old by i.p. injection (1 mg/10 g body weight, daily for five consecutive days) and killed 7 weeks after tamoxifen induction.
2.3 |. Histology analysis
We dissected lumbar spine from Runx2Agc1ER mice and Cre− control mice. Samples were fixed in 10% formalin, decalcified, and processed as previously described (B. Wang et al., 2014; Liao et al., 2017; M. Wang et al., 2012; Shen et al., 2013; T. Wang et al., 2018). And then, 3-μm mid-sagittal sections at three different levels (15-μm apart) were cut from the medial compartment of the L3-L5 intervertebral bodies. These sections were stained with Alcian blue/H&E Orange G. Three slides per mouse, six mice per group were analyzed in the experiment.
2.4 |. Microcomputed tomography
Before histologic processing, we evaluated formalin-fixed mouse spines by μCT-35 cone-beam scanner (Scanco Medical, Switzerland) with a 55 kVp source and a 145 μAmp current. We scanned the mouse spines at a resolution of 10.5 μm. The scanned images from each group were evaluated at the same thresholds to allow 3-dimensional structural rendering of each sample.
2.5 |. Immunohistochemistry
Three μm thick paraffin sections were baked at 60°C overnight. And then deparaffinized, rehydrated and heated at 95°C with Antigen Unmasking solution (Vector Laboratories, Burlingame, CA) for 5−10 min. Slices were then treated with 3% hydrogen peroxide for 10 min at room temperature for 10 min, incubated with 0.5% Triton X-100 (Sigma-Aldrich, St. Louis, MO) for 1 hr, and then blocked with Avidin/Biotin Blocking Kit (Invitrogen, CA, 004303).
Slides were then blocked with the blocking serum at 10% normal goat serum (Vector Laboratories) in 1% bovine serum albumin for 30 min at room temperature and incubated with primary antibodies against Runx2 (1:200 dilution) (Mouse IgG; MBL, Woburn, MA) or Ihh (1:500 dilution; Rabbit IgG; Abcam, Cambridge, MA) at 4°C overnight. The next day, secondary biotinylated goat anti-mouse antibody (Vector Laboratories) was added to slices for 30 min, followed by the incubation with VECTASTAIN Elite ABC HRP Kit (Vector Laboratories) for 30 min. Afterwards, positive staining was detected by ImmPACT DAB Peroxidase (HRP) Substrate (Vector Laboratories). Slides were then counterstained with CAT Hematoxylin (Biocare Medical, Pacheco, CA), dehydrated with graded ethanol and cleared with three changes of Xylene and cover-sliped finally.
2.6 |. Real-time PCR analysis
Twelve-week-old Runx2Agc1ER mice and their Cre− control mice were killed. Disc tissues were collected from freshly killed mice to obtain total RNA. Total mRNA was extract with Trizol (Invitrogen Life Technologies). 1 μg total RNA was used to synthesize complementary DNA (cDNA) using an iScripts cDNA Synthesis kit (Quanta Biosciences, MD). Subsequently, real-time PCR amplification was performed using specific primers of target genes and a SYBR Green real-time PCR kit (Quanta Biosciences). The primer names and sequences were listed in Table 1. Data were collected from cartilage of three independent mice (n = 3).
TABLE 1.
The names of sequences of primers used in this project
| Genes | Primer sequence (forward primers) | Primer sequence (reverse primers) |
|---|---|---|
| Runx2 | GACTGTGGTTACCGTCATGGC | ACTTGGTTTTTCATAACAGCGGA |
| Col10a1 | TTCTGCTGCTAATGTTCTTGACC | GGGATGAAGTATTGTGTCTTGGG |
| Mmp9 | GCAGAGGCATACTTGTACCG | TGATGTTATGATGGTCCCACTTG |
| Mmp13 | CTTCTTCTTGTTGAGCTGGACTC | CTGTGGAGGTCACTGTAGACT |
| Actin | GGCTGTATTCCCCTCCATCG | CCAGTTGGTAACAATGCCATGT |
Note. Runx2: runt-related transcription factor.
2.7 |. Statistical analysis
Data are presented as the mean ± standard deviation. For experiments comparing two groups of data, unpaired Student’s t test was performed. A value of p < 0.05 was considered to be significant.
3 |. RESULTS
3.1 |. Agc1-CreER mice target disc tissue with high recombination efficiency
To evaluate Agc1-Cre expression and recombination efficiency in disc tissue, we bred Agc1-CreER transgenic mice (Henry et al., 2009) with ROSAmT/mG reporter mice (Chelberg, Banks, Geiger, & Oegema, 1995; Dahia, Mahoney, Durrani, & Wylie, 2009; Liao et al., 2018) to generate Agc1-CreER; ROSAmT/mG mice. Mice express red fluorescence in all cell types before Cre recombination and green fluorescence after breeding with Agc1-CreER mice and administered with tamoxifen. The red fluorescent image of disc in Agc1-CreER; ROSAmT/mG mice indicates the absence of Agc1-CreER-xpressing cells (Figure 1, left panel) and the green-labeled cells marked Agc1-CreER targeting cells (Figure 1, right panel).
FIGURE 1.
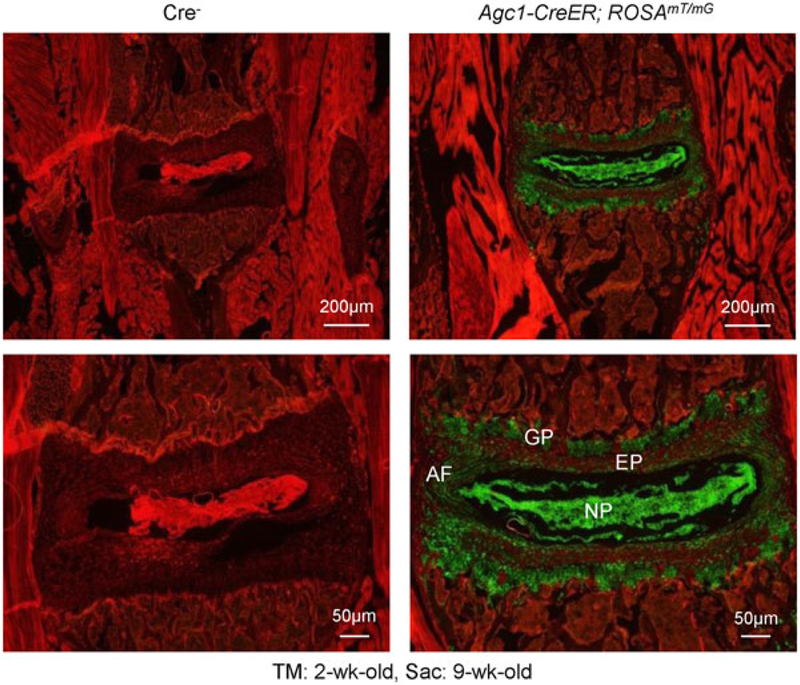
Agc1 -CreER mice efficiently target disc cells. To determine the Cre-mediated targeting specificity and recombination efficiency, we have bred Agc1-CreER mice with ROSAmT/mG reporter mice. Tamoxifen was administered into 2-week-old mice and immunofluorescence analysis was performed in disc tissue of 9-week-old mice. Results of fluorescence microscope analysis showed that most disc tissues were efficiently targeted by Agc1-CreER mice, including growth plate (GP) cartilage cells, AF and NP cells (right panel). AF: annulus fibrosus; NP: nucleus pulposus [Color figure can be viewed at wileyonlinelibrary.com]
3.2 |. Deletion of Runx2 in Aggrecan-expressing disc cells led to defects in disc tissue
μCT scanning displayed that there was no obvious phenotype in spine and disc tissues in Runx2 KO mice compared with the Cre− control mice (Figure 2), suggesting that there is no changes in bone mineralization in spine and disc tissue in Runx2 KO mice. However, the histologic results showed obvious changes in disc tissue morphology. Most NP cells are chondrocyte-like cells which are characterized by their smaller size and high ratio of proteoglycan matrix to cells in the NP in 3-month-old Cre− control mice (Figures 3a,b). In contrast, large numbers of large vacuolated notochordal cells were observed in the NP in 3-month-old Runx2Agc1ER KO mice. Furthermore, less proteoglycan matrix, demonstrated by Alcian blue staining, was found in the NP of Runx2 KO mice than that of Cre− control mice (Figure 3b). In addition, results of Alcian blue staining also demonstrated that the GP cartilage tissue was expanded in Runx2 KO mice. In some mice these expanded GP tissue was protruded to the vertebral bone (Figure 3a,b).
FIGURE 2.
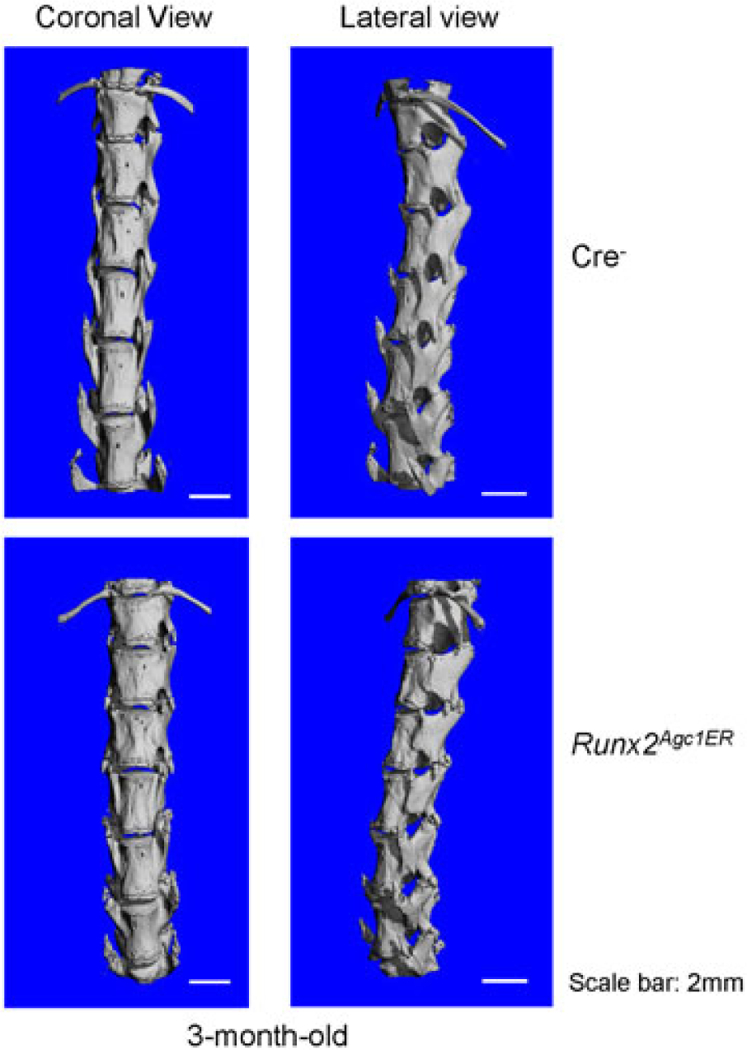
Micro-CT analysis of Runx2Agc1ER KO mice. Two-week-old Cre− control and Runx2Agc1ER KO mice were treated with tamoxifen and 3-month-old Cre− control and Runx2Agc1ER KO mice were analyzed by micro-CT (μCT). The results showed that no significant changes in the length of the spine. However, early sign of osteophyte formation was observed in Runx2Agc1ER KO mice (marked with red arrowheads). KO: knockout; Runx2: runt-related transcription factor 2 [Color figure can be viewed at wileyonlinelibrary.com]
FIGURE 3.
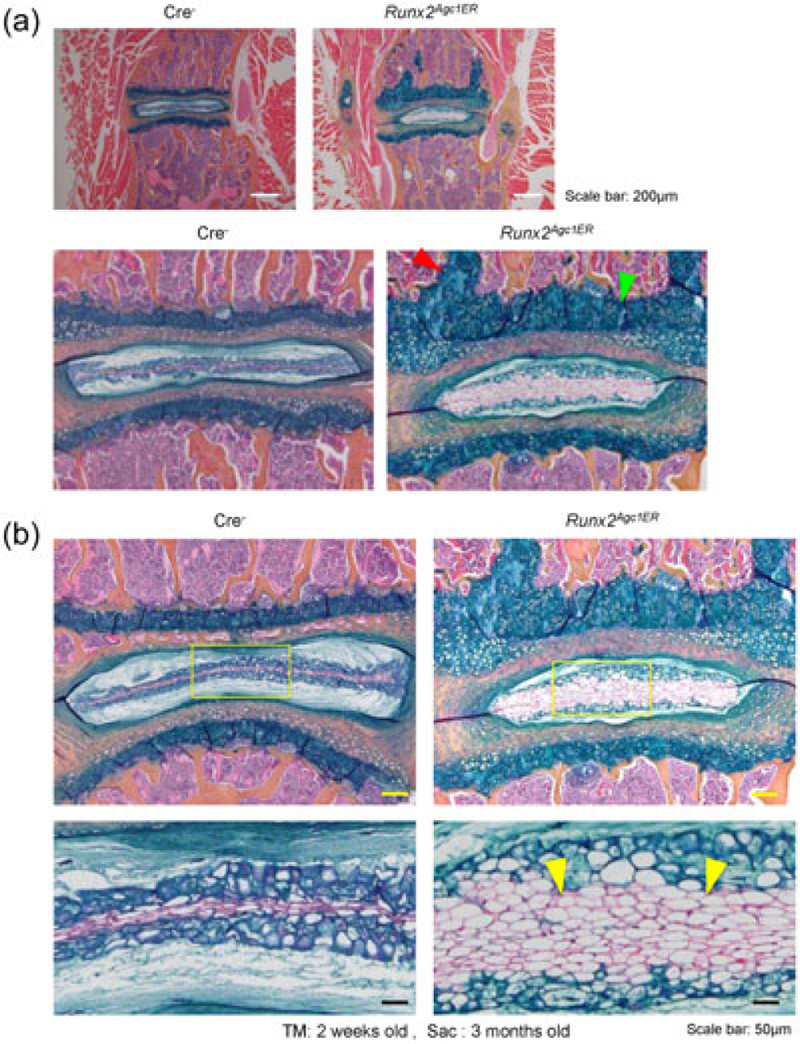
Defects in disc tissues of Runx2Agc1ER KO mice. (a) Results of histologic analysis showed that the growth plate thickness was significantly increased in Runx2Agc1ER KO mice, suggesting that Runx2 inhibits growth plate cartilage growth in normal mice (green arrowhead: increased growth plate cartilage thickness; red arrowhead: the expanded growth plate tissue was protruded to the vertebral body). (b) Increased numbers of notochordal cells in NP were observed in Runx2Agc1ER KO mice (yellow arrowheads: notochordal cells). KO: knockout; NP: nucleus pulposus; Runx2: runt-related transcription factor 2 [Color figure can be viewed at wileyonlinelibrary.com]
3.3 |. Deletion of Runx2 in Aggrecan-expressing disc cells increased Ihh expression
Runx2 is an essential transcription factor regulating endochondral ossification through control of chondrocyte proliferation and differentiation. Results of immunohistochemical (IHC) analysis showed that Runx2 expression was detected in the nucleus of chondrocyte in the Cre− control mice, whereas almost no Runx2 positive cells was detected in the disc tissue in Runx2 KO mice (Figure 4a). In contrast, Ihh protein levels were greatly increased in NP and AF cells of Runx2 KO mice (Figures 4b,c). We then further analyzed expression of other genes related to Ihh signaling, such as Ptch1 and Smo, and found that expression of both Ptch1 and Smo was significantly upregulated in the disc tissue of Runx2 KO mice (Figure 4d,e).
FIGURE 4.
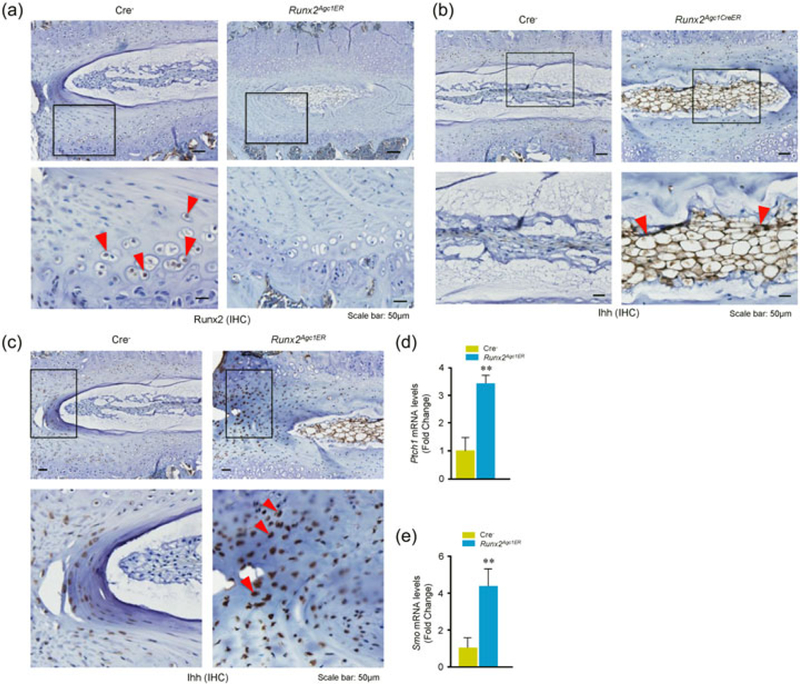
Changes in Ihh signaling in disc tissues of Runx2Agc1ER KO mice. Two-week-old Cre− control and Runx2Agc1ER KO mice were treated with tamoxifen and immunohistochemical (IHC) assay was performed using disc tissues of 3-month-old Cre− and Runx2 KO mice. (a) Results showed that Runx2 expression (red arrowheads indicate the Runx2-positive cells in Cre− mice) was significantly reduced in the annulus fibrosus (AF) cells of Runx2Agc1ER KO mice (right panels). (b) Expression of Ihh was significantly increased in the annulus fibrosus (AF) cells and nucleus pulposus (NP) cells in disc tissue of 3-month-old Runx2Agc1ER KO mice. Red arrowheads marked Ihh-positive cells. (c) Increased Ihh expression was also detected in AF cells (red arrowheads marked Ihh-positive cells) of disc tissues of Runx2Agc1ER KO mice. (d and e) Changes in mRNA expression of Ihh signaling related genes were analyzed by real-time PCR using the RNA extracted from disc tissues of 3-month-old Cre− and Runx2 KO mice. The results showed that expression of Ptch1 and Smo was significantly increased in disc tissues of Runx2 KO mice. KO: knockout; Runx2: runt-related transcription factor 2 [Color figure can be viewed at wileyonlinelibrary.com]
3.4 |. Deletion of Runx2 in disc cells altered chondrocyte maker gene expression
Total RNA was extracted from disc tissue from 3-month-old Cre− or Runx2Agc1ER KO mice followed by the real-time PCR analysis. Expression of Runx2, Osx, Sox9, Col10al, Mmp9, Mmp13, Adamts4, and Adamts5 was significantly reduced, respectively (Figures 5a–h). These observations are consistent with previous reports (Komori, 2008; Liao et al., 2017; M. Wang et al., 2012; Pratap et al., 2005). To further confirm changes in expression of Col10a1 and Mmp13, we performed IHC assays and found that Col-X and MMP13 protein levels were significantly reduced in the disc tissue of Runx2 KO mice (Figure 5i,j). Abnormal regulation of the genes encoding for matrix degradation enzymes may play an important role in the development and progression of disc degeneration.
FIGURE 5.
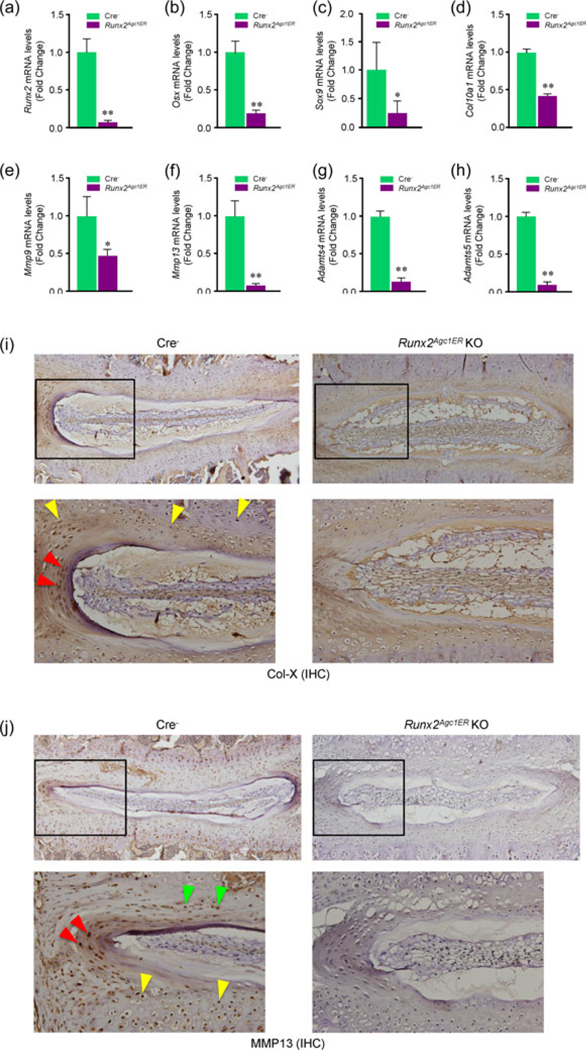
Deletion of Runx2 in Aggrecan-expressing disc tissue alters the expression of genes encoding for matrix proteins in disc tissue. (a–h) Two-week-old Cre− control and Runx2Agc1ER KO mice were treated with tamoxifen and total RNAs were extracted from disc tissue of 3-month-old Cre− and Runx2 KO mice and real-time PCR assay was performed. Expression of Runx2, Osx, Sox9, Col10al, Mmp9, Mmpl3, Adamts4, and Adamts5 was significantly decreased in Runx2Agc1ER KO mice. Statistical analysis was performed using unpaired Student’s t test (*p < 0.05, **p < 0.01, n = 3 mice per group). (i and j) Expression of Col-X and MMP13 protein was also analyzed by immunohistochemistry (IHC). The results showed that expression of both Col-X and MMP13 proteins was significantly reduced in disc tissues of Runx2 KO mice. KO: knockout; Runx2: runt-related transcription factor 2 [Color figure can be viewed at wileyonlinelibrary.com]
4 |. DISCUSSION
In this study, we found that Agc1-CreER mice could efficiently target disc cells, including GP cartilage cells, AF and NP cells (Figure 1). Using Agc1-CreER mice, we generated Runx2Agc1ER KO mice and analyzed disc phenotype of these mice. As a result, we have demonstrated an essential role of Runx2 in disc tissue development and maintaining normal disc structures at postnatal and adult stages. Our results suggest that Runx2 expression in Aggrecan-expressing cells at the postnatal stage is required for promoting the transition from notochordal cells to chondrocyte-like cells in the NP.
The NP is an aggrecan-rich gel-like tissue. In the neonate, the NP is highly cellular with relatively little proteoglycan production. In contrast, in the adult, the proportion of cells to matrix is low. As the disc matures, the composition of the NP changed. The numbers of large vacuolated cells (notochordal origin) are decreased and replaced by smaller chondrocyte-like cells. These cellular events are also accompanied with changes in the extracellular microenvironment (Risbud & Shapiro, 2011). In adult human, beginning as early as the second decade of life, notochordal cells are no longer detected within the NP, which is replaced by smaller or disperse chondrocyte-like cells (Chelberg et al., 1995). Notochordal cells survive at postnatal age, but at greatly reduced levels, at least until 48 weeks of age in mice (Dahia et al., 2009; Fujita et al., 2005). Currently, identification of these two cell types is restricted to cell morphology analysis and use of limited numbers of notochord-specified makers (Fujita et al., 2005; Gilson, Dreger, & Urban, 2010; Minogue, Richardson, Zeef, Freemont, & Hoyland, 2010). And there have been two conflicting hypotheses about the origin of the chondrocyte-like cells in the NP. Originally, it was suggested that these cells were of mesenchymal origin, a consequence of the migration of cells to the NP from the surrounding EP (Vujovic et al., 2006). Alternatively, it has been suggested that notochordal cells are progenitors for all NP cells and undergo terminal differentiation to give rise to chondrocyte-like cells (Boos et al., 2002; Liebscher, Haefeli, Wuertz, Nerlich, & Boos, 2011; Pazzaglia, Salisbury, & Byers, 1989). In these two scenarios, notochordal cells play distinct roles, either as organizer or as tissue-specific progenitors, respectively. Due to the technical limitations, no definitive conclusion has been arrived. In 2012, there is evidence that notochordal cells are the precursor cells of the NP, and undergo differentiation to generate chondrocyte-like cells during postnatal development (M.R. McCann, Tamplin, Rossant, & Seguin, 2012). This transition might be induced by a variety of factors present in the extracellular environment of the IVD. In addition, rabbit notochordal cells have been shown to be able to differentiate toward chondrocyte-like cells when maintained in culture (Kim et al., 2009). However, notochordal cells in the NP have the morphology very different from that of chondrocyte-like NP cells. Notochordal cells are markedly greater in diameter and contain large vacuoles (Guehring, Urban, Cui, & Tirlapur, 2008; Hunter et al., 2003; Mollenhauer, 2006). In this study we found that most of cells within the NP of Runx2 KO mice are notochordal cells compared with those of corresponding Cre- control mice with most of chondrocyte-like cells (Figure 3b). This observation suggests that Runx2 promotes the transition of notochordal cells into chondrocyte-like cells within the NP. Runx2 deficiency leads to the accumulation of notochordal cells in the NP. These findings suggest that Runx2 may play an important role in maintaining NP tissue normal morphology, homeostasis and normal function.
Ihh is essential for chondrocyte and osteoblast proliferation and differentiation during prenatal endochondral bone formation (Maeda et al., 2007). Hedgehog signaling is required for the formation of the notochord sheath and NP tissue patterning within the IVD (Choi & Harfe, 2011). It has been reported that deletion of Smo in Shh-expressing cells results in abnormal development of the IVD. Disc tissue with Smo removal has a smaller NP than controls and concentric lamellae were absent in the AF (Choi & Harfe, 2011). In postnatal control animals, the NP was located inside the AF throughout the vertebral column. In contrast, Smo KO animals contained small NP with the majority of notochord cells dispersed throughout the vertebral column (Choi & Harfe, 2011). In contrast, the postnatal Ihh specific deletion in Col2a1-expressing cells (P14 mice) showed loss of GP cartilage and AF tissue with enlarged NP tissue (Maeda et al., 2007). These findings clearly demonstrated the critical role of hedgehog signaling in disc tissue development, especially in the NP formation. In the current study we found that Ihh expression was significantly increased in both NP and AF tissues and Ihh signaling was activated in the disc tissue. These findings suggest that Runx2 may regulate notochordal cell transition through an Ihh-dependent mechanism. Our observations also suggest that Runx2 may play an important role in disc tissue growth and the development of degenerative disc disease. In this study, we found significant upregulation of Ihh expression in disc tissue in Runx2 KO mice, suggesting that Runx2 may suppress Ihh expression under normal conditions during postnatal disc tissue development. The detail molecular mechanism by which Runx2 regulates Ihh expression in disc tissue and the significance of this regulation in NP cell function needs to be further explored.
Our current findings suggest that Runx2 regulates multiple genes, including Osx, Col10a1, Mmp9, Mmp13, Adamts4 and Adamts5 in the disc tissue. However, we know much less about upstream regulation of Runx2 in the disc tissue. In previous studies, we found that Runx2 is regulated by β-catenin signaling in the disc tissue (M. Wang et al., 2012). We also found that microRNA miR-204/miR-211 regulate Runx2 expression in mesenchymal progenitor cells (Huang, Zhao, Xing, & Chen, 2010). To fully understand the role of Runx2 in disc tissue homeostasis, more detail mechanism studies regarding Runx2 regulation need to be further investigated.
In summary, we found that Runx2 plays an important role in regulation of Ihh expression and notochordal cell transition into chondrocyte-like cells in the NP and is required for disc tissue development.
ACKNOWLEDGMENTS
We would like to express our gratitude to Ms. Lily Yu for her help on processing and staining histologic samples. This study was supported by National Institutes of Health Grants (R01AR054465) and (R01AR070222) to D. Chen. This study was also partially supported by the grant from Shenzhen Science and Technology Innovation Committee, China (grant no. JCYJ20160331114205502) to D. Chen. This study was also partially supported by National Natural Science Foundation of China (grant no. 81700997) to L. Liao.
Funding information
National Institute of Arthritis and Musculoskeletal and Skin Diseases, Grant/Award Numbers: R01AR070222, R01AR054465; National Natural Science Foundation of China, Grant/Award Number: 81700997; Shenzhen Science and Technology Innovation Committee, China, Grant/Award Number: JCYJ20160331114205502
Footnotes
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
REFERENCES
- Aguiar DJ, Johnson SL, & Oegema TR (1999). Notochordal cells interact with nucleus pulposus cells: Regulation of proteoglycan synthesis. Experimental Cell Research, 246, 129–137. [DOI] [PubMed] [Google Scholar]
- Boos N, Weissbach S, Rohrbach H, Weiler C, Spratt KF, & Nerlich AG (2002). Classification of age-related changes in lumbar inter-vertebral discs: 2002 Volvo Award in basic science. Spine (Phila Pa 1976), 27, 2631–2644. [DOI] [PubMed] [Google Scholar]
- Chelberg MK, Banks GM, Geiger DF, & Oegema TR Jr. (1995). Identification of heterogeneous cell populations in normal human intervertebral disc. Journal of Anatomy, 186(Pt 1), 43–53. [PMC free article] [PubMed] [Google Scholar]
- Chen H, Ghori-Javed FY, Rashid H, Adhami MD, Serra R, Gutierrez SE, & Javed A (2014). Runx2 regulates endochondral ossification through control of chondrocyte proliferation and differentiation. Journal of Bone and Mineral Research, 29, 2653–2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi KS, & Harfe BD (2011). Hedgehog signaling is required for formation of the notochord sheath and patterning of nuclei pulposi within the intervertebral discs. Proceedings of the National Academy of Sciences of the United States of America, 108(23), 9484–9489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahia CL, Mahoney EJ, Durrani AA, & Wylie C (2009). Postnatal growth, differentiation, and aging of the mouse intervertebral disc. Spine (Phila Pa 1976), 34, 447–455. [DOI] [PubMed] [Google Scholar]
- Ducy P, Zhang R, Geoffroy V, Ridall AL, & Karsenty G (1997). Osf2/Cbfa1: A transcriptional activator of osteoblast differentiation. Cell, 89, 747–754. [DOI] [PubMed] [Google Scholar]
- Ellis K, Bagwell J, & Bagnat M (2013). Notochord vacuoles are lysosome-related organelles that function in axis and spine morpho-genesis. The Journal of Cell Biology, 200, 667–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Miyamoto T, Imai J, Hosogane N, Suzuki T, Yagi M,... Suda T (2005). CD24 is expressed specifically in the nucleus pulposus of intervertebral discs. Biochemical and Biophysical Research Communications, 338, 1890–1896. [DOI] [PubMed] [Google Scholar]
- Gilson A, Dreger M, & Urban JP (2010). Differential expression level of cytokeratin 8 in cells of the bovine nucleus pulposus complicates the search for specific intervertebral disc cell markers. Arthritis Research & Therapy, 12, R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guehring T, Urban JP, Cui Z, & Tirlapur UK (2008). Noninvasive 3D vital imaging and characterization of notochordal cells of the intervertebral disc by femtosecond near-infrared two-photon laser scanning microscopy and spatial-volume rendering. Microscopy Research and Technique, 71, 298–304. [DOI] [PubMed] [Google Scholar]
- Henry SP, Jang CW, Deng JM, Zhang Z, Behringer RR, & de Crombrugghe B (2009). Generation of aggrecan-CreERT2 knockin mice for inducible Cre activity in adult cartilage. Genesis, 47, 805–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Zhao L, Xing L, & Chen D (2010). MicroRNA-204 regulates Runx2 protein expression and mesenchymal stem cell differentiation. Stem Cells, 28, 357–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter CJ, Bianchi S, Cheng P, & Muldrew K (2007). Osmoregulatory function of large vacuoles found in notochordal cells of the intervertebral disc running title: An osmoregulatory vacuole. Molecular & Cellular Biomechanics, 4, 227–237. [PMC free article] [PubMed] [Google Scholar]
- Hunter CJ, Matyas JR, & Duncan NA (2003). The notochordal cell in the nucleus pulposus: A review in the context of tissue engineering. Tissue Engineering, 9, 667–677. [DOI] [PubMed] [Google Scholar]
- Kamekura S, Kawasaki Y, Hoshi K, Shimoaka T, Chikuda H, Maruyama Z, ... Kawaguchi H (2006). Contribution of runt-related transcription factor 2 to the pathogenesis of osteoarthritis in mice after induction of knee joint instability. Arthtitis and Rheumatism, 54, 2462–2470. [DOI] [PubMed] [Google Scholar]
- Kim JH, Deasy BM, Seo HY, Studer RK, Vo NV, Georgescu HI, ... Kang JD (2009). Differentiation of intervertebral notochordal cells through live automated cell imaging system in vitro. Spine (Phila Pa 1976), 34, 2486–2493. [DOI] [PubMed] [Google Scholar]
- Komori T (2008). Regulation of bone development and maintenance by Runx2. Frontiers in Bioscience, 13, 898–903. [DOI] [PubMed] [Google Scholar]
- Liao L, Zhang S, Gu J, Takarada T, Yoneda Y, Huang J, ... Chen D (2017). Deletion of Runx2 in articular chondrocytes decelerates the progression of DMM-induced osteoarthritis in adult mice. Scientific Reports, 7, 2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao L, Zhang S, Zhou G-Q, Ye L, Huang J, & Zhao L, et al. (2018). Deletion of Runx2 in condylar chondrocytes disrupts TMJ tissue homeostasis. Journal of Cellular Physiology. (Accepted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebscher T, Haefeli M, Wuertz K, Nerlich AG, & Boos N (2011). Age-related variation in cell density of human lumbar intervertebral disc. Spine (Phila Pa 1976), 36, 153–159. [DOI] [PubMed] [Google Scholar]
- Maeda Y, Nakamura E, Nguyen MT, Suva LJ, Swain FL, Razzaque MS, ... Lanske B (2007). Indian Hedgehog produced by postnatal chondrocytes is essential for maintaining a growth plate and trabecular bone. Proceedings of the National Academy of Sciences of the United States of America, 104, 6382–6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann M, & Séguin C (2016). Notochord cells in intervertebral disc development and degeneration. Journal of Developmental Biology, 4, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann MR, Tamplin OJ, Rossant J, & Séguin CA (2012). Tracing notochord-derived cells using a Noto-Cre mouse: Implications for intervertebral disc development. Disease Models & Mechanisms, 5, 73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minogue BM, Richardson SM, Zeef LA, Freemont AJ, & Hoyland JA (2010). Transcriptional profiling of bovine intervertebral disc cells: Implications for identification of normal and degenerate human intervertebral disc cell phenotypes. Arthritis Research & Therapy, 12, R22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollenhauer JA (2006). The notochord in the mammalian adult: A paradox. Arthritis and Rheumatism, 54, 3728–3729. [DOI] [PubMed] [Google Scholar]
- Pazzaglia UE, Salisbury JR, & Byers PD (1989). Development and involution of the notochord in the human spine. Journal of the Royal Society of Medicine, 82, 413–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratap J, Javed A, Languino LR, van Wijnen AJ, Stein JL, Stein GS, & Lian JB (2005). The Runx2 osteogenic transcription factor regulates matrix metalloproteinase 9 in bone metastatic cancer cells and controls cell invasion. Molecular and Cellular Biology, 25(19), 8581–8591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risbud MV, & Shapiro IM (2011). Notochordal cells in the adult intervertebral disc: New perspective on an old question. Critical Reviews in Eukaryotic Gene Expression, 21, 29–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts S (2000). Disc morphology in health and disease. Biochemical Society Transactions, 30, 864–869. [DOI] [PubMed] [Google Scholar]
- Sato S, Kimura A, Ozdemir J, Asou Y, Miyazaki M, Jinno T,... Takeda S (2008). The distinct role of the Runx proteins in chondrocyte differentiation and intervertebral disc degeneration: Findings in murine models and in human disease. Arthtitis and Rheumatism, 58, 2764–2775. [DOI] [PubMed] [Google Scholar]
- Shen J, Li J, Wang B, Jin H, Wang M, Zhang Y, ... Chen D (2013). Deletion of the type II TGF-p receptor gene in articular chondrocytes leads to progressive OA-like phenotype in mice. Arthtitis and Rheumatism, 65(12), 3107–3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takarada T, Hinoi E, Nakazato R, Ochi H, Xu C, Tsuchikane A, ... Yoneda Y (2013). An analysis of skeletal development in osteoblast- specific and chondrocyte-specific runt-related transcription factor-2 (Runx2) knockout mice. Journal of Bone and Mineral Research, 28, 2064–2069. [DOI] [PubMed] [Google Scholar]
- Vujovic S, Henderson S, Presneau N, Odell E, Jacques T, Tirabosco R, ... Flanagan A (2006). Brachyury, a crucial regulator of notochordal development, is a novel biomarker for chordomas. Journal of Pathology, 209, 157–165. [DOI] [PubMed] [Google Scholar]
- Wang B, Jin H, Zhu M, Li J, Zhao L, Zhang Y, ... Chen D (2014). Chondrocyte p-catenin signaling regulates postnatal bone remodeling through modulation of osteoclast formation in a murine model. Arthritis Rheumatol, 66(1), 107–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Tang D, Shu B, Wang B, Jin H, Hao S, ... Chen D (2012). Conditional activation of p-catenin signaling leads to severe defects in intervertebral disc tissue. Arthtitis and Rheumatism, 64(8), 2611–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Li S, Yi D, Zhou GQ, Chang Z, Ma PX,... Chen D (2018). CHIP regulates bone mass by targeting multiple TRAF family members in bone marrow stromal cells. Bone Research, 6(1), 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi SK, Sullivan AJ, van Wijnen AJ, Stein JL, Stein GS, & Lian JB (2002). Integration of Runx and Smad regulatory signals at transcriptionally active subnuclear sites. Proceedings of the National Academy of Sciences of the United States of America, 99, 8048–8053. [DOI] [PMC free article] [PubMed] [Google Scholar]


