Abstract
γδ T cells function as sentinels in early host responses to infections and malignancies. Specifically, γδ T cells recognize tumor-associated stress antigens via T-cell receptor (TCR) γδ and play important roles in the antitumor immune response. In this study, we characterized the pattern of the human TCR γδ complementary determinant region 3 (CDR3) repertoire in patients with lung carcinoma (LC) via high-throughput sequencing. The results showed that the diversity of CDR3δ was significantly reduced, and that of CDR3γ was unchanged in LC patients compared with healthy individuals; in addition, LC patients shared significantly more CDR3δ sequences with each other than healthy individuals. The CDR3 length distribution and N-addition length distribution did not significantly differ between LC patients and healthy individuals. In addition, the CDR3 repertoire tended to use more Vδ2 and fewer Vδ1 germline gene fragments among LC patients. Moreover, we found a combination of four TCR γδ repertoire features that focus on CDR3δ and can be used as a biomarker for LC diagnosis. Our research suggests that the TCR γδ CDR3 repertoire changed in LC patients due to the antitumor immune response by γδ T cells in vivo, and these changes primarily focus on the amplification of certain tumor-specific CDR3δ clones among patients. This study demonstrates the role of γδ T cells from the TCR γδ CDR3 repertoire in tumor immunity and lays the foundation for elucidating the mechanism underlying the function of γδT cells in antitumor immunity.
Introduction
During the past several years, several studies have characterized human γδ T lymphocytes, and findings regarding the immune functions of these cells, particularly their natural killer cell-like lytic activity against tumor cells, have suggested that these cells may be useful in the treatment cancer.1,2,3,4 This is accomplished through the interaction of heterodimer T-cell receptor (TCR) γδ and other receptors expressed on the cell surface, such as natural killer cell-activated receptor (NKG2D).
Although primarily considered an innate immune cell, γδ T cell expresses TCR γδ due to the rearrangement of germline gene V–(D)–J–C fragments, similar to the diversity observed in TCR αβ. The antigen-binding site of TCR γδ primarily consists of three complementary determinant regions (CDRs) contributed by each Vγ or Vδ domain. Both CDR1 and CDR2 regions are encoded by germline V genes, whereas the CDR3 region is formed by the somatic rearrangement of V, (D) and J fragments, which embody TCR diversity.5 However, the antigen recognition pattern of TCR γδ significantly differs that from TCR αβ. Without major histocompatibility complex, TCR γδ is thought to be able to directly recognize and respond to various peptide or non-peptide antigens in an unrestricted manner.6 Previous studies reported the TCRγδ CDR3 repertoire of an individual change according to the state of the cells.7,8,9
Because tumor-infiltrating γδ T lymphocytes have been detected in a broad spectrum of malignancies, γδ T cells are believed to contribute to the front line of tumor surveillance and bridge the gap between innate and adaptive immunity. The most abundant subset of circulating γδ T cells, Vγ9Vδ2 cells, can be activated and expanded in vitro or in vivo following a single treatment with the phosphoantigen isopentenyl pyrophosphate and are widely used for antitumor immunotherapy.10 Nevertheless, the specificity of γδ T cells for cancer patients remains poorly understood.
High-throughput sequencing is used to analyze the immune repertoire of TCR or immunoglobulin, and a comprehensive analysis of the diversity of the immune system is closely related to treatment effectiveness and patient prognosis in different diseases. The analysis of TCR diversity in tumor patients might also elucidate the pathogenesis and development of cellular immunotherapy approaches that target specific tumors.11 However, little is known regarding the repertoire of TCR γδ. We previously characterized the TCR γδ CDR3 repertoire in 30 healthy donors using immune repertoire sequencing and found that the TCRγδ CDR3 repertoire is quite diverse and differs between individuals.12 Our findings provide a basic understanding of the diversity of the TCRγδ repertoire under physiological conditions, which helps to elucidate the mechanism by which γδ T cells recognize pathogens and tumor antigens.
In this study, we conducted a comprehensive and systematic analysis of the TCR γδ CDR3 repertoires of seven patients with lung cancer and healthy individuals using high-throughput sequencing. This study clarifies the characteristics of the tumor-specific TCR γδ CDR3 repertoire for the first time and provides a theoretical basis for understanding the tumor antigen recognition pattern of γδ T cells and its mechanism of action in antitumor immunotherapy.
Materials and methods
Study subjects
Peripheral blood samples were obtained from seven patients (age 56.9±10.7 years, male:female ratio=5:2) with lung carcinoma (LC). All of the patients in this study were presurgery and had no treatment. The clinical characteristics of these patients are summarized in Table 1. Seven age- and gender-matched healthy volunteers (age 56.0±11.0 years, male:female ratio=5:2) were included as controls (con). The features of these LC patients and healthy controls are shown in Table 1. The study was approved by the Ethical Committee of the Chinese Academy of Medical Sciences (Project No. 008-2014) before initiation. All samples were frozen in RNA protection reagent (Qiagen, Hilden, Germany) for further processing.
Table 1.
Sample characteristics
| Sample ID | Gender | Age (years) | Tumor type | Stage | Tumor size (cm) | Control sample (gender/age (years)) |
|---|---|---|---|---|---|---|
| 01 | Female | 52 | Adenocarcinoma | I | 4 × 2.7 | Female/50 |
| 02 | Male | 48 | Adenocarcinoma | III | 3.5 × 2.7 | Male/48 |
| 03 | Male | 55 | Small cell carcinoma | Diffusion | unknown | Male/55 |
| 04 | Male | 55 | Squamous cell carcinoma | III | unknown | Male/50 |
| 05 | Male | 73 | Squamous cell carcinoma | III | 9 × 7 | Male/74 |
| 06 | Male | 70 | Adenocarcinoma | III | 3.5 × 3 | Male/69 |
| 07 | Female | 45 | Adenocarcinoma | III | 7 × 6.5 | Female/46 |
RNA isolation and arm-PCR procedure
Total RNA of was extracted from whole blood using a Qiagen RNeasy Mini Kit (No. 74104, Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The RNA samples were then subjected to reverse-transcription PCR (RT-PCR) using a Qiagen OneStep RT-PCR Kit (No. 210212, Qiagen, Hilden, Germany). Arm-PCRs were performed to amplify the complementary DNAs, which were normalized to the γδ T-cell percentage in each sample according to the manufacturer’s instructions (iRepertoire Inc., Huntsville, AL, USA), as described previously.12,13,14 Each PCR primer contained a barcode to identify samples after sequencing. A second PCR was performed using a Qiagen Multiplex PCR Kit (No. 206143, Qiagen, Hilden, Germany) and Illumina communal sequence primer, again under conditions specified by iRepertoire.
TCRγδ repertoire sequencing and alignment of CDR3 sequences
The PCR products were purified and sequenced using the Illumina Mi-seq Platform with the PE150 Kit (Illumina, San Diego, CA, USA). Raw data were analyzed by iRepertoire using the IRmap program to identify CDR3s for each sample. The best matches of the germline V and J gene were identified by determining alignments between the Illumina platform product and germline sequences in the IMGT/GENE-DB database. All of the TCRγδ CDR3 repertoire analyses in this paper were limited to in-frame sequences.
Analysis of sequencing data
The rank abundance was applied to describe the total distribution of T-cell receptor δ-chain (TRD) and T-cell receptor γ-chain (TRG) repertoire between LC patients and controls. We used diversity Hill index including Richness, Shannon index and Simpson index to evaluate the diversity characteristics of total samples. Meanwhile, we used diversity 50 (D50), the unique CDR3/total CDR3, the CDR3 clonal size and the frequency of dominant sequences to comprehensively analyze differences between lung patients and healthy controls in terms of CDR3 diversity as described previously.12
We calculated the percentage of shared CDR3 and showed the overlap of TRD CDR3 sequences to evaluate common tumor-associated CDR3s among patients by Venn diagram. Then we analyzed the Jaccard index to present the results of shared sequences.
The number of CDR3 nucleotides and random insert (N-addition) for each unique CDR3 (normalized data) were recorded, and related distribution plots were prepared in accordance with the proportion to demonstrate the distribution of the entire CDR3 repertoire. Based on the length and proportion, we calculated the weighted average to evaluate the differences in CDR3 and N-addition length distribution.
We used the Smith–Waterman algorithm to align the local V and J segment sequences in CDR3γ and CDR3δ between sequencing reads and the germline gene reference (human consensus from IGMT). The percentages of each germline V and J alleles are plotted to easily identify the frequently and infrequently used V and J alleles based on normalized data.
Statistical analysis
The data were statistically analyzed using GraphPad Prism v.6 (GraphPad Software, San Diego, CA, USA). The mean values for each parameter were compared between LC patients and controls using the Bootstrapping analysis and Mann–Whitney U-test. P<0.05 was considered significant. Unexpected clustering (cluster 3.0) and a principal component analysis (Metabo Analyst) were used to analyze selected parameters.
Results
The diversity of TRD is significantly reduced in LC patients
The diversity of the TCRγδ CDR3 repertoire, which reflects the proliferation of specific γδ T-cell clones, is one of the most important features of LC patients. We represented the rank abundance distribution curve of the total seven LC patients and seven healthy controls we sequenced. The results revealed that LC patients had higher abundance at low-rank scope than controls in the TCRδ chain (TRD) (Figure 1a). Then we applied the Hill index distribution curve (Figure 1b) and analyzed three parameters about the diversity both in the TRD and TRG between LC patients and controls (Figures 1c and d). We found that LC patients had higher Simpson index than healthy controls in TRD chain (Figure 1c). However, no significant difference was observed in TRG chain (Figure 1d).
Figure 1.
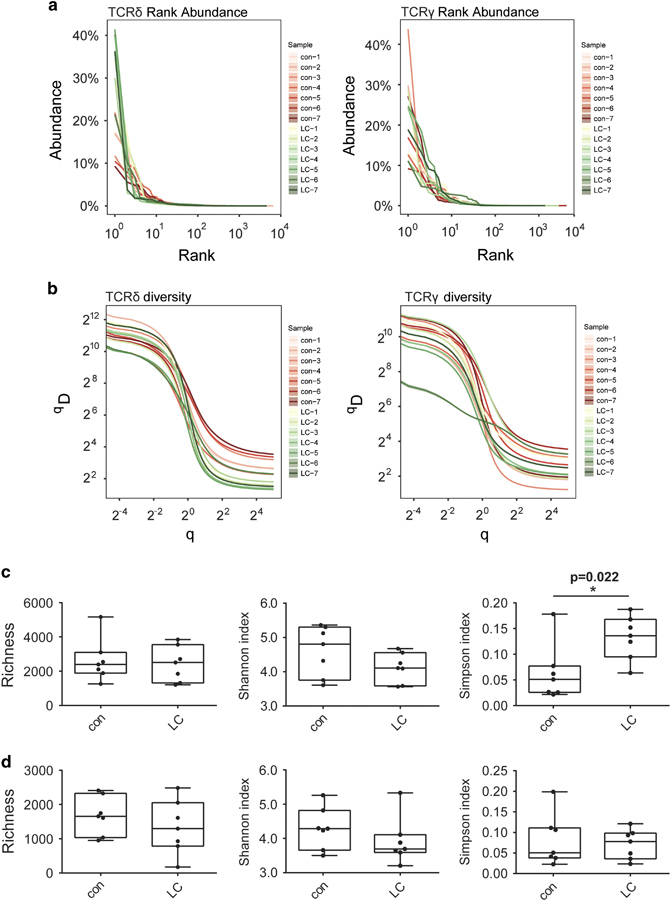
The total distribution of T-cell receptor δ-chain (TRD) and T-cell receptor γ-chain (TRG) repertoire between lung carcinoma (LC) patients and healthy controls. (a and b). The rank abundance distribution curve (a) and the diversity Hill index distribution curve (b) of 14 sequenced TRD and TRG repertoire. The left panel shows TRD, and the right panel shows TRG. Different samples are shown in different colors. The red spectrums represent healthy individuals and green represent LC patients. (c and d) Richness, Shannon index and Simpson index about the diversity Hill index distribution curve of TRD (c) and TRG (d) repertoire between LC patients and controls. *P<0.05.
We also assessed group-wise differences between LC patients and controls based on the D50, unique CDR3/total CDR3, CDR3 clonal size and frequency of the dominant sequence (Figure 2), which did not identify significant differences in the D50 of both the TRD chain and TRG chain between LC patients and controls (Figure 2a). However, the unique CDR3/total CDR3 value of TRD was 1.8-fold lower in LC patients than in controls (P=0.035) (Figure 2b). The size of each clone was calculated based on its frequency in the repertoire, and scatterplots of the top 50 clones are shown in Figure 2c. CDR3δ clones with a clonal size ranging from >20% and >15% were more common (P=0.002, P=0.039, respectively), and those with a clonal size >1% were less common (P=0.001) in the top 50 clones of the LC group than those of the control group (Figure 2d). We also analyzed the frequency of dominant sequences that most significantly contributed to the diversity of the entire repertoire, including the top 1, 5 and 10 clones for each sample (Figure 2e), which showed that the frequency of top 1 CDR3δ clones was significantly higher in the LC group than in the control group. Taken together, these results demonstrate that the diversity of TRD is significantly reduced in LC patients.
Figure 2.
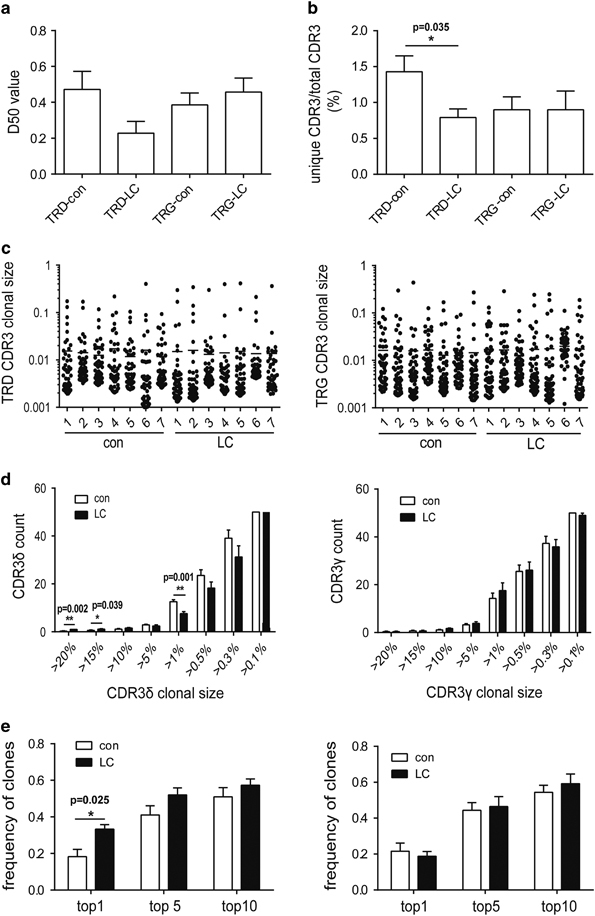
The diversity of T-cell receptor δ-chain (TRD) is significantly reduced in lung carcinoma (LC) patients. (a) The D50 of the TRD and T-cell receptor γ-chain (TRG) repertoires in LC patients. (b) The unique complementary determinant region 3 (CDR3)/total CDR3 value of the TRD and TRG repertoires in LC patients. *P<0.05. (c) The scatter plot of the distribution of the top 50 CDR3 clones in LC patients. (d) The statistical analysis of the proportion of top 50 CDR3 sequences by clone size (>20%, >15%, >10%, >5%, >1%, >0.5%, >0.3%, >0.1%). *P<0.05, **P<0.01. (e) The frequency of the top 1, top 5 and top 10 CDR3 sequences in LC patients. **P<0.01. The left panel shows TRD, and the right panel shows TRG in (c–e). Mean values with error bars representing the±standard error (s.e.m.) are plotted, and the Bootstrapping analysis and Mann–Whitney U-test was used to analyze differences between groups.
LC patients shared more CDR3δ sequences
We analyzed the differences in the shared CDR3 sequences between seven LC patients and seven healthy individuals. However, the results showed no significant difference, which may be due to the different types of LC (data not shown). Therefore, we analyzed the shared CDR3 sequences of four LC patients with adenocarcinoma and four age- and gender-matched healthy controls. The results showed that the percentage of shared CDR3δ sequences was 6.81% in LC patients, which was 4.2-fold higher than that in healthy individuals (P=0.008) (Figure 3a), and this difference remained significant when the percentage of shared CDR3δ sequences between patients and controls was set to a threshold clonal size >0.01% (P=0.013) (Figure 3b). However, the proportion of CDR3γ sequences did not significantly differ between groups, when analyzing either the entire repertoire or clones with a clonal size >0.01%. Figure 3c shows the number of CDR3δ sequences shared between four LC patients and four healthy individuals in detail. We used Jaccard index to measure the shared diversity, and the results revealed that the LC patients had significant more shared sequences than controls no matter shared by 4, 3 or 2 samples (Figure 3d). Although LC patients shared more CDR3δ sequences, the consistent proportion of the TRD repertoire remained very limited. Because LC patients appear to share more CDR3δ sequences, these sequences may be associated with specific tumor antigens. Table 2 lists the 26 common shared CDR3δ sequences among LC patients (V1: 4/26; V2: 21/26; V3: 1/26).
Figure 3.
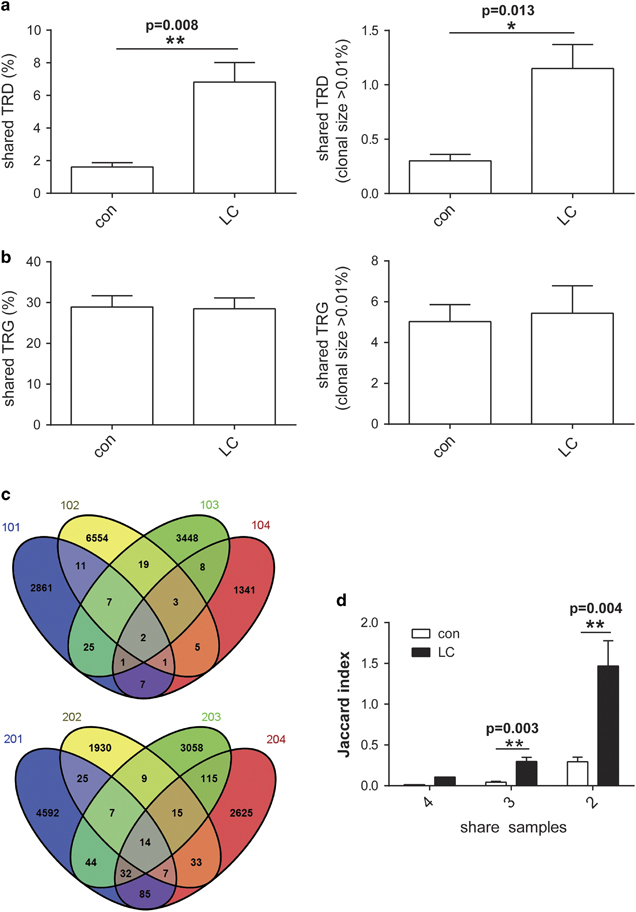
Lung carcinoma (LC) patients shared more complementary determinant region 3δ (CDR3δ) sequences than healthy individuals. (a) The proportion of total shared CDR3 T-cell receptor δ-chain (TRD) and T-cell receptor γ-chain (TRG) sequences among four LC patients and among four healthy controls. (b) The proportion of shared CDR3 TRD and TRG sequences among four LC patients patients and among four healthy patients for a threshold clonal size >0.01%. The left panel shows TRD, and the right panel shows TRG. Mean values with error bars depicting the ±s.e.m. are shown. **P<0.01 by the Bootstrapping analysis and Mann–Whitney U-test. (c) The Venn diagram shows the exact number of shared CDR3δ sequences among four LC patients and four healthy individuals. Different samples are shown in different colors. The numbers in the overlap of different colors represent the number of shared CDR3δ sequences. (d) The Jaccard index analysis of mean shared sequences shared by 4, 3 and 2 samples in groups. **P<0.01.
Table 2.
The shared CDR3δ sequences in LC patients
| No. | CDR3 sequence | Length (AA) | V–D–J usage | No. | CDR3 sequence | Length (AA) | V–D–J usage |
|---|---|---|---|---|---|---|---|
| 1 | ACDTLGSYTDKLI | 13 | V2–D3–J1 | 14 | ACEGIGTDKLI | 11 | V2–D3–J1 |
| 2 | ACDPLGDSDKLI | 12 | V2–D3–J1 | 15 | ACDTVTLQGPGDTQDKLI | 18 | V2–D3–J1 |
| 3 | ALGEAQFLVSYLGDTRSRLI | 20 | V1–D3–J1 | 16 | ACDTVTGGLKYTDKLI | 16 | V2–D3–J1 |
| 4 | ACDTMGSGDWEVDKLI | 16 | V2–D3–J1 | 17 | ACDTVLYTDKLI | 12 | V2–J1 |
| 5 | ACDTLPGTGYGDKLI | 15 | V2–D3–J1 | 18 | ACDTVGTGDRASDKLI | 16 | V2–D3–J1 |
| 6 | ACDSVLGDTRDTDKLI | 16 | V2–D3–J1 | 19 | ACDTVGPDTDKLI | 13 | V2–J1 |
| 7 | ACDSLGGPYTDKLI | 14 | V2–D3–J1 | 20 | ACDTVGDSPPGTDKLI | 16 | V2–D3–J1 |
| 8 | ACDMLDTRYTDKLI | 14 | V2–D3–J1 | 21 | ACDTLSRTDIGTDKLI | 16 | V2–J1 |
| 9 | ACDGLNTDKLI | 11 | V2–J1 | 22 | ACDSVLGPSFLDKLI | 15 | V2–D3–J1 |
| 10 | ARILGPTPPTYTDKLI | 16 | V1–D3–J1 | 23 | ACDSLLGDRTDKLI | 14 | V2–D3–J1 |
| 11 | ALGEPAPGTDKLI | 13 | V1–J1 | 24 | ACDPVLGDTPTRPPYTDKLI | 20 | V2–D3–J1 |
| 12 | ALGELIESLSDTGYTDKLI | 19 | V1–D3–J1 | 25 | ACDPLLGDRELI | 12 | V2–D3–J1 |
| 13 | AFKGGYWGRNMYTDKLI | 17 | V3–D3–J1 | 26 | ACDPLGDLPHTDKLI | 15 | V2–D3–J1 |
Abbreviations: AA, amino acid; CDR, complementary determinant region.
The CDR3 and N-addition length distributions were similar in LC patients and healthy individuals
The CDR3 region consists of V–(D)–J fragments and N-additions between the V–D, D–J and V–J fragments added during TCR rearrangement. The number of CDR3 nucleotides and N-additions for each unique CDR3 were recorded, and related distribution plots were generated in accordance with the proportion to demonstrate the distribution of the entire CDR3 repertoire. The results indicate that the average CDR3 and N-addition lengths of TRD and TRG exhibited a standard distribution in both groups, and the weighted averages of the CDR3 and N-addition lengths did not significantly differ between the two groups (Figure 4).
Figure 4.
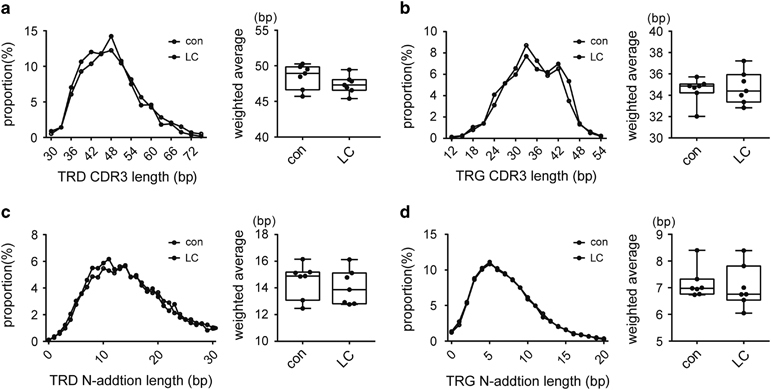
The complementary determinant region 3 (CDR3) and N-addition length distributions in lung carcinoma (LC) patients and healthy individuals were similar. (a–d) show the distributions of T-cell receptor δ-chain (TRD) CDR3 lengths, T-cell receptor γ-chain (TRG) CDR3 lengths, TRD N-addition lengths and TRG N-addition lengths, respectively. The left panel: the average length distribution curve of seven controls and LC patients. The right panel: the weighted average of each distribution curve.
The TCRγδ repertoire contained more hTRDV2 germline gene segments in LC patients than healthy individuals
The germline V–J gene usage of the TCRγδ repertoire, including three V gene segments of TRD (hTRDV1/2/3), six V gene segments of TRG (TRGV2/3/4/5/8/9), four J gene segments of TRD (hTRDJ1/2/3/4) and five J gene segments of TRD (hTRGJ1/P1/2/P2/P) were analyzed. An in-group analysis showed significant differences in the frequency of hTRDV1 and hTRDV2 (Figure 5): hTRDV2 was more common and hTRDV1 was less common in LC patients than healthy individuals, suggesting that rearrangement of Vδ2 T cells to increase tumor cytotoxicity were more common in LC patients due to the antitumor immune response.
Figure 5.
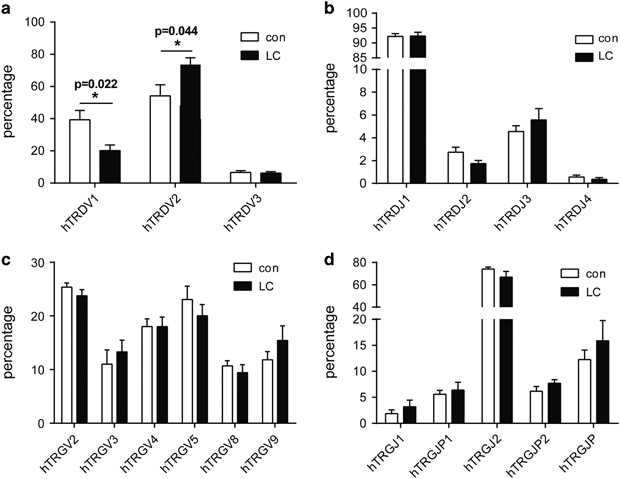
The frequency of germline V and J gene segments in the TCR γδ complementary determinant region 3 (CDR3) repertoire of lung carcinoma (LC) patients and healthy individuals. (a–d) The germline gene usage of three T-cell receptor δ-chain (TRD) V segments (hTRDV1, hTRDV2 and hTRDV3), four TRD J segments (hTRDJ1, hTRDJ2, hTRDJ3 and hTRDJ4), six T-cell receptor γ-chain (TRG) V segments (hTRGV2, hTRGV3, hTRGV4, hTRGV5, hTRGV8 and hTRDV9) and five TRG J segments (hTRGJ1, hTRGJP1, hTRGJ2 and hTRDJP2), respectively. *P<0.05.
Specific features associated with the TCRδ chain repertoire effectively identified LC patients
An in-group analysis of the TCR γδ CDR3 repertoire identified a total of four relatively independent characteristics that significantly differed between the LC and control groups, including the unique CDR3/total CDR3 ratio, top 1 CDR3 frequency, hTRDV1/hTRDV2 ratio and CDR3 clonal size 1–10%/<10% ratio of the TCR δ chain. Thus, we tested the ability of these parameters to effectively distinguish LC patients from healthy individuals. Unexpected clustering results showed that all seven LC patients and seven healthy individuals were clustered together based on these four parameters (Figure 6a), and a principal component analysis showed that the division between groups by principal component 1 based on the four indicators reached 95.9% (Figure 6b). These results show that the specific TCR repertoire features of LC patients primarily focused on the TCR δ chain, suggesting that the key sites by which TCRγδ recognizes tumor antigens may be located in the TCR δ chain.
Figure 6.
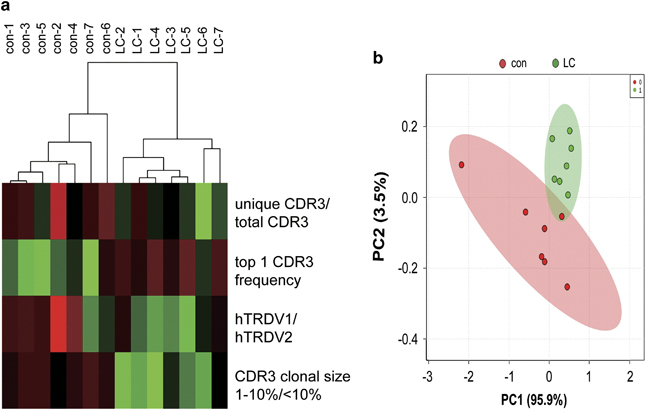
Specific features associated with the TCRδ chain repertoire effectively distinguished lung carcinoma (LC) patients. (a) Unexpected clustering results from the four selected parameters of the complementary determinant region 3δ (CDR3δ) repertoire that significantly differ between LC patients and healthy individuals. (b) Principal component analysis for the four selected parameters. Sample con-1 to con-7 represent seven control samples, and LC-1 to LC-7 represent samples from seven patients with lung cancer. Four selected parameters are unique: CDR3/total CDR3, top 1 CDR3 frequency, hTRDV1/hTRDV2 and top 50 CDR3 clonal size 1–10%/<10%.
Discussion
Since the discovery of γδ T cells in the late 1980s, a significant amount of knowledge has accumulated regarding human γδ T lymphocytes.15 Although they represent only ~5% of peripheral T cells in the blood, epithelial tissues are rich in γδ T cells. The unconventional immune functions of γδ T cells, notably, their histocompatibility leukocyte antigen-unrestricted cytotoxic activity against malignant cells, have implicated them as possible therapeutic targets in cancer. Although they are primarily considered innate immune cells, γδ T cells exhibit a diverse TCR γδ repertoire due to V–(D)–J–C gene rearrangement in the thymus. However, the role of TCR γδ diversity in tumor antigen recognition and the antitumor immune responses of γδ T cells remain unclear. In an earlier study, we characterized the TCR γδ CDR3 repertoire in the normal population via high-throughput sequencing, which showed a wide variety in the TCR γδ CDR3 repertoire and interindividual differences, but this repertoire was significantly less diverse than the TCR αβ and Ig repertoires.12 In this study, we profiled the pattern of the human TCR γδ CDR3 repertoire in LC patients using the same method to clarify the mechanism by which γδ T cells recognize tumor antigens based on TCR γδ.
We found that the CDR3δ repertoire was significantly less diverse in LC patients, and the LC patients shared significantly more CDR3δ sequences, but not CDR3γ sequences, with each other than healthy individuals. This result indicates that CDR3δ has a more important role in the recognition of and response to tumor-associated antigens than CDR3γ. TCR γδ is structurally similar to the B-cell receptor on B lymphocytes and directly binds to its antigens to trigger a rapid response. Heavy-chain CDR3 is the key determinant of the specificity of antigen binding to the B-cell receptor. Because the gene composition of the CDR3δ of TCR γδ and heavy-chain CDR3 of B-cell receptor are similar, CDR3δ has been considered to play a key role in the recognition of tumor antigens by TCR γδ. Our previous studies have demonstrated this hypothesis.16,17,18,19 This study provides substantial evidence focus on TCR γδ repertoire to support this hypothesis.
However, unlike the ‘one-to-one’ specific identification of TCR αβ and Ig, TCR γδ exhibited oligoclonal expansion after antigen stimulation. Human MutS homolog 2 is a tumor-associated antigen recognized by TCR γδ that we identified in 2008.18 We investigated the CDR3δ diversity of human MutS homolog 2-specific γδ T cells and found that the diversity of CDR3δ sequences was limited.9 Although the recognition of antigens by TCR γδ is not strictly specific, LC patients shared more CDR3δ sequences than healthy individuals. These results suggest that specific γδ T cells in the peripheral blood of LC patients undergo clonal expansion in response to an unknown stimulus. These CDR3 sequences shared among LC patients, which are likely related to tumor-associated antigens, can be used to identify more tumor antigens recognized by γδ T cells.
Human γδ T cells are divided into two subtypes, Vδ1 and Vδ2 γδ T cells, whose locations and functions differ. Vδ2 γδ T cells, which are often paired with Vγ9, are a major subset of γδ T cells in the peripheral blood and regarded as a potential candidate for tumor immunotherapy because they exhibit strong antitumor activity against different types of cancer cells. However, Vδ1 cells, which are mainly found in tissues, are reported to regulate immune regulation.20 We used both raw and normalized data to analyze the frequency of specific germline V–J mutations in the TCR γδ repertoire of LC patients, but these data were counted differently. The raw data consist of directly observed read count data, whereas the normalized data count each distinct CDR3 as one, irrespective of how it is observed. In other words, the raw data show the actual subset composition of γδ T cells, whereas normalized data better reflect the TCR γδ repertoire but do not consider the different clone sizes of each CDR. Interestingly, we found that Vδ2 germline gene fragments tended to be even more common in the CDR3 repertoire of LC patients than healthy controls based on the normalized data. This finding suggests that besides being stimulated by certain tumor-related antigens and subsequent expansion in the peripheral blood, more Vδ2 γδ T-ell clones are generated during TCR rearrangement in the thymus of LC patients.
Tumor markers are produced by tumor cells or during tumor development, and a good tumor marker is important for the diagnosis, therapy and monitoring of the tumor. Traditional tumor marker studies have focused on the search for tumor-associated protein antigens, such carcinoembryonic antigen and serum amyloid A protein. Studying the immune repertoire primarily helps to understand the role of T and B lymphocytes in the occurrence and progression of disease and provides a new, immune-based strategy for the diagnosis and treatment of these diseases.21 Our results suggest that some of the features of the TCR γδ repertoire, especially on the TCR δ chain, can also be used as tumor biomarkers. However, this conclusion is only based on the analysis of the TCR γδ repertoire of seven pairs of LC patients and healthy individuals. Given the significant interindividual differences, we are planning to analyze a larger cohort in our follow-up studies.
Compared to γδ T cells in the peripheral blood, tumor-infiltrating γδ T cells are known to mediate potent antitumor activity. Thus, understanding the characteristics of a successful antitumor γδ T-cell response and exploiting this knowledge for patient stratification is increasingly important.22 Several recent study used next-generation sequencing technology to demonstrate that the tumor-infiltrating αβ T-cell repertoire of cancer patients is distinct from that of non-tumor sites and peripheral blood.23,24 Therefore, the characteristics of the repertoire of tumor-infiltrating γδ T cells help to answer biological questions and define predictive biomarkers for cancer immunotherapy.
In this study, we analyzed the characteristics of the TCR γδ CDR3 repertoire of LC patients using high-throughput sequencing and found that the diversity of CDR3δ was significantly reduced in this population. Specifically, these patients shared significantly more CDR3δ sequences with each other, and Vδ2 germline gene fragments tended to be more common, whereas Vδ1 germline gene fragments tended to be less common in this group compared with the control group. In addition, we identified a combination of four TCR γδ repertoire features focused on CDR3δ that may be used as a biomarker for LC diagnosis. Moreover, the CDR3δ sequence shared by LC patients will be used to identify ligands recognized by TCR γδ, which may serve as new therapeutic targets for personalized treatment.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31500725, 81673010, 91542117, 81471574, 31471016), CAMS Central Public Welfare Scientific Research Institute Basal Research Expenses (2016ZX310180-5 and 2017PT31004), the CAMS Initiative for Innovative Medicine (2016-I2M-1-008), the National Key Research and Development Program of China (2016YFA0101001, 2016YFC0903900) and Peking Union Medical College Foundation (No. 3332015111). The funders had no role in the study design, data collection and analysis, decision to publish or preparation of the manuscript. We thank Dr Austin Cape at ASJ Editors for the editing and comments.
Conflict of interest
The authors declare no conflict of interest.
Contributor Information
Jianmin Zhang, Email: jzhang42@gmail.com.
Wei He, Email: hewei@ngd.org.cn.
References
- 1.Fisher JP, Heuijerjans J, Yan M, Gustafsson K, Anderson J. gammadelta T cells for cancer immunotherapy: a systematic review of clinical trials. Oncoimmunology. 2014;3:e27572. doi: 10.4161/onci.27572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zou C, Zhao P, Xiao Z, Han X, Fu F, Fu L. gammadelta T cells in cancer immunotherapy. Oncotarget. 2017;8:8900–8909. doi: 10.18632/oncotarget.13051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Legut M, Cole DK, Sewell AK. The promise of gammadelta T cells and the gammadelta T cell receptor for cancer immunotherapy. Cell Mol Immunol. 2015;12:656–668. doi: 10.1038/cmi.2015.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen H, He W. Human regulatory γδT cells and their functional plasticity in the tumor microenvironment. Cell Mol Immunol. 2017;14:1–3. doi: 10.1038/cmi.2016.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chien YH, Bonneville M. Gamma delta T cell receptors. Cell Mol Life Sci. 2006;63:2089–2094. doi: 10.1007/s00018-006-6020-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vantourout P, Hayday A. Six-of-the-best: unique contributions of gammadelta T cells to immunology. Nat Rev Immunol. 2013;13:88–100. doi: 10.1038/nri3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xi X, Han X, Li L, Zhao Z. Gammadelta T cells response to Mycobacterium tuberculosis in pulmonary tuberculosis patients using preponderant complementary determinant region 3 sequence. Indian J Med Res. 2011;134:356–361. [PMC free article] [PubMed] [Google Scholar]
- 8.Spencer CT, Abate G, Blazevic A, Hoft DF. Only a subset of phosphoantigen-responsive gamma9delta2 T cells mediate protective tuberculosis immunity. J Immunol. 2008;181:4471–4484. doi: 10.4049/jimmunol.181.7.4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen H, Ji X, Cui L, Zhang J, He W. Characterization of complementary determinant region 3delta in human MutS homologue 2-specific gammadelta T cells. Scand J Immunol. 2015;81:121–128. doi: 10.1111/sji.12256. [DOI] [PubMed] [Google Scholar]
- 10.Tyler CJ, Doherty DG, Moser B, Eberl M. Human Vgamma9/Vdelta2 T cells: Innate adaptors of the immune system. Cell Immunol. 2015;296:10–21. doi: 10.1016/j.cellimm.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 11.Friedensohn S, Khan TA, Reddy ST. Advanced methodologies in high-throughput sequencing of immune repertoires. Trends Biotechnol. 2017;35:203–214. doi: 10.1016/j.tibtech.2016.09.010. [DOI] [PubMed] [Google Scholar]
- 12.Chen H, Zou M, Teng D, Zhang J, He W. Characterization of the diversity of T cell receptor gammadelta complementary determinant region 3 in human peripheral blood by immune repertoire sequencing. J Immunol Methods. 2017;443:9–17. doi: 10.1016/j.jim.2017.01.009. [DOI] [PubMed] [Google Scholar]
- 13.Yang Y, Wang C, Yang Q, Kantor AB, Chu H, Ghosn EE, et al. Distinct mechanisms define murine B cell lineage immunoglobulin heavy chain (IgH) repertoires. Elife. 2015;4:e09083. doi: 10.7554/eLife.09083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang C, Sanders CM, Yang Q, Schroeder HW, Jr, Wang E, Babrzadeh F, et al. High throughput sequencing reveals a complex pattern of dynamic interrelationships among human T cell subsets. Proc Natl Acad Sci USA. 2010;107:1518–1523. doi: 10.1073/pnas.0913939107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayday AC. [gamma][delta] cells: a right time and a right place for a conserved third way of protection. Annu Rev Immunol. 2000;18:975–1026. doi: 10.1146/annurev.immunol.18.1.975. [DOI] [PubMed] [Google Scholar]
- 16.Xu C, Zhang H, Hu H, He H, Wang Z, Xu Y, et al. Gammadelta T cells recognize tumor cells via CDR3delta region. Mol Immunol. 2007;44:302–310. doi: 10.1016/j.molimm.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 17.Xi X, Guo Y, Chen H, Xu C, Zhang H, Hu H, et al. Antigen specificity of gammadelta T cells depends primarily on the flanking sequences of CDR3delta. J Biol Chem. 2009;284:27449–27455. doi: 10.1074/jbc.M109.011684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen H, He X, Wang Z, Wu D, Zhang H, Xu C, et al. Identification of human T cell receptor gammadelta-recognized epitopes/proteins via CDR3delta peptide-based immunobiochemical strategy. J Biol Chem. 2008;283:12528–12537. doi: 10.1074/jbc.M708067200. [DOI] [PubMed] [Google Scholar]
- 19.Chen H, You H, Wang L, Zhang X, Zhang J, He W. Chaperonin-containing T-complex protein 1 Subunit zeta serves as an autoantigen recognized by human Vdelta2 gammadelta T cells in autoimmune diseases. J Biol Chem. 2016;291:19985–19993. doi: 10.1074/jbc.M115.700070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mao Y, Yin S, Zhang J, Hu Y, Huang B, Cui L, et al. A new effect of IL-4 on human γδ T cells: promoting regulatory Vδ1 T cells via IL-10 production and inhibiting function of Vδ2 T cells. Cell Mol Immunol. 2015;13:217–228. doi: 10.1038/cmi.2015.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Page DB, Yuan J, Redmond D, Wen YH, Durack JC, Emerson R. Deep sequencing of T-cell receptor DNA as a biomarker of clonally expanded TILs in breast cancer after immunotherapy. Cancer Immunol Res. 2017;5:269. doi: 10.1158/2326-6066.CIR-17-0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poschke I, Flossdorf M, Offringa R. Next-generation TCR sequencing—a tool to understand T-cell infiltration in human cancers. J Pathol. 2016;240:384–386. doi: 10.1002/path.4800. [DOI] [PubMed] [Google Scholar]
- 23.Chen Z, Zhang C, Pan Y, Xu R, Xu C, Chen Z, et al. T cell receptor beta-chain repertoire analysis reveals intratumour heterogeneity of tumour-infiltrating lymphocytes in oesophageal squamous cell carcinoma. J Pathol. 2016;239:450–458. doi: 10.1002/path.4742. [DOI] [PubMed] [Google Scholar]
- 24.Tamura K, Hazama S, Yamaguchi R, Imoto S, Takenouchi H, Inoue Y, et al. Characterization of the T cell repertoire by deep T cell receptor sequencing in tissues and blood from patients with advanced colorectal cancer. Oncol Lett. 2016;11:3643–3649. doi: 10.3892/ol.2016.4465. [DOI] [PMC free article] [PubMed] [Google Scholar]


