Abstract
The immune system is composed of a complex hierarchy of cell types that protect the organism against disease and maintain homeostasis. Identifying heterogeneity of immune cells is the key to understanding the immune system. Advanced single-cell RNA sequencing (scRNA-seq) technologies are revolutionizing our ability to study immunology. By measuring transcriptomes at the single-cell level, scRNA-seq enables identification of cellular heterogeneity in far greater detail than conventional methods. In this review, we introduce the existing scRNA-seq technologies and present their strengths and weaknesses. We also discuss potential applications and future innovations of scRNA-seq in immunology.
Subject terms: Immunology, Biomarkers, Cancer
Introduction
The immune system is a host defense system comprising many immune cells. Technical developments in microscopy and flow cytometry have accelerated the classification of immune cells over the years. However, these methods are still limited by the number of parameters for cell-type definition and the prerequisite of prior knowledge. The classical system is facing the challenge of understanding the complexity of the immune system, including the heterogeneity, development, differentiation, and microenvironment of immune cells in health and disease.1
Recently, the advancement of single-cell RNA sequencing (scRNA-seq) has revolutionized our ability to study the immune system and break through the bottleneck of immunology studies. Individual single cells are classified by transcriptome analysis rather than surface markers. The redefined cell types show the extreme heterogeneity of immune cells, which is an important feature of immunology.2 Now we are in the “Age of Discovery.” Using scRNA-seq, many new cell types and differentiation pathways can be identified.
These findings inspire researchers to improve scRNA-seq technology throughput, sensitivity, precision, cost, and convenience. Several cutting-edge scRNA-seq methods and platforms have been established to satisfy different applications that have distinct requirements.2,3 In this review, we present an overview of existing scRNA-seq technologies and discuss their different strengths and weaknesses. We also describe the main applications of scRNA-seq in immunology and discuss potential future innovations.
Technical advances in scRNA-seq
When studying embryology, immunology, physiology, and pathology, valuable information may be missed with traditional bulk analyses. scRNA-seq provides a solution to comprehensively study multicellular tissues by identifying heterogeneity and characterizing novel cell types in health and disease samples. These single-cell characterizations are important to reconstruct developmental trajectories and cell–cell interactions in tissues. The first scRNA-seq protocol was established by Tang et al.4 in 2009. A large number of technical breakthroughs have leveraged advances in single-cell capture, sample barcoding, cDNA amplification, library preparation, sequencing, etc. They paved the way for the development and optimization of a large variety of scRNA-seq platforms. It is now possible to choose the most suitable technique for a specific scientific question. Here we review several widely used options and discuss their workflow, strengths, weaknesses, and applications.
Principle of scRNA-seq
scRNA-seq is a powerful method for analyzing the cell-specific transcriptome at the single-cell level. The workflow of scRNA-seq consists of single-cell capture, mRNA reverse transcription, cDNA amplification, cDNA library preparation, high-throughput sequencing, and data analysis. The number of sequenced reads, which represents the gene expression level, makes up a digital gene expression matrix for bioinformatic analysis. Each cell type possesses a unique transcriptome that can be presented as a data matrix. Remarkably, current scRNA-seq methods combined with a distinct single-cell capture platform can meet the diverse needs of various types of immunological research.
scRNA-seq methods
There are approximately 10 pg of total RNA (1–5% mRNA) in a typical mammalian cell. Among all the scRNA-seq, synthesis of cDNA from a minute amount of mRNA is obtained by reverse transcription with poly(T) primers. Approximately 10–20% of mRNA is reverse transcribed at this stage.5 The efficiency of reverse transcription determines the sensitivity and precision of scRNA-seq. Three mainstream strategies are used to perform reverse transcription (Table 1). One uses poly(A) tailing followed by PCR, as in the Tang-seq.4,6 Another method uses second-strand synthesis followed by in vitro transcription (IVT), such as CEL-seq/CEL-seq27,8 and MARS-seq.9 However, the premature termination of reverse transcription significantly reduces transcript coverage at the 5’ end.10 A third approach uses a template-switching method, as in STRT-seq11 and Smart-seq/Smart-seq2.10,12 The third approach can reduce 3’ coverage biases originating from incomplete reverse transcription and obtain full-length transcript coverage; it also requires fewer reaction steps, which makes it more popular. However, the sensitivity of template-switching may be lower than the first two methods.13
Table 1.
Advances in single-cell RNA sequencing methods
| Protocol | mRNA reverse transcription | cDNA amplification | Coverage | Reference |
|---|---|---|---|---|
| Tang-seq | Poly(A) tailing + second-strand synthesis | PCR | Full-length mRNA | 4, 6 |
| CEL-seq/CEL-seq2 | Second-strand synthesis | In vitro transcription | 3’ end of mRNA | 7, 8 |
| MARS-seq | Second-strand synthesis | In vitro transcription | 3’ end of mRNA | 9 |
| Smart-seq/Smart-seq2 | Template-switching method | PCR | Full-length mRNA | 10, 12 |
| STRT-seq | Template-switching method | PCR | 3’ or 5’ end of mRNA | 11, 14 |
After reverse transcription, cDNA amplification can be performed using two approaches (Table 1), PCR and IVT. PCR is used in Tang-seq,4,6 STRT-seq,11 and Smart-seq/Smart-seq2.10,12 The approach may introduce amplification bias during PCR cycles. IVT is a linear amplification process that is used in CEL-seq/CEL-seq27,8 and MARS-seq.9 However, it includes additional reverse transcription of the amplified mRNA that may cause 3’ coverage biases. Smart-seq/Smart-seq2, which is widely used for single-cell full-length mRNA analysis, may provide information regarding gene alternative splicing, gene mutation, and allele-specific expression. In the general application of gene expression analysis, gene identification and quantification can be performed using either 3’ (CEL-seq/CEL-seq2 and MARS-seq) or 5’ (STRT-seq14) fragments of genes, which reduces the number of sequenced reads and cost. In a study of T cell receptors (TCRs) and B cell receptors (BCRs), 5’ end sequencing was very important for tracking immune clones.15 To estimate and control the amplification bias, unique molecular identifiers (UMIs) are added to label each individual transcript within a cell during reverse transcription.5 In saturation sequencing, considering the initial mRNA capture efficiency, the absolute copy number of a transcript in a single cell can be counted using the number of UMIs. The repeated sequencing reads that arise from PCR with the same UMIs can be removed in the data analysis process. Several scRNA-seq methods use this strategy, such as MARS-Seq,9 updated STRT-Seq,5 and CEL-seq.8
Single-cell capture methods for sequencing
The single-cell capture strategy determines the cost and scale of single-cell sequencing. Recently, we have seen significant progress in capture platform development (Table 2). When a biological sample contains few cells, such as an early embryo, simple micromanipulation with mouth pipetting4 or laser capture microdissection (LCM)16,17 can isolate single cells quickly and accurately. These manual approaches ensure that each isolation attempt can capture a single cell. The micromanipulation approach does not require expensive instruments. The LCM approach can also provide spatial information of individual cells. However, these manual approaches are technically challenging, labor intensive, time-consuming, and low throughput. To increase throughput, flow cytometry is used to place individual cells into each well of microtiter plates containing lysis buffer. Flow cytometry is able to analyze hundreds of single-cell samples, which can be enriched by fluorescent labels. However, the volume of reaction reagent cannot be scaled down to nanoliter volumes. This leads to a high molecular reagent cost per cell. To save reagent and labor costs, single-cell barcodes are introduced into each cell during reverse transcription.7 After reverse transcription, all of the cDNAs labeled with cell information, which can be traced back to their cellular origins, are pooled together for amplification reactions and library preparation. The single-cell barcoding strategy is widely used in high-throughput scRNA-seq.
Table 2.
Advances in single-cell capture methods
| Capture method | Number of cells per experiment | Operation times | Equipment | Application | Commercialization platforms | Reference |
|---|---|---|---|---|---|---|
| Micromanipulation (mouth pipetting) | Rare samples (~100) | Time-consuming | No | Tang-seq, Smart-seq/Smart-seq2 | — | 4 |
| Laser capture microdissection | Rare samples (~100) | Time-consuming | Yes | Smart-seq/Smart-seq2 | — | 16, 17 |
| Flow cytometry | Hundreds of cells | Fast | Yes | Smart-seq/Smart-seq2, CEL-seq/CEL-seq2, MARS-seq, STRT-seq | — | 8, 9, 11, 12 |
| Integrated microfluidic circuits | Hundreds of cells | Fast | Yes | Smart-seq/Smart-seq2, CEL-seq/CEL-seq2, STRT-seq | E.g., Fluidigm C1 system | 8, 11, 12, 18 |
| Microwell platform | Thousands of cells | Fast | No | Cyto-seq, Seq-well, Microwell-seq | E.g., Rhapsody (BD) | 20– 22 |
| Microdroplet platform | Thousands of cells | Fast | Yes | Drop-seq, inDrop | E.g., Chromium (10x Genomics), Nadia (Dolomite Bio) | 23, 24, 31 |
| In-situ barcoding | Tens of thousands | Fast | No | SPLit-seq, Sci-RNA-seq | — | 34, 35 |
Recently, several commercial platforms have been invented for convenient use. The Fluidigm C1 system, which currently can analyze up to 800 cells at a time, applies integrated fluidic circuits (IFCs) for single-cell capture and mRNA amplification reactions. The highly automated microfluidic chip (nanoliter volumes) saves labor and molecular reagents. The Fluidigm C1 system had high-quality gene expression readouts in our study.18 However, high-throughput platforms always make small compromises in sensitivity and precision,19 which are indicated by gene number and distribution. The capture efficiency of IFC chips is easily affected by cellular characteristics. For cells with substantial differences in size or high viscosity, the Fluidigm C1 system may bias the population examined and have a low capture rate. Moreover, the high cost for the automated device and microfluidic chip also limits its large-scale application.
Cyto-seq, Seq-well, and Microwell-seq20–22 use a microwell array to capture single cells with high-throughput and at a low cost. In these microwell platforms, individual single cells are settled into individual wells by gravity, and then each well is covered by a barcoded magnetic bead that can capture mRNA after cell lysis. Our Microwell-seq platform uses agarose plates with 105 microwells to capture 5–10 thousand individual cells. The cell quality and capture efficiency are estimated under the microscope. Barcoded magnetic beads carrying uniquely oligonucleotides are placed on the plate at a saturated concentration. Because the beads are sized, only one is trapped into each well. After removing excess beads, the cells are lysed, and released mRNA is captured by the beads. The beads are then pooled, followed by reverse transcription (template switching), cDNA amplification (PCR), library preparation, and sequencing. Our Microwell-seq platform has advantages in throughput, convenience, and cost. Using Microwell-seq, we finished the first mammalian single-cell atlas,20 the Mouse Cell Atlas (http://bis.zju.edu.cn/MCA/). The microwell platform is easy to set up without requirements for expensive instruments. It is easily scalable by using larger microwell plates. Recently, BD released the Rhapsody system based on arrays of 200,000 microwells.
Currently, the most popular high-throughput platform is based on droplet-based microfluidics (microdroplets). This strategy uses microfluidic and reverse emulsion devices to isolate individual single cells into thousands of nanoliter droplets that contain lysis buffer and barcoded beads. In 2015, this approach was first applied by two academically developed technologies, known as inDrop23 and Drop-seq.24 Both of them use oil to surround individual aqueous microdroplets (volume of ~2 nl) that contain lysis buffer, barcoded beads, and cells. After cell lysis, the barcoded beads capture released mRNA by the poly(A) tails. For inDrop, the reverse transcription reactions (second-strand synthesis) are performed within the drops, and then cDNAs are collected and amplified by IVT. For Drop-seq, the beads are all released from the drops and pooled for reverse transcription (template-switching mechanism), and then the cDNAs are amplified using PCR. The microdroplet-based methods use massive parallelization to increase throughput and minimize labor and reagent costs, so they have gained popularity, especially in mapping single-cell atlases for multicellular organisms.25–30
Several commercial platforms based on microdroplets are available, including Chromium System (10x Genomics)31 and Nadia (Dolomite Bio, based on Drop-seq). Several comparative analyses of droplet-based high-throughput scRNA-seq systems (Drop-seq, inDrop and 10x Genomics) showed that they perform comparably32,33 and offer satisfactory transcript detection efficiency. Compared with 10x Genomics, both Drop-seq and inDrop have a cost advantage, which is typically the main concern in high-throughput analysis. As a commercialized platform, 10x Genomics, which has a high price tag, has been extensively optimized to show higher molecular sensitivity and precision with less technical noise.
An additional promising platform on the horizon is based on “In situ barcoding.” It adopts several rounds of split-pool to barcode individual single cells with combinatorial indexing in situ. This strategy is implemented in Sci-RNA-seq34 and SPLiT-seq.35 The methods use fixed single cells as the reaction vessels, which reduces equipment requirements. After fixing, cells are randomly divided into a 96-well plate for reverse transcription, which is carried out inside cells with well-specific primers. Then the cells of the plate are pooled. After sufficient mixing, these cells are randomly divided into a new 96-well plate with short, well-specific oligonucleotides. cDNAs from one cell have the same barcode order, which they receive from the split-pool process of barcoding. By increasing the number of split-pool rounds and using 384-well plates, the number of barcode combinations can hit ten million. This platform greatly facilitates ultra-high-throughput single-cell analysis and cuts the cost of library construction for sequencing.
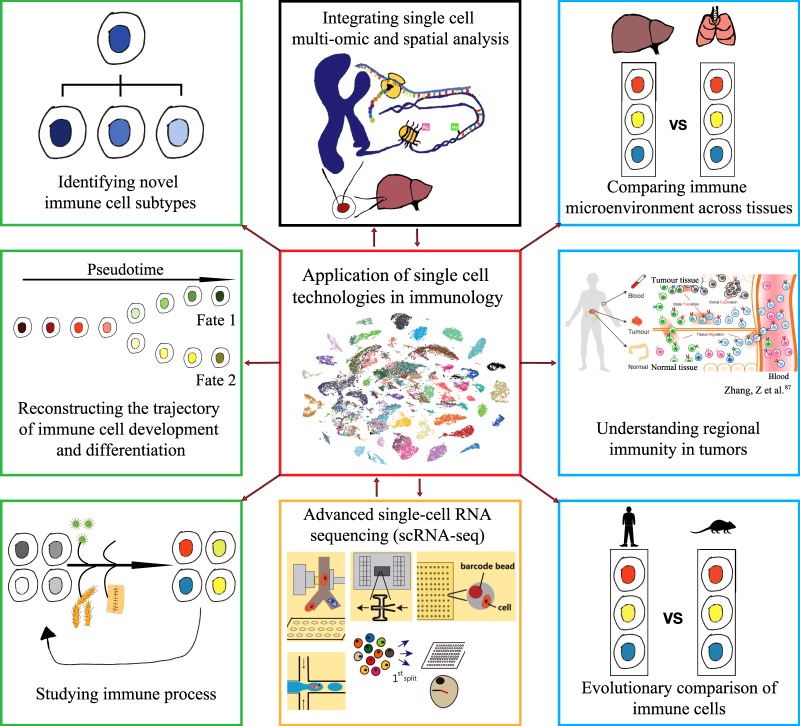
Applications of scRNA-seq in immunology
These scRNA-seq protocols, which show distinct characteristics in throughput, sensitivity, precision, cost, and operability, can meet the needs of different disciplines, such as neurobiology, stem cell biology, and immunology. In this review, we focus on applications in immunology (Fig. 1).
Fig. 1.
Mapping the immune cell atlas by single-cell RNA sequencing (scRNA-seq). The advanced technologies in scRNA-seq allows construction of an immune cell atlas at the single-cell level. The immune cell atlas contains the detailed cellular and molecular signatures of immune cells from different physiological as well as pathological contexts, tissues, individuals, and species. scRNA-seq can also be combined with single-cell multi-omic analysis, and spatial gene expression analysis to promote our understanding of the immune system
Development of the immune system
Our understanding of immune system development has important consequences for both basic research and clinical applications. For example, using the principle that drives immune system development, pluripotent stem cells36 and somatic cells37 can be induced to form immunocompetent hematopoietic stem cells (HSCs), which are widely used to treat disorders of the blood and immune system. The earliest definitive hematopoiesis, which produces HSCs with multilineage potential and long-term reconstitution ability, is detected at mouse embryonic day (E) 10.5 in the aorta–gonad–mesonephros (AGM) region. Recently, Zhou et al. applied scRNA-seq to analyze endothelial cells, pre-HSCs in the AGM region, and HSCs in the fetal liver.38 They found that pre-HSCs have unique features regarding transcription factor regulation, signaling pathways, metabolism, and cell cycle status. They identified a new molecular signature (CD20139,40) for pre-HSC isolation and revealed the importance of mammalian target of rapamycin (mTOR) for the emergence of HSCs. In a similar study, Baron et al. used scRNA-seq to study dynamic gene expression during endothelial hemogenic specification, intra-aortic hematopoietic cluster formation, and pre-HSC maturation.41 These studies pave the way for the dissection of complex molecular changes occurring in the successive steps leading to HSC formation in vivo and will inform future efforts of HSC production in vitro for clinical applications.
Several groups also describe the new use of scRNA-seq in the study of physiological hematopoiesis.42–48 In the classical model of hematopoiesis, a highly organized and hierarchical hematopoietic lineage tree starts with long-term HSCs, followed by multipotent, oligopotent, bipotent, and unipotent progenitors. scRNA-seq provides gene expression profiling to define molecular phenotypes of cells that extend beyond surface markers. The existence of these oligopotent and bipotent progenitors has become controversial.49 Paul et al.45 combined index sorting with MARS-seq to analyze bone marrow cells (KIT+SCA1−Lineage−) and found a high heterogeneity of gene expression within the common myeloid progenitors (CMPs). These CMPs are largely transcriptionally committed toward a single distinct myeloid fate but are not progenitors with a mixed state. The comprehensive data challenge traditional CMPs, which are defined by cell surface markers. Nestorowa et al.47 used a similar approach to map a single-cell resolution expression atlas of early blood stem cell differentiation in mouse bone marrow and provided new insights into dynamic gene expression during HSC differentiation. Notta et al.48 combined a single-cell functional assay and single-cell transcriptome analysis to study human hematopoiesis. They found that the hematopoietic hierarchies are distinct across human development. Prenatally, the megakaryocytic lineage is derived from HSCs and multipotent progenitors. By adulthood, the megakaryocytic activity only comes from HSCs. In a recent scRNA-seq study of human hematopoiesis, Velten et al.44 found that individual HSCs gradually acquire lineage biases without passing through oligopotent or bipotent progenitors. This indicates a continuous process of hematopoiesis downstream of HSCs. In a similar human hematopoiesis study, Karamitros et al.43 suggest the existence of bipotent and rare multilineage progenitors among lymphoid-primed multipotential progenitors, granulocyte-macrophage progenitors, and multilymphoid progenitors. In a more recent study, Jason et al.42 integrated single-cell transcriptomics and chromatin accessibility analysis to identify continuous differentiation trajectories in early human hematopoiesis. The trajectory of “HSC–CMP–MEP” provides evidence for common progenitors of erythroid and myeloid lineages. scRNA-seq is also widely used to study hematopoiesis in zebrafish.50,51 Altogether, single-cell analysis helps to identify the heterogeneity of progenitors and rebuild the hematopoietic lineage hierarchy, which indicates a more sophisticated way of immune cell differentiation.
The trajectory analysis is also used to study the development and differentiation of individual immune lineages, including the early myeloid,52–56 lymphoid,57–60 and megakaryocytic/erythroid lineage.61,62 Ginhoux et al.53,54 combined scRNA-seq and CyTOF to unravel the developmental pathways of the human dendritic cell (DC) lineage. In their model, common DC progenitors in the bone marrow diverge at the point of emergence of pre-DCs and plasmacytoid DC potential. The pre-DCs are heterogeneous and contain one early pre-DC subset and two functionally and phenotypically distinct lineage-committed subsets (pre-cDC1 and pre-cDC2). The discovery of multiple committed pre-DC populations and the mechanism of lineage differentiation open new avenues for therapeutic application. Jaitin et al.55 applied CRISP-seq, an integrated method of CRISPR-Pooled Screens and scRNA-seq, to track the development of myeloid cells and identify the regulatory circuits for lineage differentiation. Olsson et al.56 used scRNA-seq to delineate the myeloid developmental hierarchy in mice. They identified the mixed-lineage states in myeloid progenitors and proposed that these progenitors are obligatory during cell-fate specification.
scRNA-seq is also used in the study of immune cell aging. Grover et al.63 compared the transcriptomes between young and old HSCs at the single-cell level. They observed that aged HSCs are highly biased toward megakaryocyte and platelet differentiation, which can be rescued by deletion of the platelet transcription factor FOG1. Recently, Martinez et al.64 used scRNA-seq to analyze unstimulated and stimulated CD4+ T cells in young and old mice. They found that the lack of coordination in aged CD4+ T cells is responsible for impaired immune performance in old mice. Aged CD4+ T cells with increased cell-to-cell transcriptional variability respond to immune stimulation more variably, which weakens their collective effectiveness. scRNA-seq can better explain mechanisms behind aging that have not been studied before.
Cellular heterogeneity in the immune system
To fight different pathogens and disorders efficiently, immune cells have extreme heterogeneity. The heterogeneity is typically defined by a panel of surface markers, which can be measured using flow cytometry or mass cytometry. However, these approaches are limited by the number of chosen markers. Now, a more powerful approach to perform cell-type identification is based on single-cell transcriptomic techniques, which can dissect cellular heterogeneity in far greater detail without prior knowledge of the genes of interest. In human peripheral blood mononuclear cells, Villani et al.65 found new types of DCs, monocytes, and progenitors using unbiased scRNA-seq. In addition, non-immune organs (such as the liver66) also have specialized immunologic features, including immune cell types and their distinct subsets, localization, and function. It is time to map a single-cell atlas for immune cells across all mammalian tissues that can identify all regional immune cells and systematically characterize their localization, gene expression, and function in distinct organs. Moreover, an atlas would be a valuable resource for both basic research and clinical applications in immunology.
In our recent work, we used our Microwell-seq platform to construct a single-cell mouse cell atlas that covers most immune cells from major tissues and organs.20 We analyzed the cross-tissue heterogeneity of tissue-resident macrophages and found that tissue-resident macrophages increase their specialization to adapt to the microenvironment and immune functions in different tissues. Mass et al.67 used scRNA-seq to study the early development of tissue-resident macrophages and found heterogeneity of transcriptional regulation in tissue-resident macrophages across different tissues. De et al.68 combined single-cell gene expression and imaging studies to identify a self-maintaining population of tissue-resident macrophages in the gut that support enteric neurons and the submucosal vasculature. Cohen et al.69 mapped the single-cell atlas of lung cells during development (E 12.5–Day 7) and found three different subsets of macrophages. Moreover, using ligand–receptor interaction analysis, they found that lung resident basophils are important regulators of alveolar macrophage imprinting. The immune cells of the placenta play important regulatory roles in reproductive success. Vento et al.70 finished the first comprehensive single-cell atlas of the placenta between 6 and 14 weeks of gestation. Three major subsets of decidual natural killer cells (CD39+ dNK1, ANXA1+ dNK2, and CD160+ dNK3) were defined. dNK1, with high expression of glycolytic enzymes and HLA I receptors, may be responsible for the different reproductive outcomes between a first pregnancy and subsequent pregnancies. In both human tonsils71 and mouse small intestine,72 a heterogeneity of CD127+ innate lymphoid cells was also identified using scRNA-seq. These results provide critical insights in the heterogeneity of tissue-resident immune cells and demonstrate distinct roles of immune cells in development, homeostasis, and physiology across tissues.
Immune cells in diseases
One important role of the immune cell is to fight diseases, such as cancer, microbial infection, and cell damage. The preliminary single-cell studies of the composition and development of the immune system in healthy organisms paved the way for profiling immune cells in pathological tissues, which may allow for the identification of molecular drivers of disease, the characterization of regional immune escape, and a better understanding of the generation and progression of various diseases.
Infiltrated immune cells in pathological tissues directly contact pathogens. Single-cell studies on infiltrated lymphoid and myeloid cells have provided new insights into diagnosis and treatment of disease. Both Gaublomme et al.73 and Karmaus et al.74 combined scRNA-seq and mouse experimental autoimmune encephalomyelitis models (a model for human multiple sclerosis) to identify the heterogeneity of pathogenic T helper 17 (Th17) cells. Gaublomme et al. found that pathogenic Th17 have a wide spectrum of pathogenicity, spanning from more regulatory to more pathogenic cells. They also identified several candidate genes (Gpr65, Plzp, Toso, and Cd5l) related to Th17 pathogenicity. Karmaus et al. found that Th17 are phenotypically, transcriptionally, and metabolically heterogeneous, including CD27+ and CD27− subsets. CD27+ Th17 cells with inferred stemness features and low anabolic metabolism highly express the transcription factor TCF-1. The reciprocal CD27− subset specifically expresses T-bet. The transition from CD27+ Th17 to CD27− Th17 cells is mediated by mTORC1. These results highlight that metabolism regulates the stability and plasticity of Th17 cells. From a therapeutic perspective, identification of heterogeneous Th17 cells and candidate genes and signaling pathways provides new targets for treatment of excessive inflammation. In breast cancer, Savas et al.75 found significant heterogeneity in the infiltrating T cell population. A subset of CD8+ T cells with features of tissue-resident memory T (TRM) cells express high levels of immune checkpoint molecules and effector proteins. These cells are significantly associated with patient survival and prognosis. Azizi et al.76 also used scRNA-seq to map the breast cancer immune cell atlas. They observed the phenotypic expansion of intratumoral immune cells, both lymphoid and myeloid cell lineages. Using mouse cancer models (colon carcinoma and melanoma), Singer et al.77 defined distinct gene expression modules (dysfunction, activation, act/dys, and neither) that are enriched in different subsets of CD8+ tumor-infiltrating lymphocytes. Tirosh et al.78 analyzed tumor-infiltrating T cells to reveal exhaustion programs in human metastatic melanoma. Sade et al.79 profiled single-cell transcriptomes of immune cells from melanoma patients treated with checkpoint inhibitors. They found that the transcription factor TCF7 can predict positive clinical outcomes of immunotherapy. Using scRNA-seq, Lonnberg et al.80 reconstructed the developmental trajectories of Th1 and Tfh (T follicular helper) cells in a mouse malaria model. Pathological macrophages are studied in infection81,82 and mouse Alzheimer models.83 Using scRNA-seq and mass cytometry, Winkels et al.84 mapped the immune cell repertoire in mouse atherosclerosis.
The diverse repertoire of TCRs and BCRs, assembled by random V(D)J recombination, can be used to illustrate T and B cell85 clonal expansion patterns and lineage tracing. The combination of full-length (or 5’ complementarity determining regions) TCR or BCR sequencing86 and high-throughput scRNA-seq is a nontrivial challenge. Based on both single-cell transcriptome and TCR sequencing, Zhang et al.87–89 developed an integrated approach, single T cell analysis by RNA sequencing and TCR tracking (STARTRAC), to track the dynamic T cell subsets identified in liver cancer,88 non-small-cell lung cancer,87 and colorectal cancer.89 In liver cancer, they identified specific markers for regulatory T cells (Tregs; CCR8 and LAYN) and exhausted CD8+ T (LAYN) cells. Based on clonal TCRs, they found that tumor-infiltrating exhausted CD8+ T cells mainly evolve from other types of CD8+ T cells inside the tumor, whereas Tregs are more likely recruited from the periphery. Moreover, they identified a GZMK+CD8+ T cell cluster as a transition state from effector to exhausted T cells. In non-small-cell lung cancer, they identified two clusters of possible “pre-exhausted” CD8+ T cells, which are associated with a better prognosis of lung adenocarcinoma. The tumor Tregs also show heterogeneity with a bimodal distribution of TNFRSF9, a marker for activated tumor Tregs. TNFRSF9+ Tregs highly express IL1R2, which is correlated with poor prognosis. In colorectal cancer, they identified two IFNγ+ Th1-like cell clusters in tumors, including a GZMK+ effector memory T cell cluster and a CXCL13+BHLHE40+ Th1-like cell cluster. The latter cluster is highly enriched in microsatellite-instable patients and may contribute to the favorable response to immune-checkpoint blockade therapies. Moreover, they compared T cell populations across cancers and found that T cell patterns were distinct in both tumors and adjacent normal tissues. In colorectal and liver cancers, exhausted CD8+ T cells and CD4+ Treg cells are enriched, whereas non-small-cell lung cancer enriches PDCD1−CTLA4−ZNF683+ TRM cells. This suggests the influence of organ-specific immune characteristics on tumor immunity and immunotherapies. In a similar study, Li et al.90 combined scRNA-seq and TCR-seq to analyze tumor-infiltrating immune cells in human melanoma. They identified the “bystander” cytotoxic T cells and established the trajectories of CD8+ T cell dysfunction. Dysfunctional CD8+ T cells, previously defined as exhausted CD8+ T cells, do not form a specific cell population but are highly proliferating, clonal, and dynamically differentiating. Neal et al.15 combined 5’ scRNA-seq and 5’ V(D)J-seq to determine the tumor-immune microenvironment in tumor organoid models. These applications, which link repertoire and transcriptome information at the single-cell level, open new avenues for investigation of tumor regional immunity (Fig. 1).
Both leukemia and myeloma are tumors of the blood system. scRNA-seq is also widely used to identify the heterogeneity of cancerous immune cells. De et al.91 analyzed four T cell acute lymphoblastic leukemia (T-ALL) samples, in which T-ALL cells show transcriptional uniformity. They illuminated the mutational hierarchy and found that, in half of the cases, T-ALL development starts from a multipotent progenitor cell. Giustacchini et al.92 combined single-cell transcriptome analysis with high-sensitivity mutation detection to reveal the heterogeneity of cancer stem cells in chronic myeloid leukemia. They also identified a blast-crisis-specific stem cell population. Ledergor et al.93 applied scRNA-seq to study the heterogeneity of plasma cells from multiple myeloma. They found a distinct molecular characterization of tumor cells in symptomatic and asymptomatic patients.
Conclusions and perspectives
Recent studies in immunology have indicated that scRNA-seq analysis is a powerful tool for reconstructing the trajectory of development, differentiation, and identification of heterogeneity in health and disease. By combining single-cell analysis of the proteome,94,95 genome,96,97 epigenome,98,99 and spatial location,16,17,100 scRNA-seq will continue to promote our understanding of the immune system (Fig. 1). The ultimate application for scRNA-seq is to map the immune cell atlas, which contains the detailed cellular and molecular signatures of immune cells from different physiological and pathological contexts, tissues, and species (Fig. 1). Single-cell transcriptional profiling of dynamic immune cells can characterize transcriptional transition states and reconstruct a high-resolution trajectory. A molecular map for the specific immune cell types and gene regulation may provide robust cellular and molecular targets for disease diagnosis and treatment. Understanding the differences in the immune microenvironment across tissues will guide and accelerate both basic research and clinical applications for cancer. Moreover, we can compare the immune cell atlas of different species to explore the evolution of the immune system.
Acknowledgements
We thank Z Tian, X Cao, and F Gao for suggestions. This work was supported by the Natural Science Foundation of China (91842301, 31722027, and 81770188).
Competing interests
The authors declare no competing interests.
References
- 1.Stubbington MJT, Rozenblatt-Rosen O, Regev A, Teichmann SA. Single-cell transcriptomics to explore the immune system in health and disease. Science. 2017;358:58–63. doi: 10.1126/science.aan6828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Papalexi E, Satija R. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat. Rev. Immunol. 2018;18:35–45. doi: 10.1038/nri.2017.76. [DOI] [PubMed] [Google Scholar]
- 3.Proserpio V, Mahata B. Single-cell technologies to study the immune system. Immunology. 2016;147:133–140. doi: 10.1111/imm.12553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tang F, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods. 2009;6:377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 5.Islam S, et al. Quantitative single-cell RNA-seq with unique molecular identifiers. Nat. Methods. 2014;11:163–166. doi: 10.1038/nmeth.2772. [DOI] [PubMed] [Google Scholar]
- 6.Tang F, et al. RNA-Seq analysis to capture the transcriptome landscape of a single cell. Nat. Protoc. 2010;5:516–535. doi: 10.1038/nprot.2009.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hashimshony T, Wagner F, Sher N, Yanai I. CEL-Seq: single-cell RNA-Seq by multiplexed linear amplification. Cell Rep. 2012;2:666–673. doi: 10.1016/j.celrep.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 8.Hashimshony T, et al. CEL-Seq2: sensitive highly-multiplexed single-cell RNA-Seq. Genome Biol. 2016;17:77. doi: 10.1186/s13059-016-0938-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jaitin DA, et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science. 2014;343:776–779. doi: 10.1126/science.1247651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramsköld D, et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat. Biotechnol. 2012;30:777–782. doi: 10.1038/nbt.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Islam S, et al. Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq. Genome Res. 2011;21:1160–1167. doi: 10.1101/gr.110882.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Picelli S, et al. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat. Methods. 2013;10:1096–1098. doi: 10.1038/nmeth.2639. [DOI] [PubMed] [Google Scholar]
- 13.Streets AM, et al. Microfluidic single-cell whole-transcriptome sequencing. Proc. Natl. Acad. Sci. USA. 2014;111:7048–7053. doi: 10.1073/pnas.1402030111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Islam S, et al. Highly multiplexed and strand-specific single-cell RNA 5’ end sequencing. Nat. Protoc. 2012;7:813–828. doi: 10.1038/nprot.2012.022. [DOI] [PubMed] [Google Scholar]
- 15.Neal JT, et al. Organoid modeling of the tumor immune microenvironment. Cell. 2018;175:1972–1988 e1916. doi: 10.1016/j.cell.2018.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peng G, et al. Spatial transcriptome for the molecular annotation of lineage fates and cell identity in mid-gastrula mouse embryo. Dev. Cell. 2016;36:681–697. doi: 10.1016/j.devcel.2016.02.020. [DOI] [PubMed] [Google Scholar]
- 17.Chen J, et al. Spatial transcriptomic analysis of cryosectioned tissue samples with Geo-seq. Nat. Protoc. 2017;12:566–580. doi: 10.1038/nprot.2017.003. [DOI] [PubMed] [Google Scholar]
- 18.Han X, et al. Mapping human pluripotent stem cell differentiation pathways using high throughput single-cell RNA-sequencing. Genome Biol. 2018;19:47. doi: 10.1186/s13059-018-1426-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ziegenhain C, et al. Comparative analysis of single-cell RNA sequencing methods. Mol. Cell. 2017;65:631–643 e634. doi: 10.1016/j.molcel.2017.01.023. [DOI] [PubMed] [Google Scholar]
- 20.Han X, et al. Mapping the mouse cell atlas by microwell-seq. Cell. 2018;172:1091.e17–1107.e17. doi: 10.1016/j.cell.2018.02.001. [DOI] [PubMed] [Google Scholar]
- 21.Gierahn TM, et al. Seq-well: portable, low-cost RNA sequencing of single cells at high throughput. Nat. Methods. 2017;14:395–398. doi: 10.1038/nmeth.4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fan HC, Fu GK, Fodor SP. Combinatorial labeling of single cells for gene expression cytometry. Science. 2015;347:1258367. doi: 10.1126/science.1258367. [DOI] [PubMed] [Google Scholar]
- 23.Klein AM, et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161:1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Macosko EZ, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Briggs, J. A., et al. The dynamics of gene expression in vertebrate embryogenesis at single-cell resolution. Science360, eaar5780 (2018). [DOI] [PMC free article] [PubMed]
- 26.Wagner, D. E. et al. Single-cell mapping of gene expression landscapes and lineage in the zebrafish embryo. Science360, 981–987 (2018). [DOI] [PMC free article] [PubMed]
- 27.Tosches MA, et al. Evolution of pallium, hippocampus, and cortical cell types revealed by single-cell transcriptomics in reptiles. Science. 2018;360:881–888. doi: 10.1126/science.aar4237. [DOI] [PubMed] [Google Scholar]
- 28.Plass, M. et al. Cell type atlas and lineage tree of a whole complex animal by single-cell transcriptomics. Science360, eaaq1723 (2018). [DOI] [PubMed]
- 29.Fincher, C. T., Wurtzel, O., de Hoog, T., Kravarik, K. M. & Reddien, P. W. Cell type transcriptome atlas for the planarian Schmidtea mediterranea. Science360, eaaq1736 (2018). [DOI] [PMC free article] [PubMed]
- 30.Farrell, J. A. et al Single-cell reconstruction of developmental trajectories during zebrafish embryogenesis. Science360, eaar3131 (2018). [DOI] [PMC free article] [PubMed]
- 31.Zheng GX, et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 2017;8:14049. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang, X. et al. Comparative analysis of droplet-based ultra-high-throughput single-cell RNA-seq systems. Mol. Cell73, 130.e5–142.e5 (2018). [DOI] [PubMed]
- 33.Magella B, et al. Cross-platform single cell analysis of kidney development shows stromal cells express Gdnf. Dev. Biol. 2018;434:36–47. doi: 10.1016/j.ydbio.2017.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cao J, et al. Comprehensive single-cell transcriptional profiling of a multicellular organism. Science. 2017;357:661–667. doi: 10.1126/science.aam8940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosenberg AB, et al. Single-cell profiling of the developing mouse brain and spinal cord with split-pool barcoding. Science. 2018;360:176–182. doi: 10.1126/science.aam8999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sugimura R, et al. Haematopoietic stem and progenitor cells from human pluripotent stem cells. Nature. 2017;545:432–438. doi: 10.1038/nature22370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lis R, et al. Conversion of adult endothelium to immunocompetent haematopoietic stem cells. Nature. 2017;545:439–445. doi: 10.1038/nature22326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou F, et al. Tracing haematopoietic stem cell formation at single-cell resolution. Nature. 2016;533:487–492. doi: 10.1038/nature17997. [DOI] [PubMed] [Google Scholar]
- 39.Fares I, et al. EPCR expression marks UM171-expanded CD34(+) cord blood stem cells. Blood. 2017;129:3344–3351. doi: 10.1182/blood-2016-11-750729. [DOI] [PubMed] [Google Scholar]
- 40.Martin GH, Park CY. EPCR: a novel marker of cultured cord blood HSCs. Blood. 2017;129:3279–3280. doi: 10.1182/blood-2016-09-737700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baron CS, et al. Single-cell transcriptomics reveal the dynamic of haematopoietic stem cell production in the aorta. Nat. Commun. 2018;9:2517. doi: 10.1038/s41467-018-04893-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buenrostro JD, et al. Integrated single-cell analysis maps the continuous regulatory landscape of human hematopoietic differentiation. Cell. 2018;173:1535.e16–1548.e16. doi: 10.1016/j.cell.2018.03.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karamitros D, et al. Single-cell analysis reveals the continuum of human lympho-myeloid progenitor cells. Nat. Immunol. 2018;19:85–97. doi: 10.1038/s41590-017-0001-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Velten L, et al. Human haematopoietic stem cell lineage commitment is a continuous process. Nat. Cell Biol. 2017;19:271–281. doi: 10.1038/ncb3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paul F, et al. Transcriptional heterogeneity and lineage commitment in myeloid progenitors. Cell. 2015;163:1663–1677. doi: 10.1016/j.cell.2015.11.013. [DOI] [PubMed] [Google Scholar]
- 46.Haghverdi L, Buttner M, Wolf FA, Buettner F, Theis FJ. Diffusion pseudotime robustly reconstructs lineage branching. Nat. Methods. 2016;13:845–848. doi: 10.1038/nmeth.3971. [DOI] [PubMed] [Google Scholar]
- 47.Nestorowa S, et al. A single-cell resolution map of mouse hematopoietic stem and progenitor cell differentiation. Blood. 2016;128:e20–e31. doi: 10.1182/blood-2016-05-716480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Notta F, et al. Distinct routes of lineage development reshape the human blood hierarchy across ontogeny. Science. 2016;351:aab2116. doi: 10.1126/science.aab2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haas S, Trumpp A, Milsom MD. Causes and consequences of hematopoietic stem cell heterogeneity. Cell Stem Cell. 2018;22:627–638. doi: 10.1016/j.stem.2018.04.003. [DOI] [PubMed] [Google Scholar]
- 50.Macaulay IC, et al. Single-cell RNA-sequencing reveals a continuous spectrum of differentiation in hematopoietic cells. Cell Rep. 2016;14:966–977. doi: 10.1016/j.celrep.2015.12.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Athanasiadis EI, et al. Single-cell RNA-sequencing uncovers transcriptional states and fate decisions in haematopoiesis. Nat. Commun. 2017;8:2045. doi: 10.1038/s41467-017-02305-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Drissen R, et al. Distinct myeloid progenitor-differentiation pathways identified through single-cell RNA sequencing. Nat. Immunol. 2016;17:666–676. doi: 10.1038/ni.3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schlitzer A, et al. Identification of cDC1- and cDC2-committed DC progenitors reveals early lineage priming at the common DC progenitor stage in the bone marrow. Nat. Immunol. 2015;16:718–728. doi: 10.1038/ni.3200. [DOI] [PubMed] [Google Scholar]
- 54.See, P. et al. Mapping the human DC lineage through the integration of high-dimensional techniques. Science356, eaag3009 (2017). [DOI] [PMC free article] [PubMed]
- 55.Jaitin DA, et al. Dissecting immune circuits by linking CRISPR-pooled screens with single-cell RNA-seq. Cell. 2016;167:1883–1896 e1815. doi: 10.1016/j.cell.2016.11.039. [DOI] [PubMed] [Google Scholar]
- 56.Olsson A, et al. Single-cell analysis of mixed-lineage states leading to a binary cell fate choice. Nature. 2016;537:698–702. doi: 10.1038/nature19348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kristiansen TA, et al. Cellular barcoding links B-1a B cell potential to a fetal hematopoietic stem cell state at the single-cell level. Immunity. 2016;45:346–357. doi: 10.1016/j.immuni.2016.07.014. [DOI] [PubMed] [Google Scholar]
- 58.Bendall SC, et al. Single-cell trajectory detection uncovers progression and regulatory coordination in human B cell development. Cell. 2014;157:714–725. doi: 10.1016/j.cell.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stubbington MJT, et al. T cell fate and clonality inference from single-cell transcriptomes. Nat. Methods. 2016;13:329–332. doi: 10.1038/nmeth.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu Y, et al. Single-cell RNA-seq identifies a PD-1(hi) ILC progenitor and defines its development pathway. Nature. 2016;539:102–106. doi: 10.1038/nature20105. [DOI] [PubMed] [Google Scholar]
- 61.Psaila B, et al. Single-cell profiling of human megakaryocyte-erythroid progenitors identifies distinct megakaryocyte and erythroid differentiation pathways. Genome Biol. 2016;17:83. doi: 10.1186/s13059-016-0939-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tusi BK, et al. Population snapshots predict early haematopoietic and erythroid hierarchies. Nature. 2018;555:54–60. doi: 10.1038/nature25741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Grover A, et al. Single-cell RNA sequencing reveals molecular and functional platelet bias of aged haematopoietic stem cells. Nat. Commun. 2016;7:11075. doi: 10.1038/ncomms11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Martinez-Jimenez CP, et al. Aging increases cell-to-cell transcriptional variability upon immune stimulation. Science. 2017;355:1433–1436. doi: 10.1126/science.aah4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Villani, A. C. et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science356, eaah4573 (2017). [DOI] [PMC free article] [PubMed]
- 66.Gao B, Jeong WI, Tian Z. Liver: an organ with predominant innate immunity. Hepatology. 2008;47:729–736. doi: 10.1002/hep.22034. [DOI] [PubMed] [Google Scholar]
- 67.Mass, E. et al. Specification of tissue-resident macrophages during organogenesis. Science353, aaf4238 (2016). [DOI] [PMC free article] [PubMed]
- 68.De Schepper, S. et al. Self-maintaining gut macrophages are essential for intestinal homeostasis. Cell175, 400.e13–415.e13 (2018). [DOI] [PubMed]
- 69.Cohen M, et al. Lung single-cell signaling interaction map reveals basophil role in macrophage imprinting. Cell. 2018;175:1031.e18–1044.e18. doi: 10.1016/j.cell.2018.09.009. [DOI] [PubMed] [Google Scholar]
- 70.Vento-Tormo R, et al. Single-cell reconstruction of the early maternal-fetal interface in humans. Nature. 2018;563:347–353. doi: 10.1038/s41586-018-0698-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bjorklund AK, et al. The heterogeneity of human CD127(+) innate lymphoid cells revealed by single-cell RNA sequencing. Nat. Immunol. 2016;17:451–460. doi: 10.1038/ni.3368. [DOI] [PubMed] [Google Scholar]
- 72.Gury-BenAri M, et al. The spectrum and regulatory landscape of intestinal innate lymphoid cells are shaped by the microbiome. Cell. 2016;166:1231.e13–1246.e13. doi: 10.1016/j.cell.2016.07.043. [DOI] [PubMed] [Google Scholar]
- 73.Gaublomme JT, et al. Single-cell genomics unveils critical regulators of Th17 cell pathogenicity. Cell. 2015;163:1400–1412. doi: 10.1016/j.cell.2015.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Karmaus, P. W. F. et al. Metabolic heterogeneity underlies reciprocal fates of Th17 cell stemness and plasticity. Nature565, 101–105 (2018). [DOI] [PMC free article] [PubMed]
- 75.Savas P, et al. Single-cell profiling of breast cancer T cells reveals a tissue-resident memory subset associated with improved prognosis. Nat. Med. 2018;24:986–993. doi: 10.1038/s41591-018-0078-7. [DOI] [PubMed] [Google Scholar]
- 76.Azizi E, et al. Single-cell map of diverse immune phenotypes in the breast tumor microenvironment. Cell. 2018;174:1293.e36–1308.e36. doi: 10.1016/j.cell.2018.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Singer M, et al. A distinct gene module for dysfunction uncoupled from activation in tumor-infiltrating T cells. Cell. 2017;171:1221–1223. doi: 10.1016/j.cell.2017.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tirosh I, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352:189–196. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sade-Feldman M, et al. Defining T cell states associated with response to checkpoint immunotherapy in melanoma. Cell. 2018;175:998.e20–1013.e20. doi: 10.1016/j.cell.2018.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lonnberg, T. et al. Single-cell RNA-seq and computational analysis using temporal mixture modelling resolves Th1/Tfh fate bifurcation in malaria. Sci. Immunol. 2, eaal2192 (2017). [DOI] [PMC free article] [PubMed]
- 81.Avraham R, et al. Pathogen cell-to-cell variability drives heterogeneity in host immune responses. Cell. 2015;162:1309–1321. doi: 10.1016/j.cell.2015.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Saliba AE, et al. Single-cell RNA-seq ties macrophage polarization to growth rate of intracellular Salmonella. Nat. Microbiol. 2016;2:16206. doi: 10.1038/nmicrobiol.2016.206. [DOI] [PubMed] [Google Scholar]
- 83.Keren-Shaul H, et al. A unique microglia type associated with restricting development of Alzheimer’s disease. Cell. 2017;169:1276.e17–1290.e17. doi: 10.1016/j.cell.2017.05.018. [DOI] [PubMed] [Google Scholar]
- 84.Winkels H, et al. Atlas of the immune cell repertoire in mouse atherosclerosis defined by single-cell RNA-sequencing and mass cytometry. Circ. Res. 2018;122:1675–1688. doi: 10.1161/CIRCRESAHA.117.312513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Croote D, Darmanis S, Nadeau KC, Quake SR. High-affinity allergen-specific human antibodies cloned from single IgE B cell transcriptomes. Science. 2018;362:1306–1309. doi: 10.1126/science.aau2599. [DOI] [PubMed] [Google Scholar]
- 86.De Simone M, Rossetti G, Pagani M. Single cell T cell receptor sequencing: techniques and future challenges. Front. Immunol. 2018;9:1638. doi: 10.3389/fimmu.2018.01638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Guo, X. et al. Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nat. Med. 24, 978–985 (2018). [DOI] [PubMed]
- 88.Zheng C, et al. Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell. 2017;169:1342.e16–1356.e16. doi: 10.1016/j.cell.2017.05.035. [DOI] [PubMed] [Google Scholar]
- 89.Zhang L, et al. Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature. 2018;564:268–272. doi: 10.1038/s41586-018-0694-x. [DOI] [PubMed] [Google Scholar]
- 90.Li, H. et al. Dysfunctional CD8 T cells form a proliferative, dynamically regulated compartment within human melanoma. Cell176, 775.e18–789.e18 (2018). [DOI] [PMC free article] [PubMed]
- 91.De Bie, J. et al. Single-cell sequencing reveals the origin and the order of mutation acquisition in T-cell acute lymphoblastic leukemia. Leukemia32, 1358–1369 (2018). [DOI] [PMC free article] [PubMed]
- 92.Giustacchini A, et al. Single-cell transcriptomics uncovers distinct molecular signatures of stem cells in chronic myeloid leukemia. Nat. Med. 2017;23:692–702. doi: 10.1038/nm.4336. [DOI] [PubMed] [Google Scholar]
- 93.Ledergor G, et al. Single cell dissection of plasma cell heterogeneity in symptomatic and asymptomatic myeloma. Nat. Med. 2018;24:1867–1876. doi: 10.1038/s41591-018-0269-2. [DOI] [PubMed] [Google Scholar]
- 94.Peterson VM, et al. Multiplexed quantification of proteins and transcripts in single cells. Nat. Biotechnol. 2017;35:936–939. doi: 10.1038/nbt.3973. [DOI] [PubMed] [Google Scholar]
- 95.Stoeckius, M. et al. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods14, 865–868 (2017). [DOI] [PMC free article] [PubMed]
- 96.Macaulay IC, et al. G&T-seq: parallel sequencing of single-cell genomes and transcriptomes. Nat. Methods. 2015;12:519–522. doi: 10.1038/nmeth.3370. [DOI] [PubMed] [Google Scholar]
- 97.Dey SS, Kester L, Spanjaard B, Bienko M, van Oudenaarden A. Integrated genome and transcriptome sequencing of the same cell. Nat. Biotechnol. 2015;33:285–289. doi: 10.1038/nbt.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bian S, et al. Single-cell multiomics sequencing and analyses of human colorectal cancer. Science. 2018;362:1060–1063. doi: 10.1126/science.aao3791. [DOI] [PubMed] [Google Scholar]
- 99.Hu Y, et al. Simultaneous profiling of transcriptome and DNA methylome from a single cell. Genome Biol. 2016;17:88. doi: 10.1186/s13059-016-0950-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang, X. et al. Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science361, eaat5691 (2018). [DOI] [PMC free article] [PubMed]