Abstract
In the present study, we identified and characterized the apricot (Prunus armeniaca L.) homologs of three dormancy-related genes, namely the ParCBF1 (C-repeat binding factor), ParDAM5 (dormancy-associated MADS-BOX) and ParDAM6 genes. All highly conserved structural motifs and the 3D model of the DNA-binding domain indicate an unimpaired DNA-binding ability of ParCBF1. A phylogenetic analysis showed that ParCBF1 was most likely homologous to Prunus mume and Prunus dulcis CBF1. ParDAM5 also contained all characteristic domains of the type II (MIKCC) subfamily of MADS-box transcription factors. The homology modeling of protein domains and a phylogenetic analysis of ParDAM5 suggest its functional integrity. The amino acid positions or small motifs that are diagnostic characteristics of DAM5 and DAM6 were determined. For ParDAM6, only a small part of the cDNA was sequenced, which was sufficient for the quantification of gene expression. The expression of ParCBF1 showed close association with decreasing ambient temperatures in autumn and winter. The expression levels of ParDAM5 and ParDAM6 changed according to CBF1 expression rates and the fulfillment of cultivar chilling requirements (CR). The concomitant decrease of gene expression with endodormancy release is consistent with a role of ParDAM5 and ParDAM6 genes in dormancy induction and maintenance. Cultivars with higher CR and delayed flowering time showed higher expression levels of ParDAM5 and ParDAM6 toward the end of endodormancy. Differences in the timing of anther developmental stages between early- and late-flowering cultivars and two dormant seasons confirmed the genetically and environmentally controlled mechanisms of dormancy release in apricot generative buds. These results support that the newly identified apricot gene homologs have a crucial role in dormancy-associated physiological mechanisms.
Keywords: apricot, CBF, DAM, dormancy, flower development, gene expression, microsporogenesis
Introduction
Many important fruit tree species belong to the Prunus genus of the Rosaceae family. Flower bud development is one of the most critical stages in their reliable production. Spring frost injury is the most common reason for yield-loss in producing countries at the Northern Hemisphere under temperate climate (Kaya et al., 2018). The annual growing cycle of temperate woody plants forms an integrated system with subsequent phases of active growth and dormancy (Perry, 1971; Lloret et al., 2018). During dormancy plants exhibit little or no growth and their metabolic activity decreases for a period of time. It is an essential strategy for perennial plants to survive harmful environmental conditions during winter (Hanninen and Tanino, 2011). Dormancy can be divided into two phases, the endo- and ecodormancy (Lloret et al., 2018). Endodormancy is a genetically controlled mechanism that is triggered in early autumn by external factors and inhibits bud development even under growth-promoting conditions. Plants require a certain amount of chill for endodormancy-release to enter ecodormancy phase when bud growth is only prevented by unfavorable climatic conditions.
The alternation of phenological stages in Prunus is chiefly regulated by the ambient air temperature (Welling and Palva, 2006; Heide, 2008). During autumn, low temperature induces dormancy; after dormancy break during winter warming in spring causes bud burst and the rapid development of flower organs. Plants give different responses to the seasonal change of environmental conditions, depending on species or even cultivar/eco-type (Hanninen and Tanino, 2011). In case of Prunus species, chilling-accumulation during winter plays a key role in the induction of budbreak and flowering (Ruiz et al., 2007; Alburquerque et al., 2008). Over dormancy progression, characteristic histological and physiological alterations occur in flower buds. The anther developmental stages (archesporium, string, pollen mother cell, tetrad and pollen stages) are most often distinguished to follow microsporogenesis (Szalay et al., 2019). Dormancy is considered to mark a boundary between the development of the sporogenous tissue in the anther and the occurrence of pollen meiosis (Julian et al., 2011).
The genetic factors responsible for the cold- and frost stress responses first have been identified in Arabidopsis thaliana (Baker et al., 1994). The CBF transcription factors belong to the DREB1 subfamily within the AP2/ERF protein family. They are able to bind to the CRT/DRE (C-repeat/dehydration responsive element) sequence motif in the promoter region of the regulated gene, which contains a conserved CCGAC sequence as a binding site for the DNA-binding domain of CBF proteins (Stockinger et al., 1997; Liu et al., 1998). On low temperature, the Arabidopsis CBF genes can be induced within 15 min and in 2 h they activate the “CBF-regulon,” i.e., the cold-regulated genes which contain the CRT/DRE regulatory-element (Yang et al., 2005).
In the Rosaceae family, DREB/CBF-type genes have been first described in sweet-cherry (Prunus avium L.) and their amino acid sequences were approximately 50% identical to the Arabidopsis CBFs (Kitashiba et al., 2002; Owens et al., 2002). The expression of one sweet-cherry CBF homolog (CGI-B) caused increased frost resistance in transgenic Arabidopsis lines (Kitashiba et al., 2004). A full-length cDNA of a peach CBF gene (PpCBF1) has been isolated by Wisniewski et al. (2011) and its ectopic expression in apple resulted in induced dormancy and increased cold hardiness. The CBF gene sequences of apple and peach seemed to be very similar, especially concerning their AP2 binding domain (Wisniewski et al., 2011). In almond (Prunus dulcis Mill.), two CBF sequences were identified and their transcription level was in correlation with the expression of a dehydrin gene, PdDHN1 (Barros et al., 2012b). Dehydrins are encoded by a group of genes related to abiotic stress response, such as frost or drought in many species. CBF genes were identified in many other plant genera, such as Vitis (Xiao et al., 2006), Populus (Benedict et al., 2006), Betula (Welling and Palva, 2008), Eucalyptus (Gamboa et al., 2007) and Triticum (Galiba et al., 2009; Soltész et al., 2013).
Previous studies on Arabidopsis, snapdragon (Antirrhinum majus L.) and petunia hybrids shed light on the role of MADS-BOX genes in controlling flower-organogenesis (Theissen et al., 2000; Horvath, 2009). Among Rosaceae species, the role of MADS-BOX genes in bud dormancy was first proved in the “evergrowing” (evg) peach mutant that is incapable of forming terminal vegetative buds in response to dormancy-inducing conditions. Bielenberg et al. (2008) made a comparative mapping to a 132-kb region of the wild-type genome where six genes were found to be missing from mutant plant tissues. These six MADS-BOX-type genes form a cluster in the wild-type genome, while there is a 41,746-bp deletion in the same region of the evg genome. This six DAM (DORMANCY-ASSOCIATED MADS-BOX) genes are strong candidates for the regulation of growth cessation and dormancy induction in peach. PpDAMs were suggested to be orthologs of the Arabidopsis SHORT VEGETATIVE PHASE (SVP) and AGAMOUS-LIKE 24 (AGL24) (Yamane et al., 2011a), two genes that are involved in flowering (Gregis et al., 2009).
The expression of DAM genes seems to be tissue-specific, moreover, each of them has a clear seasonal expression-pattern presumably regulated by photoperiod (Li et al., 2009). The seasonal expression level of DAM5-6 genes shows correlation with bud dormancy induction and break in case of peach, P. persica L. (Yamane et al., 2011a,b). Previous studies revealed that both peach and apple DAM-like genes contain a conserved element in their promoter region that is highly homologous to the consensus CBF-binding sites (Wisniewski et al., 2011; Mimida et al., 2015). The temporal arrest of floral organ enlargement is presumably controlled by PmDAM5-6 (Jiménez et al., 2010; Yamane et al., 2011a). Specific period of low temperature exposure is necessary to reduce the expression of PmDAM5-6 and to break this inhibition; therefore, they possibly work as dose-dependent growth inhibitors in dormant buds (Yamane et al., 2011a,b). Several DAM-like genes have been found in other Rosaceae species such as Japanese apricot (Prunus mume Sieb. et Zucc.) (Yamane et al., 2008), almond (P. dulcis) (Prudencio et al., 2018), Japanese pear (Pyrus pyrifolia Nakai) (Saito et al., 2013), apple (Malus × domestica Borkh.) (Mimida et al., 2015) and some additional plant genera including Euphorbia (Horvath et al., 2010), Actinidia (Wu et al., 2012) and Betula (Elo et al., 2001).
Apricot (P. armeniaca L.) productivity is much affected by its early-flowering time since unfavorable environmental conditions in late winter/early spring frequently result in a considerable yield loss in many producing countries. The aim of the present study was to identify apricot CBF and DAM gene sequences that may contribute to bud dormancy regulation. Sequence homology and phylogenetic analysis were used to confirm the putative biological function of the newly identified apricot gene sequences. In addition, we used quantitative real-time PCR to follow the changes in gene expression over the developmental stages of flower buds and during two consecutive dormant periods of field-grown trees. The regulatory role of the identified genes was further tested by comparing the gene expression profiles of apricot cultivars differing in their flowering time.
Materials and Methods
Plant Material
Four apricot (Prunus armeniaca L.) cultivars were sampled in the apricot germplasm collection of the Department of Genetics and Plant Breeding, Szent István University (Budapest, Hungary) from November to March in 2015/2016 and 2016/2017. The North American cultivars, “Aurora” (syn. “Early Blush” and NJA 53) and “Goldrich” (syn. “Sun Giant”) are two economically important early-flowering cultivars, while the late-flowering North American “Stella” and Central Asian “Zard” could be potentially used in breeding programs due to favorable characteristics including resistance to devastating diseases. According to previous studies, “Aurora” and “Goldrich” show limited resistance to chilling conditions while both “Stella” and “Zard” have better frost tolerance (Szalay et al., 2006; Milatovic et al., 2013). For the isolation of the intron-less ParCBF1 gene, genomic DNA was extracted from the leaves of an additional apricot cultivar, “Korai zamatos,” while ParDAM5 and ParDAM6 sequences containing several introns were amplified from cDNA obtained from ‘Zard’ flower buds. In addition, flower buds were collected from all four cultivars for gene expression analysis.
Determination of Chilling Requirements and Flowering Dates
Hourly air temperatures were measured by a PT100 1/3 Class B temperature sensor with ± 1°C accuracy as implemented in the iMETOS® IMT200 (Pessl Instruments, Weiz, Austria) automatic weather station. The instrument was located in an open area within 400 m of the orchard. The amount of cold received by the plants were quantified using the chill units of the Utah model (Richardson et al., 1974) and portions of the Dynamic model (Fishman et al., 1987). The date of breaking endodormancy was determined by forcing apricot branches with approximately 100 buds/cultivar according to the method described by Ruiz et al. (2007). Flowering date was recorded when 50% of 500 tested flower buds were open.
DNA Extraction
The total genomic DNA was extracted from the leaves of “Korai zamatos” cultivar using the DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). The quantity and quality of DNA were analyzed by NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies Inc., Wilmington, DE, United States).
RNA Extraction and Reverse Transcription
Flower buds of the four tested apricot cultivars were collected 8 and 7 times in two consecutive dormant periods, 2015/2016 and 2016/2017, respectively, from leaf fall (November) to the beginning of bloom (mid of March). Samples have been frozen immediately in liquid nitrogen and stored at −80°C. Total RNA from approx. 100 mg of bud tissue was extracted using the protocol of Jaakola et al. (2001). After DNase I treatment (Thermo Fisher Scientific) to eliminate the possible genomic DNA contamination, approx. 2 ng of total RNA were reverse-transcribed using an oligo (dT)20 primer with RevertAid H Minus First Strand cDNA Synthesis Kit (Thermo Fisher Scientific) following the manufacturer’s instructions.
Polymerase Chain Reaction (PCR) Analysis and Sequencing of PCR Products
For each sample, PCR amplification was performed in a reaction volume of 25 μL containing 20–50 ng of DNA, 10X DreamTaq Green Buffer (Thermo Fisher Scientific, Waltham, MA, United States) with final concentrations of 4.5 mM MgCl2, 0.2 mM of dNTPs, and 0.75 U of DreamTaq DNA Polymerase (Thermo Fisher Scientific). Specific published primers, designed for related Prunus species, were applied for the amplification of target sequences (Table 1). The PCR products were separated on 1% TBE agarose gels at 80 V for 30 min and DNA bands were stained with ethidium bromide. Fragment sizes were estimated by comparison with the 100-bp DNA ladder (Promega, Mannheim, Germany).
Table 1.
Primers used for the isolation of Prunus armeniaca CBF1 and DAM5-6 sequences.
| Species | Primer | Sequence (5′ → 3′) | References |
|---|---|---|---|
| P. dulcis | CBF-F | GCCCCAGTCGAGTTTGTTGTC | Barros et al., 2012b |
| CBF-R | AGCATTGCGATGGAGAAAGAAG | ||
| P. persica | CBF1-F | AGGGCTTCTTCTTTCTCCAC | Wisniewski et al., 2011 |
| CBF1-R | AAATCTTTATGTTCGACTCACTCA | ||
| DAM5/1-F | ATCTCCACCACCTGCAACAGT | Yamane et al., 2011a | |
| DAM5/1-R | CTTCTTAACGCCCCAGTTTGAG | ||
| DAM5/2-F | CCCCGAAACCCACCAACGAAGATG | Bielenberg et al., 2008 | |
| DAM5/2-R | CAGCACTGTTGCAGGTGGTG | ||
| DAM6-F | CCAACAACCAGTTAAGGCAGAAGA | Bielenberg et al., 2008 | |
| DAM6-R | GGAAGCCCCAGTTTGAGAGA |
PCR products were cloned into the pTZ57R/T plasmid vector using the InsTAclone PCR Cloning Kit (Thermo Fisher Scientific) and JM109 competent cells, isolated with GeneJETTM Plasmid Miniprep Kit (Thermo Fisher Scientific) and sequenced using an ABI 3500 XL Genetic Analyzer (Applied Biosystems, Foster City, CA, United States).
Gene Expression Analysis by Quantitative PCR
The expression level of P. armeniaca CBF and DAM5-6 genes was determined by means of real-time PCR (ABI 7500 Fast Real-Time PCR instrument, Applied Biosystems) using PB20.17-05 qPCRBIO SyGreen Blue Mix (Nucleotest Bio Ltd., Budapest, Hungary). The relative fold change (FC) values were calculated with the ΔΔCt method (Bookout and Mangelsdorf, 2003). The quantitative PCR primers were designed in this study from specific regions of the newly identified ParCBF1, ParDAM5, and ParDAM6 genes (Table 2).
Table 2.
Primers designed in this study to quantify the expression of ParCBF1, ParDAM5-6 and the reference gene.
| Primer | Sequence (5′ → 3′) | Annealing temperature (°C) | Amplicon length (bp) |
|---|---|---|---|
| qACT-F | GTGCCTGCCATGTATGTTGCCA | 60 | 226 |
| qACT-R | CAGTGGTGGTGAACATGTACCCYC | ||
| qCBF-F | GGCTACTTGAACTGGGATGACATG | 60 | 104 |
| qCBF-R | ACACAAACAAATACATGATTGAC | ||
| qDAM5-F | GCTTATGGATCCGGAGAGGCTGAATA | 60 | 101 |
| qDAM5-R | CAGCACTGTTGCAGGTGGTGGAGATA | ||
| qDAM6-F | GTTTGTGGAGCCGGAGACGTTGATT | 60 | 100 |
| qDAM6-R | GCAGCTGGTGGAGGTGGCAATTTGG |
Evaluation of Male Gametophyte Developmental Stages
The male gametophyte development (microsporogenesis) was studied in 2016 and 2017 from January to March. Samples were collected six times in both years. For each of the four cultivars, 8–10 flower buds were studied. Harvested buds were kept in liquid nitrogen during transportation. Anthers removed from the buds, were squashed in aceto-carmine (2%) for 5 min and then analyzed using a Zeiss Axio Imager 2 (Carl Zeiss, Thornwood, NY, United States) optical microscope at 400 × magnification. Pictures were taken with a Zeiss Axio Cam digital camera.
Bioinformatics
Prunus armeniaca CBF and DAM5-6 sequences were used as query sequences for the NCBI MegaBLAST (Morgulis et al., 2008) analysis. An alignment of sequences was carried out using MEGA6 (Tamura et al., 2011) and presented using BioEdit v. 7.2.0. (Hall, 1999). Primers were designed manually and then checked using the Oligo Analyzer 3.1 software 1.
To demonstrate the molecular structure of the typical protein-domains of CBF and DAM transcription factors, sequence homology was used in the SWISS-MODEL server (Arnold et al., 2006) and Global Model Quality Estimates and Qualitative Model Energy Analysis were also determined as quality estimators of the models (Benkert et al., 2008).
The evolutionary history of CBF and DAM sequences was inferred using the Maximum Likelihood method based on the JTT matrix-based model (Jones et al., 1992). Initial tree(s) for the heuristic search were obtained automatically using the Neighbor-Joining and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model, and then selecting the topology with superior log likelihood value. The analysis involved 27 amino acid sequences. There were a total of 280 positions in the final dataset. The bootstrap consensus tree inferred from 1,000 heuristic replicates was taken to represent the evolutionary history of the operational taxonomic units (Felsenstein, 1985). Branches corresponding to partitions reproduced in less than 80% bootstrap replicates were not considered statistically supported clades. Evolutionary analyses were conducted using MEGA5.1 (Tamura et al., 2011).
Statistical Analyses
Real-time quantitative PCR data presented for each sample represent the mean values determined for three independent replicates. After tested for normal distribution and equality of variances, one-way analysis of variance (ANOVA) and Duncan’s multiple range test with P < 0.05 was carried out to determine significant differences. Statistical analyses were carried out using SPSS 13.0 (SPSS Inc., Chicago, IL, United States).
Results
Identification and Characterization of ParCBF1
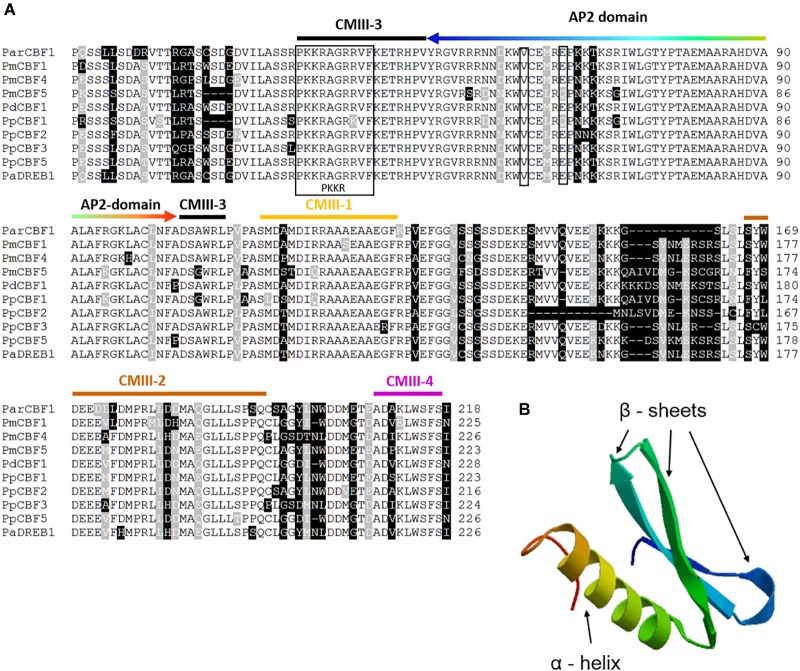
An apricot CBF-like sequence (758 bp) has been isolated from the genomic DNA of P. armeniaca “Korai zamatos” (the sequence was deposited into GenBank under the accession number of MH464453). A sequence homology test was performed using the NCBI MegaBLAST algorithm that detected 29 Prunus sequences with E-values of 0. The significant matches to the query included CBF/DREB1 gene sequences. The deduced amino acid sequences were aligned with homologous sequences from several Prunus species available in the NCBI database (Figure 1A). The ParCBF1 sequence contains the highly conserved AP2 DNA-binding domain and four CMIII domains (CMIII1-4) that are also called CBF signatures. CMIII-1, 2, and 4 are located downstream of AP2 domain, while CMIII-3 includes the PKKR/PAGR and DSAWR motifs flanking the AP2 domain on the 5′ and 3′ ends, respectively. These results show that ParCBF1 shares significant homology with other Prunus CBFs and has the structural motifs required for its biological function.
FIGURE 1.
Sequence alignment of Prunus C-repeat Binding Factors (CBF) and the structure of Prunus armeniaca CBF1 protein. (A) Multiple alignment of the deduced amino acid sequence of P. armeniaca CBF and other Prunus CBF sequences. Dashes indicate gaps introduced to optimize the alignment. Identical amino acids and conserved substitutions are shaded black and gray, respectively. The total number of amino acids for each deduced protein is indicated at the end of each sequence. The PKKR/PAGR nuclear localization signal and the DSAWR motif are indicated by a black frame. Further CBF signature sequences are marked by orange (CMIII-1), brown (CMIII-2), black (CMIII-3) and purple (CMIII-4) colored stripes over the sequence motifs. The DNA-bindig AP2 domain is indicated by a rainbow-colored arrow (the order of colors indicate identical amino acid positions in the alignment and ribbon diagram in B), two narrow frames point to the two important amino acids responsible for DNA-binding specificity (a valine and a glutamic acid in the positions 14th and 19th, respectively). The CBF sequences and their GenBank accession numbers or references are as follows: ParCBF1 (MH464453) from P. armeniaca L.; PmCBF1, PmCBF4 and PmCBF5 from P. mume published in Zhao et al., 2018; PdCBF1 (KJ818900) is from P. dulcis; PpCBF1 (HM992943), PpCBF2 (KC543498), PpCBF3 (KC543499), and PpCBF5 (KC543501) from P. persica; and PaDREB1 (AB121674) from P. avium. (B) Molecular model of the AP2 domain (http://swissmodel.expasy.org). AP2 domain consists of three β-sheets and an α-helix.
To further support unimpaired activity of the identified gene, the molecular structure (ribbon diagram) of its AP2-domain was modeled (Figure 1B). For AP2 molecular modeling, Arabidopsis AtERF1-DNA-binding domain (1gcc) was used as template, since this is the only plant protein belonging to the AP2 family with determined 3D structure (Allen et al., 1998). The sequence identity between ParCBF1 and AtERF1-DNA-binding domain was 51.7%, and the model was characterized by a GMQE value of 0.76 and a QMEAN value of −2.74, indicating major alterations are unlikely to occur in the native 3D structure of the AP2-domain in ParCBF1.
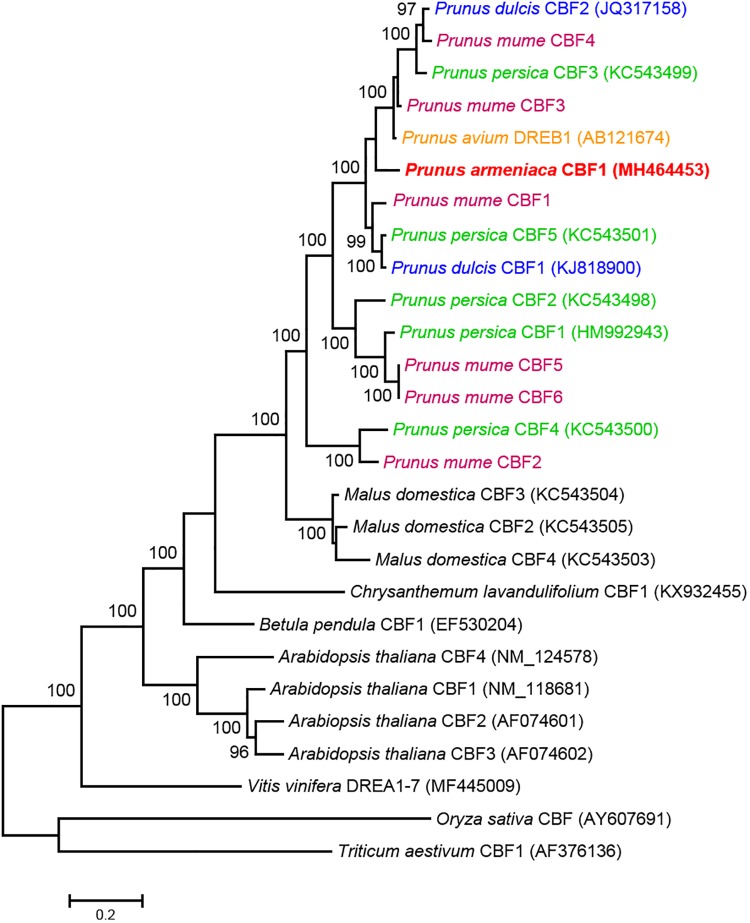
The CBF amino acid sequences previously described in monocotyledonous (Oryza sativa and Triticum aestivum) and several dicotyledonous (Arabidopsis thaliana, Betula pendula, Vitis vinifera, and Malus domestica) species were included in a phylogenetic analysis to support homology and functionality of ParCBF1. Most of the genes chosen for the phylogenetic analysis were described to be responsive to low temperature. The maximum likelihood tree showed that monocotyledonous sequences formed an outgroup, while each of the Arabidopsis, Malus and Prunus CBFs clustered together within dicotyledonous sequences and all the clusters received 100% bootstrap support (Figure 2). This pattern demonstrates a high-level similarity among Prunus CBF genes. The ParCBF1 was found within a complex sub-structured cluster that received 100% bootstrap support and included P. dulcis, P. persica, and P. mume CBF sequences. The sequences did not form species-specific clusters but were scattered among statistically supported (bootstrap ≥ 97%) sub-clusters and mixed with sequences from other species.
FIGURE 2.
Molecular phylogenetic analysis of the amino acid sequences of C-repeat binding factor (CBF) genes using the Maximum Likelihood method. The bootstrap confidence values (%) exceeding 80% from 1000 replicates are indicated on the branches. The sequence name in red refers to the ParCBF1 identified in the present study. The values presented above or below the specific nodes are the percentage of replicates in which relationships were recovered. CBF sequences from different Prunus species are indicated by colors.
Identification and Characterization of ParDAM5 and ParDAM6
DAM5- (729 bp) and DAM6-like (230 bp) sequences were determined from cDNA obtained from flower buds of “Zard” and were deposited into GenBank under the accession numbers MH464454 and MH464455, respectively. A sequence homology test was performed using the NCBI MegaBLAST algorithm. Among the homologous sequences, DAM5 sequences from other Prunus species including P. persica, were the most similar to ParDAM5 with E-values of 0. Our ParDAM6 sequence was significantly similar to P. pseudocerasus, Japanese apricot and peach sequences with E-values ranging from 2 × 10−101 to 10−83.
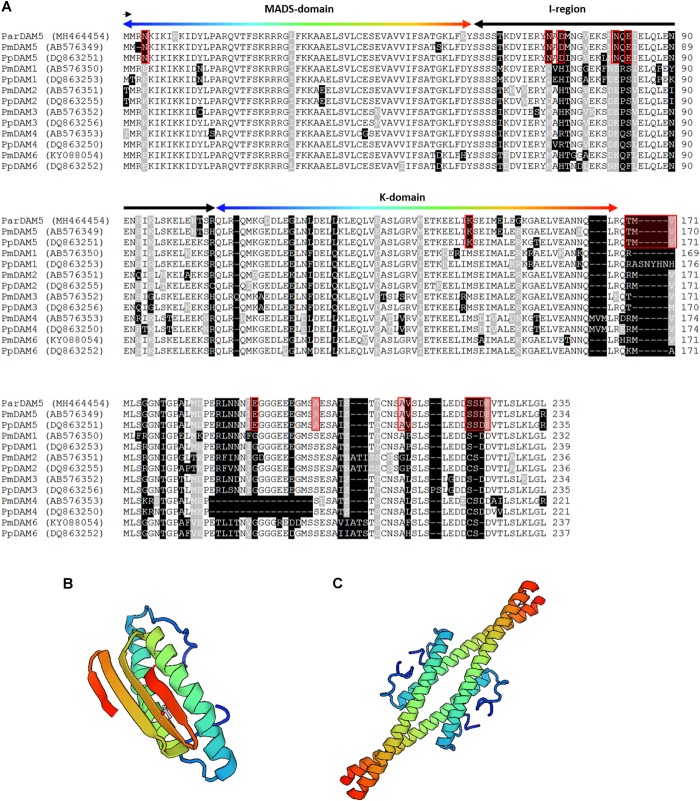
The analysis of the functional motifs gave further support for the identification of apricot sequences as DAM5 and DAM6 homologs. In case of ParDAM5, three main protein domains could have been identified in the deduced amino acid sequence: the MADS box domain (M-domain) that is the highly conserved dimerization domain at the N-terminal end, the K-domain whose function is probably to participate in protein-protein interactions, finally the variable intervening (I-) region that connects the two others (Figure 3A). The molecular structures of the MADS- and keratin-like (K-) domains are demonstrated in Figure 3B,C. For MADS-domain molecular modeling, the human MEF2 (myocyte enhancer factor 2) proteins (1c7u) was used as template, as the 3D structure of plant homologs are currently not available. The MADS domain of MEF2 is responsible for DNA-binding (Huang et al., 2000) and sequence identity between human MEF2 domain and the MADS-domain of ParDAM5 was 53.6% The reliability of the model was characterized by 0.76 and −2.68 GMQE and QMEAN values, respectively. SEPALLATA 3 MADS transcription factor of Arabidopsis (Puranik et al., 2014) was used as template for K-domain and its homo-tetramer model is characterized by GMQE and QMEAN values of 0.73 and 0.14, respectively. Although the length of the cloned ParDAM6 sequence did not allow a similar structural analysis, many characteristic amino acid positions and motifs were detected in the deduced amino acid sequence of ParDAM6 that are exclusively shared with DAM6 sequences from other Prunus species (Supplementary Figure S3).
FIGURE 3.
Sequence alignment of Prunus Dormancy-Associated MADS-BOX proteins and the structure of Prunus armeniaca DAM5 protein. (A) Multiple alignment of P. armeniaca DAM5 amino acid sequence with other related Prunus DAM sequences. Dashes indicate gaps introduced to optimize the alignment. Identical amino acids and conserved substitutions are shaded black and gray, respectively. The total number of amino acids for each deduced protein is indicated at the end of each sequence. MADS-domain, K-domain and I-region are indicated by arrows. The order of colors in the arrow over the MADS- and K-domains indicate identical amino acid positions in the alignment and ribbon diagram in (B,C), respectively. Red frames indicate the amino acid positions or small motifs exclusively occurring in DAM5 sequences. (B) Molecular model of the MADS-domain and (C) the K-domain (http://swissmodel.expasy.org).
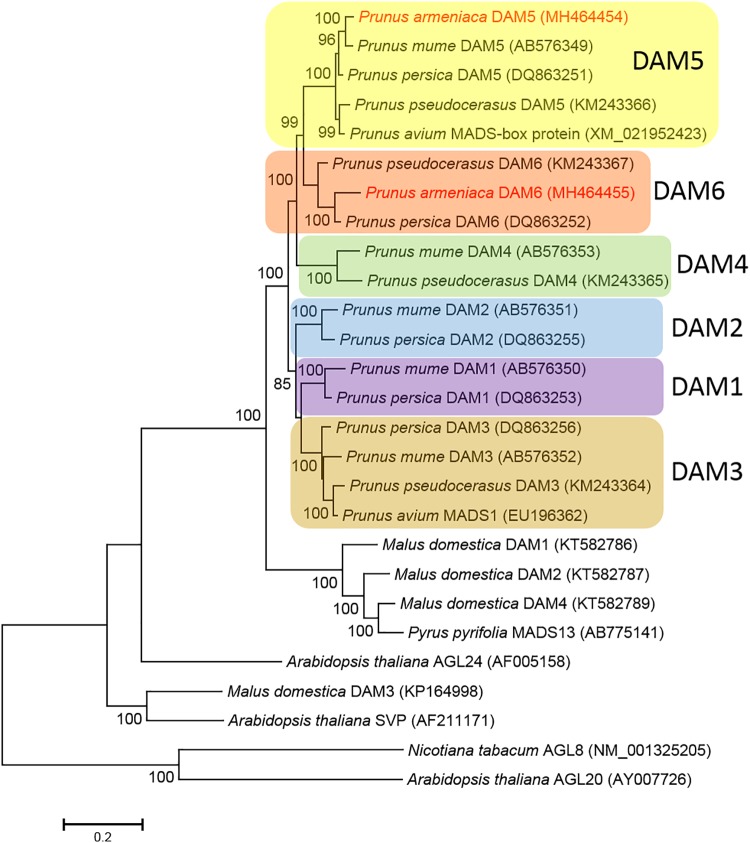
The phylogenetic analysis of DAM predicted protein sequences indicated that Prunus and Malus DAM sequences formed a sister group to Arabidopsis thaliana AGL24. Malus and Pyrus sequences formed a sister group to the clade encompassing sequences from all analyzed Prunus species. The Prunus clade received a 100% bootstrap support and was divided into six sub-clades according to the six different (DAM1 to DAM6) genes. Each of the gene-specific sub-clades also received 100% bootstrap support. Prunus DAM5 and DAM6 sequences were clustered within the respective sequences from other Prunus species. The ParDAM5 sequence showed the closest relationship with P. mume DAM5, while ParDAM6 clustered with the corresponding P. persica sequence, with both groups receiving a 100% bootstrap support (Figure 4). Our analysis confirmed that ParDAM5 and ParDAM6 are evolutionarily related to DAM5 and DAM6 genes in other Prunus species, pointing to a putatively identical function in P. armeniaca.
FIGURE 4.
Molecular phylogenetic analysis of the deduced amino acid sequences of dormancy-associated MADS-BOX (DAM) genes using the Maximum Likelihood method. The bootstrap confidence values (%) exceeding 80% from 1000 replicates are indicated on the branches. The sequence names in red refer to ParDAM5 and 6 identified in the present study. The values presented above specific nodes are the percentage of replicates in which relationships were recovered. Colors indicate different DAM genes.
Chilling Accumulation and Anther Meiosis Time in Two Consecutive Seasons
The chilling accumulation under field conditions were followed over two consecutive dormant seasons using the chill units (CU) according to the Utah model and portions of the Dynamic model. Supplementary Figures S1, S2 show the chilling accumulation from 3 October to 30 March according to the Utah and Dynamic models, respectively. In the first part of the dormant season, the chilling accumulation was more intensive in 2016/2017 compared to 2015/2016. From 21 December, the amount of accumulated CU was higher in 2015/2016 than in 2016/2017. From 21 December to 9 January, the difference in CU between seasons was small (within 25 CU). In terms of chill portions, 2016/2017 had higher values than the preceding season only after 26 January. However, toward the end of the dormant seasons, considerably more (approx. 500 CU) and cill portions (10) accumulated in 2015/2016 than in 2016/2017.
The tendency of portion accumulation over the two dormant seasons was very similar to that shown in case of CU accumulation but results were more homogenous between dormant seasons. The variation coefficient for the chilling accumulation was smaller with the Dynamic model compared to the value of the Utah model. The variation of coefficient (cv) for the chilling accumulation until 1 March was 19.7% with the Utah model and 4.6% with the Dynamic model.
The chilling requirements (CR) for breaking dormancy are shown in Table 3 for all four apricot cultivars and in both seasons. The two early-flowering cultivars, “Aurora” and “Goldrich” had lower CR compared to the late-flowering “Stella” and “Zard.” The difference between the CR of early- and late flowering cultivars ranged between 122 and 224 CU and 7-11 portions. All cultivars were characterized by smaller CR in 2016/2017 evaluated by the Utah model, the differences between years ranged from 8.4 to 11.8%. However, the Dynamic model provided more homogenous results with variations between years equal to or less than 1.4%. The flowering of “Aurora” and “Goldrich” occurred on the same day in both seasons, while the flowering of “Stella” and “Zard” was delayed by 1 day in 2016/2017 compared to 2015/2016.
Table 3.
Chilling requirements of the studied apricot cultivars for breaking dormancy in two consecutive dormant periods (2015/2016 and (2016/2017).
| Cultivar | Season | Breaking dormancy | Chill units (Utah model) |
Portions (Dynamic model) |
Flowering time | ||||
|---|---|---|---|---|---|---|---|---|---|
| Value | Mean | cv (%) | Value | Mean | cv (%) | ||||
| Aurora | 2015/2016 | January 31 | 1203 | 1136 | 8.4 | 70 | 70 | 1.4 | March 23 |
| 2016/2017 | February 02 | 1068 | 69 | March 23 | |||||
| Goldrich | 2015/2016 | February 05 | 1299 | 1213 | 10.1 | 74 | 74 | 1.4 | March 24 |
| 2016/2017 | February 06 | 1127 | 73 | March 24 | |||||
| Stella | 2015/2016 | February 13 | 1439 | 1335 | 11.0 | 80 | 81 | 1.2 | March 30 |
| 2016/2017 | February 21 | 1231 | 81 | March 31 | |||||
| Zard | 2015/2016 | February 15 | 1473 | 1360 | 11.8 | 81 | 81 | 0.0 | April 01 |
| 2016/2017 | February 22 | 1247 | 81 | April 02 | |||||
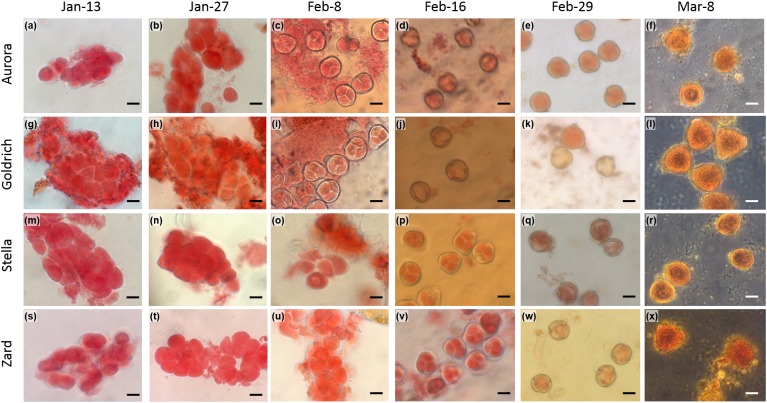
We determined the anther developmental changes over the two tested dormant seasons since chilling temperatures contribute to flower bud formation in dormant trees. Figure 5 shows anther developmental stages in 2016. Four stages of microsporogenesis were distinguished in both years: archesporium – undifferentiated sporogenic tissue; premeiotic conditions (development and separation of pollen mother cells); tetrad (after meiosis) and microspores (development of pollen grains). In 2016, development and separation of pollen-mother cells started from the beginning of January, tetrad stage could be first observed at the beginning of February, the microspores could have been detected from mid-February and matured pollen grains occurred in the end of February. In 2017, the whole process was delayed and the earliest time of tetrad stage was observed in the middle of February 2017 (Figure 6). This one-week delay in male gametophyte development could have been caused by low temperatures at the beginning of February (Supplementary Table S1).
FIGURE 5.
Light microscopy images of the male gametophyte development in apricot (Prunus armeniaca) flower buds. Two early-flowering cultivars, “Aurora” (a–f) and “Goldrich” (g–l) and two late-flowering cultivars, “Stella” (m–r) and “Zard” (s–x) were sampled in 2016 from January to March. Scale bars correspond to 20 μm.
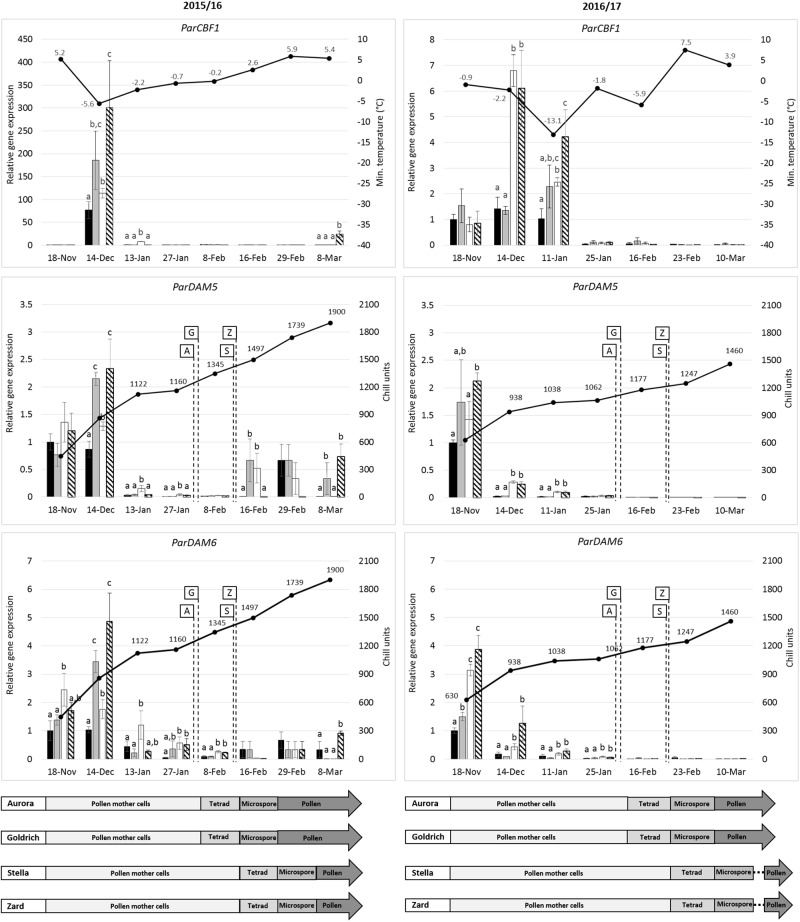
FIGURE 6.
Seasonal gene expression patterns of ParCBF1, ParDAM5 and ParDAM6 in apricot (Prunus armeniaca) flower buds. Two early-flowering cultivars, “Aurora” (black) and “Goldrich” (gray) and two late-flowering cultivars, “Stella” (white) and “Zard” (hatched) were sampled in two consecutive dormant seasons. Real-time PCR data are means of three replicates, means followed by the same letters were not significantly different at P < 0.05 according to a Duncan’s multiple range test. Minimum temperatures or accumulating chill units determined for each date are indicated as black spots. An interpretative diagram shows anther developmental stages of the four assayed cultivars over sample collection dates.
The late-flowering cultivars, “Stella” and “Zard” showed one-week delay in the timing of meiosis and the development of pollen grains in both years compared to the respective developmental stages of the early-flowering “Aurora” and “Goldrich.” The difference in timing of the anther development of early- and late flowering cultivars coincided with the fulfillment of the CR of apricot cultivars. All cultivars reached the tetrad stage after their CR was fulfilled.
Expression Analysis of ParCBF1 and ParDAM5-6 Genes
Figure 6 shows the changes in relative transcript levels of the ParCBF1 and ParDAM5-6 genes in apricot flower buds compared to the daily minimum temperature values and chilling units, respectively, during two consecutive dormant periods (2015/2016 and 2016/2017). During the dormant period of 2015/2016, the relative expression level of ParCBF1 was the highest in December in case of all cultivars. The transcript abundance was 4-fold higher in “Zard” flower buds than in case of “Aurora,” the most frost-sensitive genotype among the four cultivars tested in this study. In 2015 December, the increase in ParCBF1 expression level was much higher compared to 2016/2017. Regardless of the discrepancy in the extent of increase in expression rate, the increase of ParCBF1 expression in December coincided with temperatures falling below the freezing point in both seasons (−5.6°C in 2015 and −2.2°C in 2016). In addition, statistically significant differences were observed in ParCBF1 expression of “Aurora” and both “Stella” and “Zard” in each of the seasons. In December 2015, “Goldrich” was also characterized by significantly higher expression rates than “Aurora.” The expression of ParCBF1 was downregulated in all cultivars to the end of January in both seasons and remained almost zero until the time of budbreak.
A characteristic seasonal pattern was observed in the expression rate of ParDAM5-6 genes, which were quite similar in both seasons. Transcript levels were high at the beginning of the dormant period in both years. The highest gene expression levels were detected in December 2015 and in November 2016 for both ParDAM5 and ParDAM6. Then, the expression levels of both genes started to decrease gradually.
After a considerable drop in ParDAM5 and ParDAM6 expression, significant differences were detected among some cultivars in both seasons. In the 2015/2016 dormant season tested, “Stella” accumulated moderate although significantly higher ParDAM5 and ParDAM6 transcript levels in flower bud tissues in January 2016 than other cultivars. Significant differences were found between the early and late-flowering cultivars until 08 February. Correspondences among the expression rates of ParCBF1 and ParDAM5-6 were evident in December 2015 and January 2017 when cultivars showing higher expression rates of ParCBF1 also had higher transcript levels for both ParDAM5 and ParDAM6. However, in the end of January ParCBF1 expression was downregulated without significant alterations among cultivars, while “Stella” still had a higher ParDAM5 expression level and both late-flowering cultivars had higher transcript levels for ParDAM6. This difference could have been observed until 08 February 2016. During the 2016/2017 dormant season, between December 2016 and January 2017, both late-flowering apricot cultivars were characterized by significantly higher ParDAM5 and ParDAM6 expression levels compared to the early-flowering cultivars. For ParDAM6, the difference also occurred on 25 January. Similar variations were observed between the ParCBF1 expression levels of early- and late-flowering apricot cultivars until 11 January.
The expression of all three genes were definitely downregulated once the CR of the cultivars had been fulfilled. It is clearly shown by the significantly higher ParDAM6 expression rates in “Stella” and “Zard” on 08 February 2016, which resulted in a delay of 10–14 days in dormancy breaking time compared to early-flowering cultivars. Although values were smaller, significant differences in ParDAM6 transcript levels were also detected before the end of endodormancy.
Both ParDAM5-6 genes were characterized by somewhat higher expression rates over February and March in 2015/2016 than 2016/2017, however, characteristic differences between the early- and late-flowering cultivars were not detected any more.
Discussion
Identification and Structural Verification of Dormancy-Related Genes in Apricot
Apricot is one of the earliest flowering fruit trees, a fact that makes this species susceptible to yield-loss induced by unfavorable weather conditions in early spring. The initiation and release of endodormancy was shown to be regulated by a set of genes in Prunus species with CBF and DAM genes playing crucial roles in this complex regulatory network (Saito et al., 2015; Zhao et al., 2018). In the present study, we identified the apricot homologs of CBF1, DAM5, and DAM6 genes and provided in silico and experimental data to support their impaired structure and putative function in bud dormancy regulation.
Among stone fruit species, the CBF genes were first isolated in sweet cherry (Kitashiba et al., 2002) and peach (Wisniewski et al., 2011) and shown to be associated with dormancy induction and freezing tolerance. Homologous sequences were also identified in P. cerasus (Owens et al., 2002), P. dulcis (Barros et al., 2012b) and P. mume (Guo et al., 2014). This study was carried out to identify a CBF homolog in apricot, P. armeniaca. A 758-bp sequence of the intronless gene was amplified from the genomic DNA of “Korai zamatos” apricot cultivar using the primers designed by Barros et al. (2012b) and Wisniewski et al. (2011) from the conserved regions of Prunus CBF sequences and the amplicon was cloned and sequenced. The MegaBLAST analysis provided significant support of its homology with CBF/DREB1 genes of Prunus species. In addition, the alignment of the deduced amino acid sequence with P. mume, P. dulcis, P. persica and P avium CBF/DREB sequences confirmed the presence of the AP2 domain and several CBF signature motifs including CMIII1-4 (Figure 1). The PKK and AP2 domains seem to affect nuclear localization of CBF transcription factors, whereas the same domains in cooperation with the C-terminal hydrophobic amino acids affect transactivation (Carlow et al., 2017). Such domains were shown to be strongly conserved in functional CBF proteins (Jaglo et al., 2001; Nakano et al., 2006) that suggest ParCBF1 is a functionally intact homologous gene in apricot.
The 3D structure of the complex of the Arabidopsis AtERF1-DNA-binding domain and its target DNA was determined by NMR (Allen et al., 1998) and used for molecular modeling of ParCBF1. The Quality estimators, GMQE and QMEAN indicated the built model was reliable and declared the typical AP2 fold with a three-stranded beta-sheet and an alpha helix almost parallel to the beta-sheet. This topology was found to be associated with contacting eight consecutive base pairs in the major groove of DNA (Allen et al., 1998) and hence it indicates an unimpaired DNA-binding ability of ParCBF1. Since CBF transcription factors may bind specifically to the C-repeat/dehydration-responsive (CRT/DRE) element in the promoters of DAM genes, the intact AP2 domain structure indicates a possible connection between CBF and dormancy-associated MADS-box proteins.
The Prunus genome contains several CBF genes. The in silico analysis of genome sequences detected six CBF genes both in P. persica (Wisniewski et al., 2014) and P. mume (Zhao et al., 2018) genomes. According to our phylogenetic analysis, the first apricot homolog (ParCBF1) identified in the present study is the most likely homolog of other Prunus CBF sequences indicating close evolutionary relatedness and putative functional similarities. Co-clustering of PmCBF1 and PpCBF5 was also shown by Zhao et al. (2018) indicating a close relationship between those genes labeled by different numbers in two Prunus species. Considering the number of CBF genes in Prunus genome, at least five additional homologs are still likely to be identified in the apricot genome. The involvement of ParCBF1 in dormancy regulation is supported by a similar physiological role of its almond and Japanese apricot homologs (Barros et al., 2012a; Zhao et al., 2018).
The ParDAM5 and 6 cDNA sequences were isolated using the gene-specific primers designed by Yamane et al. (2011b) and Bielenberg et al. (2008), respectively. The homology searches indicated that they are closely related to the corresponding Prunus sequences in the GenBank database. All characteristic domains (including the MADS box, I region and K domain) of the type II (MIKCC) subfamily of MADS-box transcription factors (Horvath, 2015) were detected in the ParDAM5 sequence.
Six DAM genes were identified in peach and Japanese apricot (Bielenberg et al., 2008; Sasaki et al., 2011) but only two of the six (DAM5 and DAM6) genes were reported to be associated with endodormancy release in peach generative buds (Yamane et al., 2011a). Using primers specifically amplifying DAM5 and DAM6, we determined the partial sequences of their P. armeniaca homologs. For PpDAM5, the homology was confirmed by MegaBlast and phylogenetic analyses, indicating that ParDAM5 belongs to the super-clade of SVP/AGL24 Arabidopsis sequences as was previously reported for P. mume DAM6 (Yamane et al., 2008). The identification of protein domains and the homology modeling of characteristic domains suggest its functional integrity.
Since putatively functional DAM sequences were available from several species, a careful screening of the aligned sequences helped circumscribe the amino acid positions or small motifs that are consequently characteristics of DAM5 and DAM6 genes and differentiate them from the rest of the DAM genes. For DAM5, 6 of the 11 diagnostic motifs located in the conserved domains (MADS, I and K), and 5 additional in the C terminal part of the protein (Figure 3). One of those flanked a region of indels, i.e., insertions or deletions downstream of the K domain. The 75 amino acids compared in DAM6 sequences also detected six specific motifs (Supplementary Figure S3). Among the 260 amino acid positions in the alignment of all six PpDAMs (Bielenberg et al., 2008), 122 amino acid positions were invariable. The conserved amino acids are likely to be crucial to the biological function of the genes, however, the contribution of the newly identified gene-specific sequence variations to the physiological function of the corresponding genes might be an interesting scope for future studies.
Functional Verification of Dormancy-Related Genes in Apricot
The newly isolated ParCBF1 and ParDAM5 sequences show all structural features that are indispensable for the relevant biological functions. For ParDAM6, only a small part of the cDNA was sequenced that allowed the quantification of gene expression levels. CBF and DAM genes were described to have characteristic expression profiles over the dormant season and dormancy release (Jiménez et al., 2010; Sasaki et al., 2011; Yamane et al., 2011a,b; Barros et al., 2012a; Zhao et al., 2018), which makes them appropriate candidates for the internal factors controlling endodormancy of temperate trees.
The expression of ParCBF1 was associated with decreasing ambient temperatures. In 2015, the peak of ParCBF1 expression occurred in December, the coldest month in the 2015/2016 dormant period. After that, temperature was going to be increased and ParCBF1 expression immediately dropped. January was colder in 2017 than 2016 and the expression rate of ParCBF1 did not show a sharp decrease compared to the year before. A rapid increase of CBF gene expression due to low temperature was observed in many plant species including fruit trees like apple (Wisniewski et al., 2011), almond (Barros et al., 2012a,b) and Japanese apricot (Guo et al., 2014; Zhang et al., 2013). Such a characteristic expression profile of ParCBF1 validates its physiological role as an important element of the low temperature signaling cascade leading to dormancy induction and release in apricot.
The presence of a conserved core sequence motif (CCGAC) in the promoter region of DAM5 and DAM6 genes in peach and Japanese apricot suggests their participation in the CBF regulon (Yamane et al., 2011b; Zhao et al., 2018). ParDAM5 and ParDAM6 had similar expression patterns as their peach and Japanese apricot homologs (Yamane et al., 2011a,b; Zhao et al., 2018). Significant differences between the expression levels of ParDAM6 in early- and late-flowering cultivars could be also detected 12–14 days later than in case of ParDAM5, which indicates differences in their regulation. The expression levels of DAMs correlated with the number of CBF-binding sites in the promoter of Japanese apricot genes with PmDAM5 and ParDAM6 having several sites and showing the highest levels. The expression levels of ParDAM5 and ParDAM6 changed according to low temperature induced CBF1 expression rates and the fulfillment of cultivar CR. The expression levels of ParDAM5 and ParDAM6 were related to the daily minimum temperatures and CBF1 expression rates: the sampling day in December 2015 was colder than in 2016 (Supplementary Table S1) and ParCBF1 and ParDAM5 and 6 had higher expression rates for all cultivars in 2015.
There were important differences between the climatic data of the two dormant seasons studied. In the first part of the dormant season chilling accumulation was more intensive in 2016/2017, while the second part of the dormant season was more efficient in relation to chilling accumulation in 2015/2016. The CR for breaking endodormancy was lower in 2016/2017 for all cultivars. Our results fit well with those of Viti et al. (2010) who found that in warmer dormant seasons, all tested cultivars including “Goldrich” showed lower CR than in colder years. In addition, the Dynamic model showed lower variation in chill accumulation across years that the Utah model, which indicates that Dynamic model may also provide more accurate assessment in colder climates, similarly to its reliability under warm climatic conditions (Ruiz et al., 2007; Viti et al., 2010).
The early-flowering cultivars had lower CR for endodormancy release as compared to the late-flowering cultivars in both dormant seasons. The CR of “Aurora” averaged across 2 years was similar to the 1140 ± 60 CU measured by Viti et al. (2006), while we determined slightly higher CR for “Goldrich” compared to the data collected in Spain and Italy by Viti et al. (2010). For “Stella” and “Zard,” our study provides the first CR data and they were definitely higher compared to those of the early-flowering cultivars and consistent with their late-flowering dates.
The early-flowering cultivars broke endodormancy between 31 January and 06 February. The date of endodormancy release differed in one and 2 days between the two dormant seasons for “Aurora” and “Goldrich,” respectively. Although the intensity of chilling accumulation was different in the two dormant seasons, differences practically disappeared in the second half of January that explains the nearly identical date of endodormancy release of early-flowering cultivars in both years. However, late-flowering cultivars were characterized by strikingly different endodormancy release dates in the two dormant seasons ranging from 7 (“Zard)” to 8 (“Stella”) days. It might be explained by the slower chilling accumulation in 2016/2017 resulting in a longer period for late-flowering cultivars to achieve the critical amount of chilling units in this season.
Our results indicated that early- and late-flowering cultivars broke endodormancy in the beginning and the middle of February, respectively. The expression level of ParDAM5 and ParDAM6 markedly decreased in the middle of January (in the season 2015/2016) or in the middle of December (2016/2017). After the downregulation of ParDAM5 and ParDAM6, throughout the next 4 and 6 weeks in the seasons 2015/2016 and 2016/2017, respectively, a consequent and significant difference (p ≤ 0.05) occurred between the expression levels of both genes in early- (“Aurora” and “Goldrich”) and late-flowering (“Stella” and “Zard”) cultivars. Peach PpDAM5 and PpDAM6 were proposed to function as a dose-dependent growth inhibitor in dormant flower buds (Yamane et al., 2011a) and were characterized by lower expression rates in the low-chill peach cultivar compared to the high-chill cultivar (Yamane et al., 2011c). Our results suggest a similar function for ParDAM5 and ParDAM6 with higher expression rates associated with delayed endodormancy release time and consequent late-flowering time of “Stella” and “Zard” apricot cultivars.
Meiosis has been long considered a sign of endodormancy release (Szabó et al., 2002), but pollen tetrads of medium and high-chilling requirement apricot cultivars appeared earlier with respect to the end of endodormancy (Bartolini et al., 2006). In our study, the tetrad stage emerged in all four apricot cultivars after endodormancy release time (Figure 6). Our results confirm the findings of Julian et al. (2011) that winter dormancy set a boundary between the development of the sporogenous tissue and further microspore development and meiosis appears to occur around breaking of dormancy. Pollen tetrad stage occurred 7–8 days later in late-flowering apricot cultivars in both seasons. It indicates the genetically controlled mechanisms of male gametophyte development as it was also shown for other apricot cultivars (Julian et al., 2014).
In addition to the genotype-dependent difference between the assayed early- and late-flowering cultivars, timing of the anther development stages also showed a year-to-year variation with an approximate one-week delay in 2016/2017 compared to 2015/2016. Such a difference occurred in both early- and late-flowering cultivars although the flowering time was delayed by only a single day in 2017. Since chilling accumulation was more intensive in 2015/2016 after 15 January, all cultivars achieved earlier the required amount of chilling for breaking endodormancy than during the 2016/2017 dormant season. Similar differences between cold and warm winter years were also recorded for Mediterranean apricot cultivars (Julian et al., 2014). Compared to a cold winter year, cultivars with medium or high CR showed a considerable delay in reaching the tetrad stage in a warm winter year due to CR fulfilled later in the dormant period. Although flowering time was shown to be primarily influenced by the specific CR of apricot cultivars, other factors (differences in pre-blossom temperatures, irradiation etc.) may also influence flowering dates (Rodrigo and Herrero, 2002; Ruiz et al., 2007; Julian et al., 2014).
The timing of male gametophyte development in apricot flower buds is influenced by temperature to help avoid floral organ damage. The expression of the newly identified ParCBF1 was mainly induced by low temperature, while decrease of the expression of ParDAM5 and ParDAM6 apricot genes coincided with the end of endodormancy stage. The difference in ParDAM5 and ParDAM6 gene expression toward the end of endodormancy showed a clear association with the differing dates of endodormancy release and consequent flowering times in case of cultivars with differing CR. In Hungary, annual temperature has increased 0.8°C over the past century (Lakatos et al., 2011) and this warming tendency resulted in a shift of endodormancy release time by 19–23 days earlier over 24 years while flowering time occurred 3 days earlier (Szalay et al., 2019). Hence, the identification and characterization of the apricot homolog of three genes that have been associated with the onset and release of flower bud dormancy in other Prunus species are of crucial importance. ParCBF1, ParDAM5, and ParDAM6 show all typical structural features and genetically and environmentally controlled expression levels over the endodormancy stages to be important elements of the molecular network behind bud dormancy of apricot trees.
Author Contributions
JH, GG, and AH designed the study. EB and AH wrote the manuscript. All authors performed the research and critically revised the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
- AP2/ERF
Apetala2-Ethylene Responsive Factor
- CBF
C-repeat Binding Factor
- DREB1
Dehydration-Responsive Element Binding
- GMQE
Global Model Quality Estimates
- JTT
Jones–Taylor–Thornton Model
- QMEAN
Qualitative Model Energy Analysis.
Funding. This study was supported by the “NKFIH K 112554” and the Higher Education Institutional Excellence Program (1783-3/2018/FEKUTSTRAT) awarded by the Ministry of Human Capacities, Hungary, within the framework of plant breeding and plant protection researches of Szent István University.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00402/full#supplementary-material
The accumulation of chilling units (Utah model) over the 2015/16 (black) and 2016/16 (gray) dormant periods.
The accumulation of chilling portions (Dynamic model) over the 2015/16 (black) and 2016/16 (gray) dormant periods.
Multiple sequence alignment of partial P. armeniaca DAM6 amino acid sequence with other related Prunus DAM6 sequences. Non-conservative and conservative amino acid replacements are shaded black and gray, respectively. The total number of amino acids for each deduced protein is indicated at the end of each sequence. Blue frames indicate the amino acid positions or small motifs exclusively occurring in DAM6 sequences.
Daily minimum temperatures throughout the sample collection periods of the 2015/16 and 2016/17 dormant seasons.
References
- Alburquerque N., García-Montiel F., Carrillo A., Burgos L. (2008). Chilling and heat requirements of sweet cherry cultivars and the relationship between altitude and the probability of satisfying the chill requirements. Environ. Exp. Bot. 64 162–170. 10.1016/j.envexpbot.2008.01.003 [DOI] [Google Scholar]
- Allen M. D., Yamasaki K., Ohme-Takagi M., Tateno M., Suzuki M. (1998). A novel mode of DNA recognition by a β-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA. EMBO J. 17 5484–5496. 10.1093/emboj/17.18.5484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold K., Bordoli L., Kopp J., Schwede T. (2006). The swiss-model workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22 195–201. 10.1093/bioinformatics/bti770 [DOI] [PubMed] [Google Scholar]
- Baker S. S., Wilhelm K. S., Thomashow M. F. (1994). The 5′-region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol. Biol. 24 701–713. 10.1007/BF00029852 [DOI] [PubMed] [Google Scholar]
- Barros P. M., Gonçalves N., Saibo N. J. M., Oliveira M. M. (2012a). Cold acclimation and floral development in almond bud break: insights into the regulatory pathways. J. Exp. Bot. 63 4585–4596. 10.1093/jxb/ers144 [DOI] [PubMed] [Google Scholar]
- Barros P. M., Goncalves N., Saibo N. J. M., Oliveira M. M. (2012b). Functional characterization of two almond C-repeat binding factors involved in cold response. Tree Physiol. 32 1113–1128. 10.1093/treephys/tps067 [DOI] [PubMed] [Google Scholar]
- Bartolini S., Viti R., Guerriero R. (2006). Xylem differentiation and microsporogenesis during dormancy of apricot flower buds (Prunus armeniaca L.). Eur. J. Hortic. Sci. 71 84–90. [Google Scholar]
- Benedict C., Skinner J. S., Meng R., Chang Y., Bhalerao R., Hunter N. P. A., et al. (2006). The CBF1-dependent low temperature signaling pathway, regulon and increase in freeze tolerance are conserved in Populus spp. Plant Cell Environ. 29 1259–1272. 10.1111/j.1365-3040.2006.01505.x [DOI] [PubMed] [Google Scholar]
- Benkert P., Tosatto S. C. E., Schomburg D. (2008). QMEAN: a comprehensive scoring function for model quality assessment. Proteins 71 261–277. 10.1002/prot.21715 [DOI] [PubMed] [Google Scholar]
- Bielenberg D. G., Li Z., Zhebentyayeva T., Fan S., Reighard G. L. R., Scorza R., et al. (2008). Sequencing and annotation of the evergrowing locus in peach [Prunus persica (L.) batsch] reveals a cluster of six MADS-box transcription factors as candidate genes for regulation of terminal bud formation. Tree Genet. Genomes 4 495–507. 10.1007/s11295-007-0126-9 [DOI] [Google Scholar]
- Bookout A. L., Mangelsdorf D. J. (2003). Quantitative real-time PCR protocol for analysis of nuclear receptor signaling pathways. Nucl. Recept. Signal. 1:e012. 10.1621/nrs.01012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlow C. E., Faultless J. T., Lee C., Siddiqua M., Edge A., Nassuth A. (2017). Nuclear localization and transactivation by Vitis CBF transcription factors are regulated by combinations of conserved amino acid domains. Plant Physiol. Biochem. 118 306–319. 10.1016/j.plaphy.2017.06.027 [DOI] [PubMed] [Google Scholar]
- Elo A., Lemmetyinen J., Turunen M.-L., Tikka L., Sopanen T. (2001). Three MADS-box genes similar to apetala1 and fruitfull from silver birch (Betula pendula). Physiol. Plant. 112 95–103. 10.1034/j.1399-3054.2001.1120113.x [DOI] [PubMed] [Google Scholar]
- Felsenstein J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39 783–791. 10.1111/j.1558-5646.1985.tb00420.x [DOI] [PubMed] [Google Scholar]
- Fishman S., Erez A., Couvillon G. A. (1987). The temperature dependence of dormancy breaking in plants: mathematical analysis of a two-step model involving a cooperative transition. J. Theor. Biol. 124 473–483. 10.1016/S0022-5193(87)80221-7 [DOI] [Google Scholar]
- Galiba G., Vágújfalvi A., Li C. X., Soltész A., Dubcovsky J. (2009). Regulatory genes involved in the determination of frost tolerance in temperate cereals. Plant Sci. 176 12–19. 10.1016/j.plantsci.2008.09.016 [DOI] [Google Scholar]
- Gamboa M. C., Rasmussen-Poblete S., Venezuela P. D. T., Krauskopf E. (2007). Isolation and characterization of a cDNA encoding a CBF transcription factor from E. globulus. Plant Physiol. Biochem. 45 1–5. 10.1016/j.plaphy.2006.12.006 [DOI] [PubMed] [Google Scholar]
- Gregis V., Sessa A., Dorca-Fornell C., Kater M. K. (2009). The Arabidopsis floral meristem identity genes AP1, AGL24 and SVP directly repress class B and C floral homeotic genes. Plant J. 60 626–637. 10.1111/j.1365-313X.2009.03985.x [DOI] [PubMed] [Google Scholar]
- Guo C., Zhang J. Q., Peng T., Bao M. Z., Zhang J. W. (2014). Structural and expression analyses of three PmCBFs from Prunus mume. Biol. Plant. 58 247–255. 10.1007/s10535-014-0393x [DOI] [Google Scholar]
- Hall T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucl. Acid. Symp. 41 95–98. [Google Scholar]
- Hanninen H., Tanino K. (2011). Tree seasonality in warming climate. Trends Plant Sci. 16 412–416. 10.1016/j.tplants.2011.05.001 [DOI] [PubMed] [Google Scholar]
- Heide O. M. (2008). Interaction of photoperiod and temperature in the control of growth and dormancy of Prunus species. Sci. Hortic. 115 309–314. 10.1016/j.scienta.2007.10.005 [DOI] [Google Scholar]
- Horvath D. (2009). Common mechanisms regulate flowering and dormancy. Plant Sci. 177 523–531. 10.1016/j.plantsci.2009.09.002 [DOI] [Google Scholar]
- Horvath D. P. (2015). “Dormancy-associated MADS-BOX genes: a review,” in Advances in Plant Dormancy, ed. Anderson J. (Cham: Springer; ), 137–146. 10.1007/978-3-319-14451-1_7 [DOI] [Google Scholar]
- Horvath D. P., Sung S., Kim D., Chao W., Anderson J. (2010). Characterization, expression and function of dormancy associated mads-box genes from leafy spurge. Plant Mol. Biol. 73 169–179. 10.1007/s11103-009-9596-5 [DOI] [PubMed] [Google Scholar]
- Huang K., Louis J. M., Donaldson L., Lim F. L., Sharrocks A. D., Clore G. M. (2000). Solution structure of the MEF2A–DNA complex: structural basis for the modulation of DNA bending and specificity by MADS-box transcription factors. EMBO J. 19 2615–2628. 10.1093/emboj/19.11.2615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaakola L., Pirttilä A. M., Halonen M., Hohtola A. (2001). Isolation of high quality RNA from bilberry (Vaccinium myrtillus L.) fruit. Mol. Biotechnol. 19 201–203. 10.1385/MB:19:2:201 [DOI] [PubMed] [Google Scholar]
- Jaglo K. R., Kleff S., Amundsen K. L., Zhang X., Haake V., Zhang J. Z., et al. (2001). Components of the Arabidopsis C-repeat/dehydration-responsive element binding factor cold-response pathway are conserved in Brassica napus and other plant species. Plant Physiol. 127 910–917. 10.1104/pp.010548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez S., Reighard G. L., Bielenberg D. G. (2010). Gene expression of DAM5 and DAM6 is suppressed by chilling temperatures and inversely correlated with bud break rate. Plant Mol. Biol. 73 157–167. 10.1007/s11103-010-9608-5 [DOI] [PubMed] [Google Scholar]
- Jones D. T., Taylor W. R., Thornton J. M. (1992). The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8 275–282. 10.1093/bioinformatics/8.3.275 [DOI] [PubMed] [Google Scholar]
- Julian C., Herrero M., Rodrigo J. (2014). Anther meiosis time is related to winter cold temperatures in apricot (Prunus armeniaca L.). Environ. Exp. Bot. 100 20–25. 10.1016/j.envexpbot.2013.12.002 [DOI] [Google Scholar]
- Julian C., Rodrigo J., Herrero M. (2011). Stamen development and winter dormancy in apricot (Prunus armeniaca). Ann. Bot. 108 617–625. 10.1093/aob/mcr056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaya O., Kose C., Gecim T. (2018). An exothermic process involved in the late spring frost injury to flower buds of some apricot cultivars (Prunus armenica L.). Sci. Hortic. 241 322–328. 10.1016/j.scienta.2018.07.019 [DOI] [Google Scholar]
- Kitashiba H., Ishizaka T., Isuzugwa K., Nishimura K., Suzuki T. (2004). Expression of a sweet cherry DREB1/CBF ortholog in Arabidopsis confers salt and freezing tolerance. J. Plant Physiol. 161 1171–1176. 10.1016/j.jplph.2004.04.008 [DOI] [PubMed] [Google Scholar]
- Kitashiba H., Matsuda N., Ishizaka T., Nakano H., Suzuki T. (2002). Isolation of genes similar to DREB1/CBF from sweet cherry (Prunus avium L.). J. Jpn. Soc. Hortic. Sci. 71 651–657. 10.2503/jjshs.71.651 [DOI] [Google Scholar]
- Lakatos M., Szentimrey T., Bihari Z. (2011). Application of gridded daily data series for calculation of extreme temperature and precipitation indices in hungary. Idõjárás 115 99–109. [Google Scholar]
- Li Z., Reighard G. L., Abbott A. G., Bielenberg D. G. (2009). Dormancy-associated MADS genes from the EVG locus of peach [Prunus persica (L.) Batsch] have distinct seasonal and photoperiodic expression patterns. J. Exp. Bot. 60 3521–3530. 10.1093/jxb/erp195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Kasuga M., Sakuma Y., Abe H., Miura S., Yamaguchi-Shinozaki K., et al. (1998). Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10 1391–1406. 10.1105/tpc.10.8.1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloret A., Badenes M. L., Ríos G. (2018). Modulation of dormancy and growth responses in reproductive buds of temperate trees. Front. Plant Sci. 9:1368. 10.3389/fpls.2018.01368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milatovic D., Durovic D., Zec G. (2013). “Susceptibility of apricot cultivars to winter and late spring frosts,” in Proceedings of the 4th Conference Innovation in Fruit Growing, ed. Milatović D. (Beograd: FAO; ), 239–248. [Google Scholar]
- Mimida N., Saito T., Moriguchi T., Suzuki A., Komori S., Wada M. (2015). Expression of dormancy-associated mads-box (DAM)-like genes in apple. Biol. Plant. 59 237–244. 10.1007/s10535-015-0503-4 [DOI] [Google Scholar]
- Morgulis A., Coulouris G., Raytselis Y., Madden T. L., Agarwala R., Schäffer A. A. (2008). Database indexing for production megaBLAST searches. Bioinformatics 24 1757–1764. 10.1093/bioinformatics/btn322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T., Suzuki K., Fujimura T., Shinshi H. (2006). Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 140 411–432. 10.1104/pp.105.073783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens C. L., Thomasow M. F., Hancock J. F., Iezzoni A. F. (2002). CBF1 orthologues in sour cherry and strawberry and the heterologous expression of CBF1 in strawberry. J. Am. Soc. Hort. Sci. 127 489–494. 10.21273/JASHS.127.4.489 [DOI] [Google Scholar]
- Perry T. O. (1971). Dormancy of trees in winter. Science 171 29–36. 10.1126/science.171.3966.29 [DOI] [PubMed] [Google Scholar]
- Prudencio Á, Dicenta F., Martínez-Gómez P. (2018). Monitoring dormancy transition in almond [Prunus dulcis (Miller) Webb] during cold and warm Mediterranean seasons through the analysis of a DAM (Dormancy-Associated MADS-Box) gene. Horticulturae 4:41 10.3390/horticulturae4040041 [DOI] [Google Scholar]
- Puranik S., Acajjaoui S., Conn S., Costa L., Conn V., Vial A., et al. (2014). Structural basis for the oligomerization of the MADS domain transcription factor SEPALLATA3 in Arabidopsis. Plant Cell 26 3603–3615. 10.1105/tpc.114.127910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson E. A., Seeley S. D., Walker D. R. (1974). A model for estimating the completion of rest for ‘Redhaven’ and ‘Elberta’ peach trees. Hort. Sci. 9 331–332. 26849277 [Google Scholar]
- Rodrigo J., Herrero M. (2002). Effects of pre-blossom temperatures on flower development and fruit set in apricot. Sci. Hortic. 92 125–135. 10.1016/S0304-4238(01)00289-8 [DOI] [Google Scholar]
- Ruiz D., Campoy J. A., Egea J. (2007). Chilling and heat requirements of apricot cultivars for flowering. Environ. Exp. Bot. 61 254–263. 10.1016/j.envexpbot.2007.06.008 25071812 [DOI] [Google Scholar]
- Saito T., Bai S., Imai T., Ito A., Nakajima I., Moriguchi T. (2015). Histone modification and signalling cascade of the dormancy-associated mads-box gene, ppmads13-1, in japanese pear (Pyrus pyrifolia) during endodormancy. Plant Cell Environ. 38 1157–1166. 10.1111/pce.12469 [DOI] [PubMed] [Google Scholar]
- Saito T., Bai S., Ito A., Sakamoto D., Saito T., Ubi B. E., et al. (2013). Expression and genomic structure of the dormancy-associated MADS box genes MADS13 in Japanese pears (Pyrus pyrifolia Nakai) that differ in their chilling requirement for endodormancy release. Tree Physiol. 33 654–667. 10.1093/treephys/tpt037 [DOI] [PubMed] [Google Scholar]
- Sasaki R., Yamane H., Ooka T., Jotatsu H., Kitamura Y., Akagi T., et al. (2011). Functional and expressional analyses of PmDAM genes associated with endodormancy in Japanese apricot (Prunus mume). Plant Physiol. 157 485–497. 10.1104/pp.111.181982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltész A., Smedley M., Vashegyi I., Galiba G., Harwood W., Vágújfalvi A. (2013). Transgenic barley lines prove the involvement of TaCBF14 and TaCBF15 in the cold acclimation process and in frost tolerance. J. Exp. Bot. 64 1849–1862. 10.1093/jxb/ert050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger E. J., Gilmour S. J., Thomashow M. F. (1997). Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcription activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. U.S.A. 94 1035–1040. 10.1073/pnas.94.3.1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabó Z., Szalay L., Papp J. (2002). Connection between the developmental stage and the cold hardiness of peach cultivars. Acta Hortic. 592 549–552. 10.17660/ActaHortic.2002.592.74 [DOI] [Google Scholar]
- Szalay L., Froemel-Hajnal V., Bakos J., Ladányi M. (2019). Changes of the microsporogenesis process and blooming time of three apricot genotypes (Prunus armeniaca L.) in Central Hungary based on long-term observation (1994–2018). Sci. Hortic. 246 279–288. 10.1016/j.scienta.2018.09.069 [DOI] [Google Scholar]
- Szalay L., Papp J., Pedryc A., Szabo Z. (2006). Diversity of apricot varieties based on traits determining winter hardiness and early spring frost tolerance of floral buds. Acta Hortic. 701 131–134. 10.17660/ActaHortic.2006.701.17 [DOI] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theissen G., Becker A., Di Rosa A., Kanno A., Kim J. T., Münster T., et al. (2000). A short history of MADS-box genes in plants. Plant Mol. Biol. 42 115–149. 10.1023/A:1006332105728 [DOI] [PubMed] [Google Scholar]
- Viti R., Andreini L., Ruiz D., Egea J., Bartolini S., Iacona C., et al. (2010). Effect of climatic conditions on the overcoming of dormancy in apricot flower buds in two Mediterranean areas: Murcia (Spain) and Tuscany (Italy). Sci. Hortic. 124 217–224. 10.1016/j.scienta.2010.01.001 [DOI] [Google Scholar]
- Viti R., Bartolini S., Guerriero R. (2006). Apricot floral biology: the evolution of dormancy and the appearance of bud anomalies in several Italian genotypes. Adv. Hort. Sci. 20 267–274. [Google Scholar]
- Welling A., Palva E. T. (2006). Molecular control of cold acclimation in trees. Physiol. Plantarum 127 167–181. 10.1111/j.1399-3054.2006.00672.x [DOI] [Google Scholar]
- Welling A., Palva E. T. (2008). Involvement of CBF transcription factors in winter hardiness in birch. Plant Physiol. 147 1119–1211. 10.1104/pp.108.117812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski M., Nassuth A., Teulières C., Marque C., Rowland J., Cao P. B., et al. (2014). Genomics of cold hardiness in woody plants. Crit. Rev. Plant Sci. 33 92–124. 10.1080/07352689.2014.870408 [DOI] [Google Scholar]
- Wisniewski M., Norelli J., Bassett C., Artlip T., Macarisin D. (2011). Ectopic expression of a novel peach (Prunus persica) CBF transcription factor in apple (Malus×domestica) results in short-day induced dormancy and increased cold hardiness. Planta 233 971–983. 10.1007/s00425-011-1358-3 [DOI] [PubMed] [Google Scholar]
- Wu R. M., Walton E. F., Richardson A. C., Wood M., Hellens P., Varkonyi-Gasic E. (2012). Conservation and divergence of four kiwifruit SVP-like MADS-box genes suggest distinct roles in kiwifruit bud dormancy and flowering. J. Exp. Bot. 63 797–807. 10.1093/jxb/err304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H., Siddiqua M., Braybrook S., Nassuth A. (2006). Three grape CBF/DREB1 genes respond to low temperature, drought and abscisic acid. Plant Cell Environ. 29 1410–1421. 10.1111/j.1365-3040.2006.01524.x [DOI] [PubMed] [Google Scholar]
- Yamane H., Kashiwa Y., Ooka T., Tao R., Yonemori K. (2008). Suppression subtractive hybridization and differential screening reveals endodormancy-associated expression of an SVP/AGL24-type MADS-box gene in lateral vegetative buds of Japanese apricot. J. Am. Soc. Hort. Sci. 133 708–716. 10.21273/JASHS.133.5.708 [DOI] [Google Scholar]
- Yamane H., Ooka T., Jotatsu H., Sasaki R., Tao R. (2011a). Expression analysis of PpDAM5 and PpDAM6 during flower bud development in peach (Prunus persica). Sci. Hortic. 129 844–848. 10.1016/j.scienta.2011.05.013 [DOI] [Google Scholar]
- Yamane H., Ooka T., Jotatsu H., Sasaki R., Tao R. (2011b). Expressional regulation of PpDAM5 and PpDAM6, peach (Prunus persica) dormancy-associated MADS-box genes, by low temperature and dormancy-breaking reagent treatment. J. Exp. Bot. 62 3481–3488. 10.1093/jxb/err028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane H., Tao R., Ooka T., Jotatsu H., Sasaki R., Yonemori K. (2011c). Comparative analyses of dormancy-associated MADS-box genes, PpDAM5 and PpDAM6, in low-and high-chill peaches (Prunus persica L.). J. Jpn. Soc. Hortic. Sci. 80 276–283. 10.2503/jjshs1.80.276 [DOI] [Google Scholar]
- Yang T., Zhang L., Zhang T., Zhang H., Xu S., An L. (2005). Transcriptional regulation network of cold-responsive genes in higher plants. Plant Sci. 169 987–995. 10.1016/j.plantsci.2005.07.005 [DOI] [Google Scholar]
- Zhang J., Yang W. R., Cheng T. R., Pan H. T., Zhang Q. X. (2013). Functional and evolutionary analysis of two CBF genes in Prunus mume. Can. J. Plant Sci. 93 455–464. 10.4141/cjps2012-193 [DOI] [Google Scholar]
- Zhao K., Zhou Y., Li Y., Zhuo X., Ahmad S., Han Y., et al. (2018). Crosstalk of PmCBFs and PmDAMs based on the changes of phytohormones under seasonal cold stress in the stem of Prunus mume. Int. J. Mol. Sci. 19:15. 10.3390/ijms19020015 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The accumulation of chilling units (Utah model) over the 2015/16 (black) and 2016/16 (gray) dormant periods.
The accumulation of chilling portions (Dynamic model) over the 2015/16 (black) and 2016/16 (gray) dormant periods.
Multiple sequence alignment of partial P. armeniaca DAM6 amino acid sequence with other related Prunus DAM6 sequences. Non-conservative and conservative amino acid replacements are shaded black and gray, respectively. The total number of amino acids for each deduced protein is indicated at the end of each sequence. Blue frames indicate the amino acid positions or small motifs exclusively occurring in DAM6 sequences.
Daily minimum temperatures throughout the sample collection periods of the 2015/16 and 2016/17 dormant seasons.