Abstract
Hereditary hearing loss (HHL) is an extremely heterogeneous disorder with autosomal dominant, recessive, and X-linked forms. Here, we described an Italian pedigree affected by HHL but also prostate hyperplasia and increased ratio of the free/total PSA levels, with the unusual and extremely rare Y-linked pattern of inheritance. Using exome sequencing we found a missense variant (r.206A>T leading to p.Asp69Val) in the TBL1Y gene. TBL1Y is homologous of TBL1X, whose partial deletion has described to be involved in X-linked hearing loss. Here, we demonstrate that it has a restricted expression in adult human cochlea and prostate and the variant identified induces a lower protein stability caused by misfolded mutated protein that impairs its cellular function. These findings indicate that TBL1Y could be considered a novel candidate for HHL.
Subject terms: Heritable quantitative trait, Diseases
Introduction
Hearing loss is the most common sensory disorder in humans, caused by both genetic and environmental factors [1, 2]. Hereditary hearing loss (HHL) contributes to more than 60% of deafness cases, with autosomal dominant, recessive, and X-linked forms.
HHL including non-syndromic (70% NSHHL) and syndromic (30% SHHL) forms [3, 4] and, so far, more than 100 forms of SHHL presenting different patterns of inheritance have been described [5].
The Y-chromosome is one of the two sex-chromosomes in mammals and is the smallest chromosome in the karyotype. The human Y-chromosome is transmitted with a male-specific pattern of inheritance, thus traits should be readily recognizable from their male-line inheritance [6, 7]. It has been hypothesized that along the evolution, the Y-chromosome has acquired a large number of testes-specific genes responsible for spermatogenesis. Alterations in these genes have been associated with several male fertility-specific traits (i.e. spermatogenic impairment, azoospermia, sperm-fertilizing inability, etc.) both in humans and animals [8].
Recently, two families affected by HHL and showing a rare Y-linked pattern of inheritance have been described [9, 10]. In one of these families the pathogenic mechanism underlying the disease has been explained by the presence of a complex chromosomal rearrangement, while no data have been reported so far for the second family. Here, we describe a pedigree characterized by HHL showing a rare Y-linked pattern of inheritance, in which only males are affected. Patients also present benign prostate hyperplasia and increased ratio of free/total PSA levels. Whole exome sequencing (WES) analysis revealed the presence of a missense variant r.206A>T; p.Asp69Val (rs199659121) in TBL1Y (NM_134259.1) gene, which has high homology to TBL1X (NM_001139467.1) gene whose partial deletion has been described to be involved in X-linked hearing loss [11].
In vitro studies (i.e. gene transcription, protein translation, post-translational modifications, and function) revealed a restricted cochlear and prostatic expression of TBL1Y in humans, as well as the loss of protein function, caused by its premature proteasomal degradation. Present findings identify a new interesting candidate gene for HHL in males combined with some prostate signs and symptoms.
Materials and methods
Please see Supplemental Material.
Results
Clinical phenotype
A multi-generational Italian pedigree showing bilateral sensorineural hearing loss was identified and enrolled through our Clinical Genetics Service in the “IRCCS” Burlo Garofolo Hospital in Trieste, Italy (Fig. 1a). Pure tone audiometric evaluation of affected members of the family showed symmetrical bilateral hearing loss ranging from mild to severe without any vestibular dysfunction (assessed by clinical history and thorough bedside examination) (Fig. 1b). Age of onset was ranging from 20 to 40 years and accordingly only one young male (V:6, 17 years old) was showing a normal audiogram.
Fig. 1.
The five-generation pedigree of an Italian family affected by TBL1Y gene variant. a The family showing bilateral progressive hearing loss and benign prostate hyperplasia with a rare Y-linked inheritance pattern. The individuals affected by hearing loss are indicated with black symbols. All patients analyzed for PSA levels (III:4, IV:1, IV:5, IV:7, and IV:9) showed a benign prostate hyperplasia and increased ratio of the free/total PSA. b Audiometric features of male patients; the V:6 patient does not show the hearing loss phenotype may be due to its young age (17 years old). c Sanger sequencing identified a novel genetic variant r.206A>T;p.Asp69Val in the TBL1Y gene. The variant leads to the substitution of an aspartic acid to valine in position 69. The mutated sequence was identified in the proband and in all male family patients analyzed
The following patients III:4, IV:1, IV:5, IV:7, and IV:9 were also showing benign prostatic hyperplasia and an increased ratio of the free/total PSA level, ranging from 0.3 to 0.6 ng/ml (normal value PSA free/total ratio: 0.10–0.18 ng/ml), for patients V:7 and V:8 we do not have PSA exam data.
Prostate examinations were done routinely and the data here reported refer to the last evaluation carried out when patients were: III:4 (80 years), IV:1 (52 years), IV:5 (48 years), IV:7 (50 years), and IV:9 (46 years).
No other clinical signs and/or symptoms where present in the affected members.
Genetic analysis and identification of genetic variant in TBL1Y
The five-generation pedigree shows an unusual pattern of segregation only in the paternal lineage: this evidence suggested a Y-linked inheritance. A linkage study was then carried out: both two point and multipoint linkage analyses failed to identify any significant Lod Score in autosomal chromosomes, excluding the involvement of any autosomal locus. Thus, on two male patients (III:4 and IV:9) the WES was carried out. Coding variants were filtered against allele frequencies in public and internal databases. In accordance with the variants-filtering procedure described in Methods, we identified an interesting missense variant r.206A>T;p.Asp69Val (rs199659121) in TBL1Y (NM_134259.1) gene in all patients. Sanger sequencing performed on all males (III:4; IV:1; IV:4; IV:5; IV:7; IV:9; V:5; V:6; V:7; V:8) confirmed the presence of this allele in all of them including the young one aged 17 years and not showing the phenotype yet (V:6) (Fig. 1a). The male IV:4 is unaffected and does not show the mutation since he is the husband of the IV:3 component of the family. Their son (V:5) is not affected, since he inherited the Y chromosome from his healthy father.
The variant replaces a highly conserved small aspartic acid with a valine at position 69 (Fig. 1c). PolyPhen-2 [12] (score = 0.932), Mutation Taster [13] (score = 0.9999), and Provean [14] (score: −6.435) predict this variant to be highly damaging for the protein function. The Asp69Val allele is present at a very low frequency in public databases: 0.12% NHLBI Exome Sequencing Project (ESP) Exome Variant Server [15] and 0.04789% in ExAC database [16], thus being still compatible with a possible pathogenetic role, also considering the typical late onset of the disease itself (i.e. 20–40 years), as well as the fact that the ExAC database is enriched for disease-causing alleles.
TBL1Y expression analysis
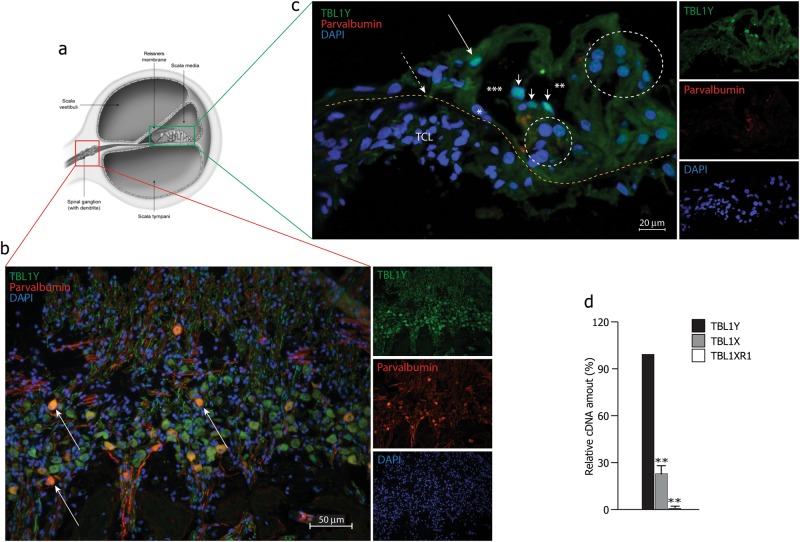
Using a TBL1Y-specific antibody, the subcellular localization of endogenous TBL1Y was investigated on human cochlea slices, counterstained using a parvalbumin-specific antibody, as neuronal marker. Figure 2b shows a peculiar expression of the TBL1Y protein in cochlear spiral ganglion neurons, while Fig. 2c shows its peculiar expression in outer and inner hair cells. Evidences in the literature report TBL1Y mRNA expression in human fetal thyroid gland, brain and kidney, while in adults its expression is limited to the prostate, an organ involved in the patients here described [6, 17]. These findings strongly support the possible role of the TBL1Y gene in both cochlea and prostate function. sqRNA-PCR showed a high expression of the TBL1Y gene in human male cochlea compared to TBL1X (known to be expressed in mouse cochlea) and TBL1XR1 (NM_024665.5) (its homologous on the autosomal chromosome—Fig. 2d).
Fig. 2.
Expression analysis of TBL1Y in human cochlea. a Schematic representation of the human inner ear. b Immunodetection of TBL1Y in the upper middle turn of the human cochlea. TBL1Y showed mainly a peculiar localization in the spiral ganglion neurons (SGNs). Parvalbumin was used to counterstain both cell bodies and neuritis (arrows indicated a few co-labeled neuron cell bodies). TBL1Y (green), parvalbumin (red) and DAPI (blue). c Immunodetection of TBL1Y in outer and inner hair cells in the organ of corti. Yellow line, upper border of the basilar membrane; TCL, tympanic cover layer; long thin arrow, inner hair cell; short arrows, outer hair cells; larger circle, Hensen’s cells; smaller circle, Deiters’ cells; *nucleus of inner pillar cell; ***tunnel of Corti; **Nuel’s space. d sqRT-PCR of TBL1Y, TBL1X, TBL1XR1 genes from adult male human cochlear cDNA. Both TBL1Y and TBL1X are expressed, while TBL1XR1 was not detectable
Impaired protein stability causes quantitative loss of the mutated TBL1Y protein
To better understand a putative causative role of r.206A>T;p.Asp69Val a series of in vitro experiments, ranging from gene transcription to protein translation, subcellular localization, protein stability, conformation, and intrinsic cellular function were carried out. Gene and protein expression studies were performed in HEK 293 cells, transiently transfected with the cDNA of TBL1Y wild-type (TBL1Y-WT) or the mutant (TBL1Y-Asp69Val) Myc-tagged protein. Figure 3a shows a lower amount of mutated protein as compared to the wild-type counterpart 48 h after transfection, despite an equally stable mRNA expression (Fig. 3b). Since missense variants might alter protein biological function, we investigated the sub-cellular localization of the mutant TBL1Y protein. We detected a strong nuclear staining for TBL1Y wild type and, consistent with the immunoblotting results, a much weaker signal in case of the mutant TBL1Y-Asp69Val isoform (Fig. 3c): these data suggest that the mutant protein shows a correct subcellular localization, despite its much lower level of expression, compared to the wild-type counterpart.
Fig. 3.
Missense variant in the TBL1Y gene decreases protein stability. a Western blot showing the amount of TBL1Y wild-type (TBL1Y-WT) and mutant (TBL1Y-Asp69Val) proteins in HEK 293 transfected cells, 24 and 48 h after transfection. The results showed a minimum amount of the mutant protein 48 h post transfection. b qRT-PCR; mRNA quantification after 24 and 48 h from transfection. The qRT-PCR results show no differences between wild-type and mutated mRNA expression. c Immunodetection of TBL1Y-WT and TBL1Y-Asp69Val in HeLa-transfected cells. The wild-type and mutated proteins show the same cellular localization, although with different levels of expression. Myc (green), beta tubulin (red), and DAPI (blue). Scale bar: 50 μm. d Western blot experiment showing the amounts of TBL1Y in lysates from TBL1Y-WT or TBL1Y-Asp69Val-transfected HEK 293 cells, collected at the indicated time points after addition of cycloheximide (CHX). Results, quantified densitometrically, are shown in the graphs below the Western blots, where the amount of TBL1Y protein upon treatment of MG132 for 4 h was set as 100%. The genetic variant decreases protein stability and triggers its premature degradation through the proteasome. e Western blot showing the amount of ubiquitinated TBL1Y-WT and TBL1Y-Asp69Val in lysates of MG132-treated or untreated HEK 293 cells, co-transfected with HA-tagged ubiquitin (HA-Ubi). Ubiquitinated TBL1Y proteins were detected after immunoprecipitation with a Myc-specific antibody followed by Western blotting with a HA-specific antibody. f TBL1Y-WT and TBL1Y-Asp69Val immune-precipitations were in vitro digested with trypsin at different time points (15, 30, 180, and 600 min) and then analyzed by LC–MS/MS. Peptides were identified using the X!Tandem search engine. The abundance of tryptic fragments was determined by the log of the intensity of all the peaks obtained from TBL1Y MS/MS spectra. Results are representative of three independent experiments (four replicates/independent experiment) at the indicated time points; shown are mean ± s.e.m.; *P < 0.05; **P < 0.001. The data were analyzed with one-way ANOVA and Bonferroni’s multiple comparison test
The intact mRNA expression and quantitative loss of the TBL1Y-mutated protein in transfected cells, suggested that post-translational changes might occur in TBL1Y-Asp69Val protein that could alter its stability. Since is it well accepted that enhanced proteolytic degradation of mutant proteins results from their improper folding [18], we hypothesized that misfolding of the mutated TBL1Y protein and its consequent increased degradation would underlie the net loss of the functional TBL1Y. In order to test this hypothesis, we compared the intracellular half-life of the wild-type and mutant TBL1Y proteins in transiently transfected HEK 293 cells, treated with cycloheximide (CHX) to inhibit protein synthesis (Fig. 3d). Whole cell extracts were prepared from transfected cells at different time points from CHX addition, and the amount of TBL1Y was detected by Western blot. We observed that after 4 h of CHX treatment almost 30% of the wild-type protein was still detectable, while a significant loss of mutant TBL1Y protein was observed. In all cases, cell treatment with MG132 resulted in protein stabilization (set as 100% of protein content in Fig. 3d). These results are consistent with the conclusion that a premature degradation occurs for the mutated protein. These evidences were further confirmed by the ubiquitination assay results, shown in Fig. 3e. HEK 293 cells were transiently transfected with either of the two TBL1Y isoforms and HA-tagged ubiquitin (HA-Ubi): ubiquitinated TBL1Y proteins were detected after immunoprecipitation with a Myc-specific antibody, followed by Western blotting with a HA-specific antibody. The wild-type TBL1Y protein ubiquitination pattern increased upon MG132 treatment, indicating its physiological turnover and degradation through the proteasome. Conversely, the TBL1Y mutant showed a much higher ubiquitin binding (by an average more than 40%; quantification is shown in Suppl. Figure 1) even in absence of MG132, suggesting its enhanced ubiquitin-proteasome-mediated degradation. Moreover, the in vitro trypsin digestion profile of the wild type and mutated TBL1Y proteins shows a completely different pattern of peptides, further confirming a putative different folding of the two proteins, possibly leading to the enhanced degradation of the mutated variant (Fig. 3f).
Mutant TBL1Y shows an altered intrinsic cellular function
Considering that the TBL1X homologous protein interacts with the thyroid nuclear receptor through the SMRT co-repressor, we verified whether TBL1Y protein belongs to the same nuclear complex and if the mutated allele impairs protein–protein interactions and therefore the TBL1Y function [19]. Both wild-type and mutated TBL1Y (Myc tagged) were transfected in HEK 293 cells together with a plasmid coding for the SMRT (Flag/Myc-tagged) co-repressor. The co-immunoprecipitation experiment shown in Fig. 4a demonstrates that TBL1Y, as its homologous TBL1X, interacts with SMRT and the interaction is detectable for both wild-type and mutated TBL1Y proteins. The binding efficiency of SMRT mirrors the total amount of the TBL1Y proteins expressed by transfected cells (quantification in Suppl. Figure 2). Suppl. Figure 3 shows the controls of the specificity of the immuno-detection of the experiment. Given these evidences, we investigated whether the genetic variant could affect the intrinsic TBL1Y function of transcriptional activation of genes downstream the thyroid hormone receptor (THR) [20]. We assessed the transcriptional activity of the natural thyroid response element (TRE) controlling the expression of the luciferase reporter gene. HEK 293 cells were transfected with the luciferase reporter together with either the TBL1Y-WT or the TBL1Y-Asp69Val mutant plasmid. Overexpression of the wild-type protein possibly favors the recruitment of co-repressors on the TRE-promoter, inducing a decrease in the transcriptional response. This effect is completely lost in case of overexpression of the TBL1Y-mutated isoform. Consistent with the decay kinetics of the TBL1Y proteins, MG132 treatment restores a comparable repression of the luciferase reporter gene, driven by both wild type and mutant TBL1Y proteins (Fig. 4b).
Fig. 4.
Functional effect of the missense variant in TBL1Y gene. a Co-immunoprecipitation assay of either TBL1Y-WT or TBL1Y-Asp69Val (Myc tagged) with the SMRT protein (Flag/Myc tagged). The results show that both isoforms are able to interact with SMRT, and the binding efficiency mirrors the total amount of the expressed TBL1Y proteins. b The missense variant in the TBL1Y gene alters TRE-luciferase reporter transcription. The results show that in HEK 293 transfected cells, the TRE-luciferase reporter activation is inhibited by the TBL1Y-WT protein, while the mutant has an opposite effect; this effect is reverted in the presence of MG132. Firefly luciferase activity was normalized using Renilla luciferase activity. The histograms show mean ± SEM; n = 3 (four replicates/independent experiment). *P < 0.05, **P < 0.01, vs. control. The data were analyzed with one-way ANOVA and Bonferroni’s multiple comparison test
Discussion
Here, we report an Italian family affected by HHL, benign prostate hyperplasia and increased ratio of the free/total PSA levels for the patrilineal relatives. The severity of hearing impairment ranged from mild to severe with an onset for the patrilineal relatives from 20 to 40 years. A series of genetic and genomic analyses led to the identification of a missense variant in TBL1Y gene (r.206A>T;p.Asp69Val).
Some years ago, it was described that partial deletion of TBL1X (an homologous gene of TBL1Y) was responsible for sensorineural hearing loss with X-linked transmission [11]. To further assess the functional role of TBL1Y in the disease, we analyzed the protein expression profile: TBL1Y has a restricted and specific expression in adult human cochlea and prostate, the only organs affected in our patients. We next investigated the possible presence of impaired gene transcription, mRNA stability alteration, impaired translation, increased protein degradation, defective cellular localization, and loss of intrinsic function of the TBL1Y-mutated protein. None of these various potential mechanisms was altered in the Asp69Val variant, apart from a enhanced protein degradation consequent to a higher ubiquitination level compared to the wild type, leading to a quantitative loss of mutant TBL1Y protein and likely to a decrease of protein function. Furthermore, since it is known that the ubiquitin–proteasome system (UPS) is a complex machinery targeting to proteasomal degradation misfolded proteins [21], we analyzed the folding of both wild type and mutated proteins by in vitro trypsin digestion, followed by mass spectroscopy. Results showed different patterns of peptides derived from the in vitro digestion between mutated and wild-type proteins, confirming the possible misfolding of the TBLY1 mutant as cause of its premature degradation. Moreover, to further investigate the possible role of TBLY1 and better understand the mechanisms underlying the disease, we tested if TBL1Y protein was able to interact, such as its homologous TBL1X, with the SMRT co-repressor, a component of the thyroid nuclear receptor complex. Our results clearly demonstrated the presence of the interaction, which is detectable for both wild-type and mutated TBL1Y proteins. However, the lower level of the mutated isoform affects its physiological function as a transcriptional regulator, fully restored by inhibiting its proteasome degradation.
Summarizing, despite the mutated TBL1Y protein maintains a normal cellular localization and it is able to assemble with the SMRT protein, likely its amount is not sufficient to carry out its function, due to its rapid degradation, impairing thyroid hormone receptor function. Thyroid hormone receptors have been described to play a key role in the development of auditory function in human and mice [22], as well as in prostate cells. For example, it is known that hypothyroidism or iodine deficiency may impair hearing and cause profound deafness [23–26] and that thyroid hormone syndrome (OMIM #188570), due to thyroid receptor beta locus (THRß) mutations, exhibits elevated thyroid hormones (THs) levels and hearing loss [27, 28]. Moreover, mice with targeted disruption of the entire THRß locus, show elevated THs levels and develop profound hearing loss associated with abnormal electrophysiological maturation of inner hair cells [29], while rodents with hypothyroidism show a hearing loss phenotype caused by alteration of the tectorial membrane, whose function is essential to transform the sound from a mechanical to an electrical signal [30]. Despite these evidences, the biological mechanisms controlling cochlear gene expression driven by thyroid hormone receptor activity are still unknown. As already mentioned, the benign prostate hyperplasia combined with the increased ratio of the free/total PSA could be the result of the interaction between TBL1Y and thyroid hormone receptors, both expressed in the prostatic tissue [31, 32].
Moreover, the homologous TBL1X belongs to genes escaping to X inactivation and thus is needed in double dose to properly exert its function in females. Therefore, we can hypothesize that in males, in the presence of a rapid degradation of TBL1Y, due to r.206A>T;p.Asp69Val variant, the activity of TBL1X alone is not enough to avoid the clinical phenotype. In support of this hypothesis, patients affected by the Turner syndrome, carrying a 45 X0 karyotype and thus only one copy of TBL1X gene, show an increased incidence of late-onset sensorineural hearing loss [33, 34]. In the last decade, two Chinese families affected by hearing loss and showing a Y-linked pattern of inheritance have been described. In one of these families the pathogenic mechanism underlying the disease has been explained by the presence of a complex chromosomal rearrangement. In order to exclude possible chromatin rearrangements, we looked for the presence of genomic deletions/insertions and microduplications using a molecular karyotyping protocols based on high density SNP-arrays. No deletions/insertions were identified in autosomal or sexual chromosomes in affected males in our family, nor duplications of genomic regions containing already known genes related to deafness.
Some contradictory data on TBL1Y are present in literature. For example, Yan et al. suggest that TBL1Y, unlike its homologous TBL1X and TBL1XR1, had no repressor activity in transcriptional repression functional assays [39]. In the same manuscript the authors describe a polymorphism c.457+1G>C (NC_000024.10) that would abolish the splicing donor site of intron 7 of TBL1Y, probably leading to a non-functional TBL1Y protein. Unfortunately no functional studies have been carried out and data are not available on the effect of this polymorphism on mRNA or protein function. Furthermore, this polymorphism is present only in the Japanese population (6.3% frequency) [35], and basic clinical and instrumental information about patients is lacking. Thus, it is very difficult to compare the possible clinical outcome of this polymorphism with that of the p.Asp69Val variant described here. In this light, it would be very interesting to look for hearing and prostate function in the before mentioned Japanese cohort.
Moreover, in gnomAD the MAF of r.206A>T variant shows a frequency of 0.063%, that is a bit higher to the disease-causing [36]. Moreover, we have to underlight, that the phenotype here described is a “late onset hearing loss” who affecting subjects aged 20 or more, for this region in contrast with frequency data reported in literature regarded hearing loss. The overall worldwide incidence of hearing loss is much higher in this age group as compared to the congenital one. As a matter of facts it has been reported that the frequency of bilateral hearing loss doubles for every 10 years over the age of 50. Thus, 15% of people between 50 and 59 years have bilateral hearing loss, rising to 31% in people between 60 and 69 years of age, and 63% in those over 70 years [37, 38].
Moreover, a higher incidence of hearing loss in males as compared to females has been also reported in these late onset forms. Both these findings are in agreement with the data described in our manuscript [39]. In addition, in gnomAD database there are no indications regarding the HL phenotype of individuals annotated in this database (i.e. we cannot exclude the inclusion of subjects with late onset phenotypes such as Age-Related Hearing loss, an extremely common complex trait). Furthermore, gnomAD database is enriched with data from many different diseases consortia (e.g. Myocardial Infarction Genetics Consortium, Inflammatory Bowel Disease, Schizophrenia Trios from Taiwan, etc.) and only individuals affected by severe pediatric diseases were removed, but still a high proportion of people affected by other late-onset or mild diseases (as HL can appear in some cases) might have been included. Regarding the possibility to collect phenotypic information of individuals present in gnomAD, the database clearly states that: “Most of the individuals who have contributed data to gnomAD were not fully consented for phenotype data sharing, and unfortunately at this time we are typically unable to provide any information about the clinical status of variant carriers” (http://gnomad.broadinstitute.org/faq).
In conclusion, in this study we present a most likely pathogenic allele in TBL1Y gene in an Italian pedigree with male to male transmission. Functional studies further support the pathogenetic mechanism and its involvement in the auditory and prostate phenotype. Present findings increase our knowledge on the molecular basis of hearing function and benign prostate hyperplasia. Additional studies are needed to further investigate the functional role of TBLY gene in other cases of hearing loss, as well as in patients showing a familial aggregation of benign prostate hyperplasia. Although this gene was only detected in one family (common in heterogeneous genetic disorders), mutational screening of this gene in a larger collection of cases may lead to the identification of other patients/families affected by HHL and benign prostate hyperplasia.
Electronic supplementary material
Acknowledgements
We thank the patients for their participation in this study. We thank the Prof. Morton C., and the Dr. Nahid G. Robertson (Boston, MA, USA) for cDNA of human cochlea. We thank the Prof. V. Perissi (University of California San Diego) for kindly donating the TRE-plasmid. Thanks to Pediatric hospital “IRCCS” Burlo Garofolo, Trieste, Italy for approving the experimental protocol.
Author contributions
M.D.S., C.C., P.G., and M.G. conceived and designed the project. P.G., G.G., and E.R. phenotyped the cases, provided clinical samples and clinical information. D.V. and A.M. were responsible for Exome Sequencing, Sanger sequencing, and D.V. for data analysis. M.D.S. and C.C. performed the functional studies, D.P. performed linkage and SNPs analysis data, L.W. performed the immunofluorescence on human cochlea. M.M. performed the trypsinization assay. The manuscript was drafted by M.D.S., C.C., and P.G. All authors discussed the results and commented on the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
The online version of this article (10.1038/s41431-018-0282-4) contains supplementary material, which is available to authorized users.
References
- 1.Morton CC, Nance WE. Newborn hearing screening—a silent revolution. N Engl J Med. 2006;354:2151–64. doi: 10.1056/NEJMra050700. [DOI] [PubMed] [Google Scholar]
- 2.Deltenre P, Van Maldergem L. Hearing loss and deafness in the pediatric population: causes, diagnosis, and rehabilitation. Handb Clin Neurol. 2013;113:1527–38. doi: 10.1016/B978-0-444-59565-2.00023-X. [DOI] [PubMed] [Google Scholar]
- 3.Morton NE. Genetic epidemiology of hearing impairment. Ann N Y Acad Sci. 1991;630:16–31. doi: 10.1111/j.1749-6632.1991.tb19572.x. [DOI] [PubMed] [Google Scholar]
- 4.ACMG statement. Genetics evaluation guidelines for the etiologic diagnosis of congenital hearing loss. Genetic Evaluation of Congenital Hearing Loss Expert Panel. Genet Med. 2002;4:162–71. doi: 10.1097/00125817-200205000-00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koffelr T, Ushakov K, Avraham BK. Genetics of hearing loss—syndormic. Otolaryngol Clin North Am. 2015;48:1041–61. doi: 10.1016/j.otc.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–37. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- 7.Jobling MA, Lo ICC, Turner DJ, et al. Structural variation on the short arm of the human Y chromosome: recurrent multigene deletions encompassing Amelogenin Y. Hum Mol Genet. 2007;16:307–16. doi: 10.1093/hmg/ddl465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Page DC. Curt Stern Award address. On low expectation exceeded; or, the genomic salvation of the Y chromosome. Am J Hum Genet. 2004;74:399–402. doi: 10.1086/382659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang QJ, Xue Y, Zhang Y, et al. Genetic basis of Y-linked hearing impairment. Am J Hum Genet. 2013;92:301–6. doi: 10.1016/j.ajhg.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu S, Yan J, Wang X, et al. The audiological characteristics of a hereditary Y-linked hearing loss in a Chinese ethnic Tujia pedigree. Int J Pediatr Otorhinolaryngol. 2011;75:202–6. doi: 10.1016/j.ijporl.2010.10.035. [DOI] [PubMed] [Google Scholar]
- 11.Bassi MT, Ramesar RS, Caciotti B, et al. X-linked late-onset sensorineural deafness caused by a deletion involving OA1 and a novel gene containing WD-40 repeats. Am J Hum Genet. 1999;64:1604–16. doi: 10.1086/302408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arora S, Huwe PJ, Sikder R, et al. Functional analysis of rare variants in mismatch repair proteins augments results from computation-based predictive methods. Cancer Biol Ther. 2017;18:519–33. doi: 10.1080/15384047.2017.1326439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwarz JM, Rödelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7:575–6. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 14.Choi Y, Chan AP. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics. 2015;31:2745–7. doi: 10.1093/bioinformatics/btv195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Auer PL, Reiner AP, Wang G, et al. Guidelines for Large-Scale Sequence-Based Complex Trait Association studies: lessons learned from the NHLBI Exome Sequencing Project. Am J Hum Genet. 2016;99:791–801. doi: 10.1016/j.ajhg.2016.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gene: TBL1Y (ENSG00000092377)—gene expression—Homo sapiens—Ensembl genome browser 84.
- 18.Bross P, Corydon TJ, Andresen BS, JØrgensen MM, Bolund L, Gregersen N. Protein misfolding and degradation in genetic diseases. Hum Mutat. 1999;14:186–98. doi: 10.1002/(SICI)1098-1004(1999)14:3<186::AID-HUMU2>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 19.Tomita A, Buchholz DR, Shi YB. Recruitment of N-CoR/SMRT-TBLR1 corepressor complex by unliganded thyroid hormone receptor for gene repression during frog development. Mol Cell Biol. 2004;24:3337–46. doi: 10.1128/MCB.24.8.3337-3346.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perissi V, Aggarwal A, Glass CK, Rose DW, Rosenfeld MG. A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell. 2004;116:511–26. doi: 10.1016/S0092-8674(04)00133-3. [DOI] [PubMed] [Google Scholar]
- 21.Li S, Izumi T, Hu J, et al. Rescue of enzymatic function for disease-associated RPE65 proteins containing various missense mutations in non-active sites. J Biol Chem. 2014;289:18943–56. doi: 10.1074/jbc.M114.552117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ng L, Kelley MW, Forrest D. Making sense with thyroid hormone—the role of T(3) in auditory development. Nat Rev Endocrinol. 2013;9:296–307. doi: 10.1038/nrendo.2013.58. [DOI] [PubMed] [Google Scholar]
- 23.Uziel A. Periods of sensitivity to thyroid hormone during the development of the organ of Corti. Acta Otolaryngol Suppl. 1986;429:23–7. doi: 10.3109/00016488609122726. [DOI] [PubMed] [Google Scholar]
- 24.Meza G, Acuña D, Escobar C. Development of vestibular and auditory function: effects of hypothyroidism and thyroxine replacement therapy on nystagmus and auditory evoked potentials in the pigmented rat. Int J Dev Neurosci. 1996;14:515–22. doi: 10.1016/0736-5748(95)00100-X. [DOI] [PubMed] [Google Scholar]
- 25.Refetoff S, DeWind LT, DeGroot LJ. Familial syndrome combining deaf-mutism, stuppled epiphyses, goiter and abnormally high PBI: possible target organ refractoriness to thyroid hormone. J Clin Endocrinol Metab. 1967;27:279–94. doi: 10.1210/jcem-27-2-279. [DOI] [PubMed] [Google Scholar]
- 26.Forrest D, Erway LC, Ng L, Altschuler R, Curran T. Thyroid hormone receptor β is essential for development of auditory function. Nat Genet. 1996;13:354–7. doi: 10.1038/ng0796-354. [DOI] [PubMed] [Google Scholar]
- 27.Lee JH, Kim EY. Resistance to thyroid hormone due to a novel mutation of thyroid hormone receptor beta gene. Ann Pediatr Endocrinol Metab. 2014;19:229–31. doi: 10.6065/apem.2014.19.4.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Slezak R, Lukienczuk T, Noczynska A, et al. A novel p.E311K mutation of thyroid receptor beta gene in resistance to thyroid hormone syndrome, inherited in autosomal recessive trait. Horm Metab Res. 2012;44:704–7. doi: 10.1055/s-0032-1312666. [DOI] [PubMed] [Google Scholar]
- 29.Forrest D, Hanebuth E, Smeyne RJ, et al. Recessive resistance to thyroid hormone in mice lacking thyroid hormone receptor beta: evidence for tissue-specific modulation of receptor function. EMBO J. 1996;15:3006–15. doi: 10.1002/j.1460-2075.1996.tb00664.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Griffith AJ, Szymko YM, Kaneshige M, et al. Knock-in mouse model for resistance to thyroid hormone (RTH): an RTH mutation in the thyroid hormone receptor beta gene disrupts cochlear morphogenesis. J Assoc Res Otolaryngol. 2002;3:279–88. doi: 10.1007/s101620010092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferreira ASS, Guerra MR, Lopes HE, Lima UTM, Vasconcelos YA, Teixeira MTB. Brachytherapy and radical prostatectomy in patients with early prostate cancer. Rev Assoc Méd Bras. 2015;61:431–9. doi: 10.1590/1806-9282.61.05.431. [DOI] [PubMed] [Google Scholar]
- 32.Aranda N, Sosa S, Delgado G, Aceves C, Anguiano B. Uptake and antitumoral effects of iodine and 6-iodolactone in differentiated and undifferentiated human prostate cancer cell lines. Prostate. 2013;73:31–41. doi: 10.1002/pros.22536. [DOI] [PubMed] [Google Scholar]
- 33.Elsheikh M, Dunger DB, Conway GS, Wass JAH. Turner’s syndrome in adulthood. Endocr Rev. 2002;23:120–40. doi: 10.1210/edrv.23.1.0457. [DOI] [PubMed] [Google Scholar]
- 34.Morimoto N, Tanaka T, Taiji H, et al. Hearing loss in Turner syndrome. J Pediatr. 2006;149:697–701. doi: 10.1016/j.jpeds.2006.06.071. [DOI] [PubMed] [Google Scholar]
- 35.Yan HT, Shinka T, Kinoshita K, et al. Molecular analysis of TBL1Y, a Y-linked homologue of TBL1X related with X-linked late-onset sensorineural deafness. J Hum Genet. 2005;50:175–81. doi: 10.1007/s10038-005-0237-9. [DOI] [PubMed] [Google Scholar]
- 36.Shearer AE, Eppsteiner RW, Booth KT, Ephraim SS, et al. Utilizing ethnic-specific differences in minor allele frequency to recategorize reported pathogenic deafness variants. Am J Hum Genet. 2014;4:445–53. doi: 10.1016/j.ajhg.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bowl MR, Dawson SJ. The mouse as a model for age-related hearing loss. Gerontology. 2015;61:149–57. doi: 10.1159/000368399. [DOI] [PubMed] [Google Scholar]
- 38.Tu NC, Friedman RA. Age-related hearing loss: unraveling the pieces. Laryngoscope Investig Otolaryngol. 2018;21:68–72. doi: 10.1002/lio2.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Agrawal Y, Platz EA, Niparko JK. Prevalence of hearing loss and differences by demographic characteristics among USadults: data from the National Health and Nutrition Examination Survey, 1999–2004. Arch Intern Med. 2008;14:1522–30. doi: 10.1001/archinte.168.14.1522. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.