Abstract
The phenotypic heterogeneity of Lamin A/C (LMNA) variants renders it difficult to classify them. As a consequence, many LMNA variants are classified as variant of unknown significance (VUS). A number of studies reported different types of visible nuclear abnormalities in LMNA-variant carriers, such as herniations, honeycomb-like structures and irregular Lamin staining. In this study, we used lamin A/C immunostaining and nuclear DAPI staining to assess the number and type of nuclear abnormalities in primary dermal fibroblast cultures of laminopathy patients and healthy controls. The total number of abnormal nuclei, which includes herniations, honeycomb-structures, and donut-like nuclei, was found to be the most discriminating parameter between laminopathy and control cell cultures. The percentage abnormal nuclei was subsequently scored in fibroblasts of 28 LMNA variant carriers, ranging from (likely) benign to (likely) pathogenic variant. Using this method, 27 out of 28 fibroblast cell cultures could be classified as either normal (n = 14) or laminopathy (n = 13) and no false positive results were obtained. The obtained specificity was 100% (CI 40–100%) and sensitivity 77% (46–95%). We conclude that assessing the percentage of abnormal nuclei is a quick and reliable method, which aids classification or confirms pathogenicity of identified LMNA variants causing formation of aberrant lamin A/C protein.
Subject terms: Disease genetics, Cardiomyopathies
Introduction
LMNA variants can cause a plethora of phenotypes that are collectively called laminopathies [1]. At least 12 different types can be distinguished [1], presenting as multi-organ systemic diseases or as tissue-specific diseases. Systemic diseases are often due to sporadic variants that lead to severe clinical symptoms such as Hutchinson Gilford Progeria Syndrome (HGPS) causing premature ageing [2]. In rare cases, compound heterozygous variants can lead to a recessive Progeria phenotype [3]. Tissue-specific laminopathies are much more common, and include Emery Dreifuss Muscular dystrophy (EDMD), Dunnigan type familial partial lipodystrophy (FPLD2), limb girdle dystrophy, and dilated cardiomyopathy (DCM). Genotype-phenotype correlations have not yet been fully resolved. On the one hand, different tissues can be affected in laminopathies arising from a single point variant, leading to overlapping syndromes [4]. Even within a single family, different phenotypes can be found. As example, patients carrying an identical LMNA variant were diagnosed as having DCM, DCM with Emery-Dreifuss muscular dystrophy (EDMD)-like symptoms and DCM with limb girdle muscular dystrophy (LGMD)-like symptoms [5].
The genetic heterogeneity of DCM in combination with phenotypic heterogeneity and variable penetrance of LMNA variants complicates LMNA variant classification. Following the guidelines of the American College of Medical Genetics and Genomics [6], LMNA variants are often classified as variant of unknown significance (VUS) in the absence of segregation and/or functional data from the literature. A VUS neither explains, nor excludes a clinical diagnosis, which poses challenges for patient counseling. In order to study the functional effect of LMNA variant, a number of groups have assessed the nuclear structure of fibroblasts from LMNA-mutated persons and linked nuclear abnormalities in dermal fibroblasts with the type of variant, and disease phenotype [7–13]. While most laminopathy cell cultures showed nuclear abnormalities, such as nuclear herniations (blebs) and/or irregularities in lamin staining, seen as honeycomb structures, it was difficult to correlate these nuclear abnormalities with the type of the variant, location in the protein or disease status [13]. In this paper we describe the development of a simple analysis procedure using lamin A/C immunostaining and DAPI staining to assess nuclear morphology. Combined with linear regression analysis, it can be determined whether a fibroblast culture qualifies as laminopathy and herewith this method will aid to the classification of LMNA variants based on nuclear morphology analysis of the patients’ dermal fibroblasts.
Materials and methods
Cell culture
Human dermal fibroblast cultures were obtained from dermal biopsies after written informed consent as described previously [14]. Anonymous control fibroblasts were obtained from healthy individuals or individuals with a variant in a gene that is not expressed in fibroblasts, e.g., SCN5A or MYH7. Whenever possible, fibroblasts were stained at a low passage number (mean 3.4 ± 1.1, range p2–p7). Cells were seeded onto 18 mm round glass coverslips (Menzel) without coating and grown for 48 h to a confluence of 50–75%. Cells were fixed in 100% methanol for 15 min at −20 °C and were stored at 4 °C in PBS containing 0.01% Na-azide until use.
Genetic analysis
Sanger sequencing or next-generation sequence analysis and interpretation of LMNA (NG_008692.2; NM_170707.3), SCN5A (NG_008934.1; NM_198056.2) and MYH7 (NG_007884.1; NM_00257.3); EMD (NG_08677.1; NM_000117.2) gene variants was performed as described previously [15]. A VUS or (likely) pathogenic variant in LMNA was excluded in control fibroblasts. Initial classification of identified LMNA variants was performed in line with the ACMG standards and guidelines [6]. All reported variants have been uploaded into the publically available LOVD database (https://databases.lovd.nl/shared/genes/LMNA), patient IDs: 165,050; 165,051; 165,052; 165,045; 165,042; 165,053; 165,054; 165,044; 164,979; 164,980; 164,981; 165,048; 165,003; 165,047; 164,804; 165,023; 164,810; 165,005–165,009; 165,029; 165,031; 165,010; 165,012; 165,011; 165,019; 165,046; 165,016; 181,217–181,221.
Immunofluorescence
All samples were pre-incubated in PBS containing 3% BSA (Roche Diagnostics, Mannheim, Germany) and then incubated for 60 min with primary monoclonal antibody JoL2 for Lamin A/C [16] (IgG1, kindly provided by C. Hutchison, University of Durham, UK) 1:50 diluted in PBS containing 3% BSA. Alternatively, a Lamin A mouse monoclonal antibody 133A2 (IgG3, Nordic-MUbio, Susteren, The Netherlands, diluted 1:1000); or Lamin C specific rabbit polyclonal antibody RalC, (Nordic-MUbio, Susteren, The Netherlands, diluted 1:500) were used.
After washing with PBS, FITC conjugated rabbit anti-mouse Ig antibody (Dakopatts, Glostrup, DK) diluted 1:100 in PBS/BSA was applied and incubated for 60 min. After another series of washing steps in PBS, cells were mounted in 90% glycerol, containing 20 mM Tris–HCl pH 8.0, 0.02% NaN3, 2% 1,4-di-azobicyclo-(2,2,2)-octane (DABCO; Merck, Darmstadt, Germany), and diamidino-2-phenylindole (DAPI; 0.5 μg/ml Sigma-Aldrich).
Detection of nuclear abnormalities
For every patient’s fibroblast culture at least 2 × 100 cells in different areas of the sample were evaluated using a Leica DMRBE fluorescence microscope (Leica, Mannheim, Germany), equipped with a 63x oil objective (Plan Apo, NA 1.32). Different aspect of the nuclear morphology were assessed, i.e., presence of nuclear shape abnormalities, seen as irregular lining of the nuclear membrane, forming nuclear blebs (herniations), extensive lobulations or donut-like invaginations of the nucleus. In addition, Lamin staining abnormalities were scored, including extranuclear staining, and the presence of so-called honeycombs. Also, the intensity of staining was registered as being weak, moderate or strongly positive. Part of the samples were re-counted independently by a second examiner, revealing that only a limited interobserver variation was found (variation 1.09 ± 0.25% (n = 7) (mean ± SEM). Finally, the presence of intranuclear aggregates was registered, after excluding intranuclear invaginations that are common also in normal fibroblasts.
Statistical analyses
A linear logistic regression model was constructed to classify the controls versus the laminopathy patients. A hierarchical model was built allowing for the percentage of normal cells, or cells with blebs, honeycombs, or donut cells present to be taken into account. The inference criterion used for comparing the models is their ability to predict the observed data, i.e., models are compared directly through their minimized minus log-likelihood. When the numbers of parameters in models differ, they are penalized by adding the number of estimated parameters, a form of the Akaike information criterion (AIC) [17]. All statistical analysis presented were performed using the freely available program R [18]. The formula: round(1−(1/(1 + exp(−145.1 + 1554.9 × total abnormal count/total cCount))),2) was generated in order to classify a fibroblast cell-line as “laminopathy” or “normal”.
Results
Determination of the “nuclear morphology classifier“
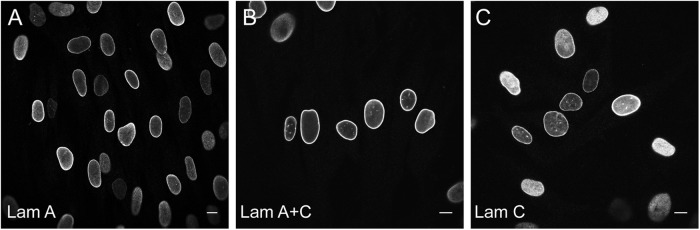
For morphological analysis, different LMNA antibodies detecting lamin A only; lamin A and lamin C; and lamin C only, were initially tested. As shown in Fig. 1, similar results were obtained with all three antibodies. Lamin A showed in general a relative homogeneous staining reaction in all nuclei, while the lamin C antibody showed a more heterogeneous labeling. The lamin A + C antibody shows an intermediate staining pattern. While some variations in intensity levels occurred, none of the patient samples examined showed a clear differential expression between lamin A and C. Nuclei, not showing any lamin staining were not detected. Therefore, for further development of the nuclear morphology classifier, only the JoL-2 antibody (recognizing both lamin A and lamin C) and DAPI for nuclear counterstaining were used.
Table 1.
Overview of published laminopathy fibroblasts used in this study
| ID | LMNA nucleotidea | Predicted potein change | Zygosity | Phenotypeb | Referencesc |
|---|---|---|---|---|---|
| 1 | c.94_96del | p.(Lys32del) | Heterozygous | EDMD | [13, 29] |
| 2 | c.777T > A | p.(Tyr259*) | Homozygous | Died shortly after birth | [30–32] |
| 3 | c.992G > A | p.(Arg331Gln) | Heterozygous | DCM | [33–35] |
| 4 | c.1315C > T | p.(Arg439Cys) | Heterozygous | FPLD | [20, 31, 36] |
| 5 | c.1444C > T | p.(Arg482Trp) | Heterozygous | FPLD | [10, 36, 37] |
| 6 | c.1583C > T | p.(Thr528Met) | Heterozygous | DCM | [3, 31, 38] |
| 7 | c.[1583C > T];[1619T > C] | p.[(Thr528Met)];[(Met540Thr)] | Compound heterozygous | HGPS | [3, 31, 38] |
| 8 | c.1609-12T > G | p.(Glu537Val_fs*14) | Heterozygous | Heart-Hand syndrome | [39] |
| 9 | c.1824C > T | cryptic splice donor: p.(Gly607_656del) | Heterozygous | HGPS | [40–42] |
aLMNA NG_008692.2 NM_170707.3
bPhenotype abbreviations: EDMD Emery Dreifuss muscular dystrophy, DCM dilated cardiomyopathy, FPLD familial partial lipodystrophy, HGPS Hutchinson Gilford Progeria syndrome
cReferences are limited to maximal three per variant; additional publications are available via HGMD website [58]
Fig. 1.
Immunofluorescence labeling of control fibroblast cells with different A-type lamin antibodies. a Lamin A antibody 133A2; b lamin A + C antibody JoL2; c lamin C antibody RalC
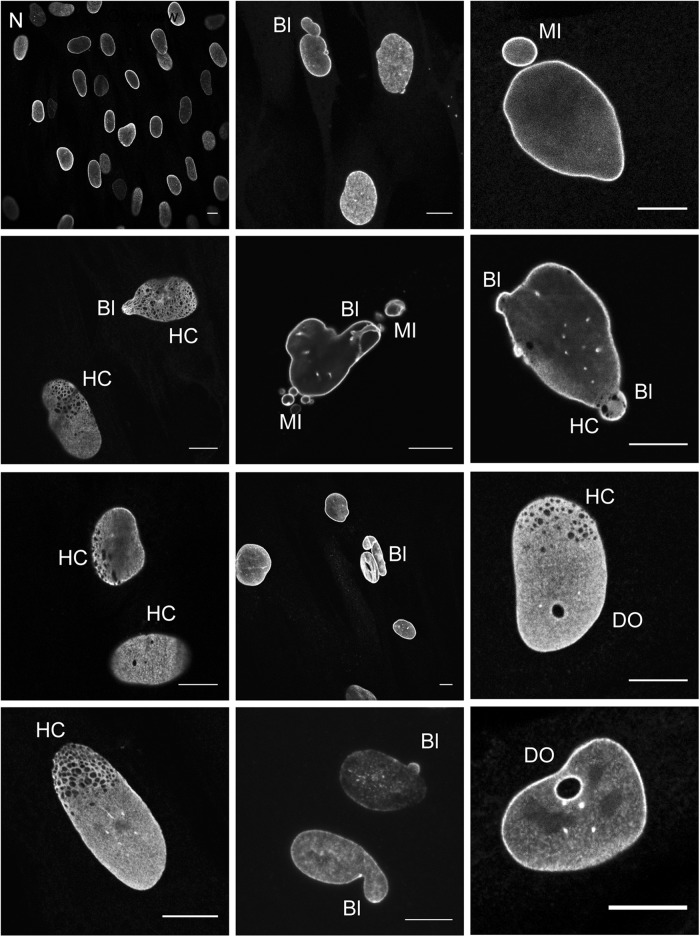
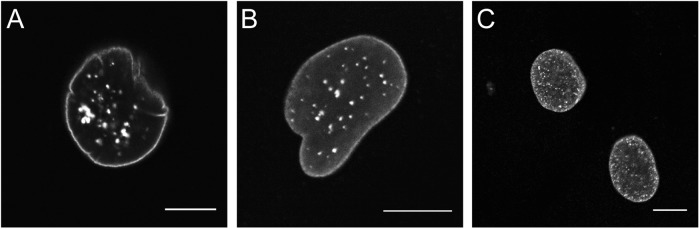
The morphology of ≥200 nuclei per cell culture was analyzed for all 8 controls and 9 fibroblast cultures with a LMNA variant that were published previously and designated as pathogenic and laminopathy based on clinical phenotype, segregation data and/or functional analyses (Table 1). In addition to classifying nuclei as normal or abnormal, the types of nuclear malformations were noted: blebs (herniations, including micronuclei), honeycomb structures, donut-like structures, or combinations of aforementioned categories (see Fig. 2; Table 2). In some cultures, intra-nuclear aggregates occurred next to a normal lamina staining (Fig. 3). However, since these aggregates varied dramatically in size as well as in number between cells within the same culture and were even noted in some normal cells, cells with aggregates were not included in the classifier. Statistical analyses of these parameters for the healthy and laminopathy cell-lines were conducted to determine the most discriminating parameter. As shown in Table 3, the parameter “percentage of abnormal nuclei” resulted in the lowest AIC, and was therefore most discriminative and used for subsequent analyses. Using this parameter, the eight control cell-lines showed on average 4.8 ± 1.0% abnormal nuclei (range 3.3–6%) and the nine laminopathy samples showed 26.3 ± 13.6% abnormal nuclei (range 10.6–53%).
Fig. 2.
Types of scored nuclear abnormalities in laminopathy cell cultures N, normal; BL, bleb; HC, honeycombs; DO, donuts, MI, micronuclei. Note that some cells contain more than 1 nuclear abnormality
Table 2.
Results nuclear morphology analysis of control- and published laminopathy fibroblasts
| ID | LMNA varianta | Other varianta | % abnorm. nuclei | # total nuclei | # norm. nuclei | # abnorm. nuclei | type of abnormalityb | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| B | HC | D | B+HC | B+D | |||||||
| 1 | p.(Lys32del) | 10.67 | 300 | 268 | 32 | 19 | 4 | 8 | 1 | 0 | |
| 2 | p.(Tyr259*) | 53.00 | 300 | 141 | 159 | 6 | 145 | 1 | 7 | 0 | |
| 3 | p.(Arg331Gln) | 22.00 | 500 | 390 | 110 | 31 | 68 | 10 | 0 | 1 | |
| 4 | p.(Arg439Cys) | 24.50 | 200 | 151 | 49 | 31 | 10 | 6 | 1 | 1 | |
| 5 | p.(Arg482Trp) | 24.50 | 200 | 151 | 49 | 43 | 0 | 4 | 1 | 1 | |
| 6 | p.(Thr528Met) | 13.50 | 800 | 692 | 108 | 29 | 50 | 18 | 11 | 0 | |
| 7 | p.(Thr528Met) + p.(Met540Thr) | 39.80 | 1000 | 602 | 398 | 46 | 202 | 10 | 140 | 0 | |
| 8 | p.(Glu537Val_fs*14) | 30.00 | 200 | 140 | 60 | 11 | 13 | 26 | 7 | 3 | |
| 9 | p.(Gly607_656del) | 14.12 | 500 | 438 | 62 | 38 | 10 | 11 | 2 | 1 | |
| 10 | None | 3.33 | 300 | 290 | 10 | 4 | 4 | 1 | 1 | 0 | |
| 11 | None | 4.33 | 300 | 287 | 13 | 11 | 0 | 2 | 0 | 0 | |
| 12 | None | 4.67 | 300 | 286 | 14 | 11 | 0 | 3 | 0 | 0 | |
| 13 | None | SCN5A p.(Phe1617del) | 6.00 | 300 | 282 | 18 | 8 | 3 | 3 | 4 | 0 |
| 14 | None | SCN5A p.(Phe1617del) | 5.67 | 300 | 283 | 17 | 3 | 7 | 6 | 1 | 0 |
| 15 | None | 5.33 | 300 | 284 | 16 | 11 | 0 | 5 | 0 | 0 | |
| 16 | None | MYH7 p.(Ala161Pro) | 5.33 | 300 | 284 | 16 | 9 | 1 | 6 | 0 | 0 |
| 17 | None | 3.33 | 300 | 290 | 10 | 5 | 1 | 4 | 0 | 0 | |
aProtein change identified variant according to LMNA NG_008692.2 NM_170707.3; SCN5A NG_008934.1 NM_198056.2; MYH7 NG_007884.1 NM_00257.3)
bType of abnormality: B, Blebs/herniations; HC, honeycomb-like structure; D, donut-like structures, or combinations of aforementioned
Fig. 3.
Differently sized intranuclear aggregates in laminopathy cells detected with the three lamin antibodies. a Antibody to lamin C; b antibody to lamin A; c antibody to lamin A+C
Table 3.
Quality of statistical model using different parameters
| Nuclear morphology parameter | AICa |
|---|---|
| Percentage of blebs | 19.32 |
| Percentage of honeycombs | 16.46 |
| Percentage of donuts | 22.92 |
| Percentage of blebs and honeycombs | 22.01 |
| Percentage of blebs and donuts | 19.28 |
| Percentage of all abnormal nuclei (blebs, donuts, and honeycombs) | 4.00 |
aAIC Akaike information criterion; Each row of the table represents the fit of a model containing the morphological parameter mentioned in the first column. A smaller AIC denotes a better fitting model
Validation study “nuclear morphology classifier”
As noted above, the percentage of abnormal nuclei was identified as the most discriminating parameter between laminopathy and control cells. Subsequently, a validation study comprising nuclear morphology analysis of 28 fibroblast cell-lines was performed and verified by second opinion from expert (Fig. 4). These cell-lines contained a LMNA variant that was classified by molecular genetics criteria as (likely) benign, variant of unknown significance (VUS), likely pathogenic variant or pathogenic variant (Table 4). The number of normal and abnormal nuclei per cell culture was determined according to the “nuclear morphology classifier”. A cell culture with a classifier score ≥0.95 was considered as “laminopathy” and a value of ≤0.05 was considered as “normal”. If the classifier value ranged between 0.05 and 0.95, the cell-line was considered as “unclassified”. As shown in Table 4, 27 out of the 28 cell-lines (96%) could be classified with ≥95% probability as laminopathy or normal nuclear morphology. Only ID 37, containing a LMNA VUS c.1634G>A ((p.(Arg545His)), had a score of 51% and could therefore not be classified. Out of the 27 classified cell-lines, 13 were defined as laminopathy and 14 as normal. The laminopathy group contained fibroblasts with either a pathogenic variant (n = 1), a likely pathogenic variant (n = 8) or a VUS (n = 4). The group classified as normal mainly contained VUS cell cultures (n = 8), two cell cultures with a (likely) benign polymorphism, one recessive pathogenic variant, one likely pathogenic variant and two cell cultures containing a pathogenic variant. Fibroblasts containing either a (likely) benign polymorphism or recessive pathogenic LMNA c.892C>T (p.(Arg298Cys)) variant [19] (n = 3) and fibroblasts containing a (likely) pathogenic variant (n = 13) were used to calculate sensitivity and specificity of the nuclear morphology classifier. Specificity was found to be 100% (CI 29–100%) and the sensitivity was 77% (CI 46–95%). With respect to reproducibility, for two LMNA variants, nuclear morphology of two unrelated individuals was analyzed. Both fibroblast cell cultures with the LMNA c.1930C>T (p.(Arg644Cys)) variant displayed normal nuclear morphology, and nuclear morphology of two carriers of LMNA c.313_314delinsTT (p.(Glu105Leu)) both presented as laminopathy.
Fig. 4.
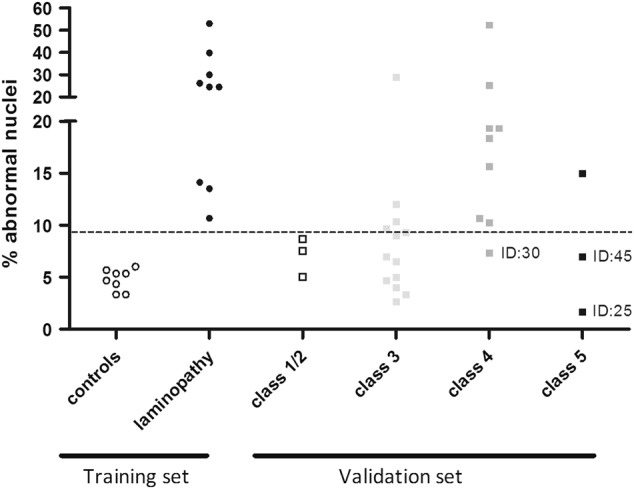
Distribution of the percentage abnormal nuclei identified per class LMNA variant. The percentage abnormal nuclei in controls (open circles) and laminopathy patients (black circles) were used as training set to generate the classifier. The percentage of nuclear abnormalities identified in the validation set are shown grouped based on their initial clinical genetic classification: class 1/2 ((likely) benign or recessive pathogenic variants in heterozygous state) in open squares; class 3 (VUS) light gray squares; class 4 (likely pathogenic) dark gray squares; class 5 (pathogenic) black squares. The three false negative variants are indicated by their ID number
Table 4.
Results nuclear morphology analysis in validation study
| ID | LMNA varianta | Predicted effect on protein level | LMNA domainb | References | Initial classificationc | % abnormal nuclei | Total # nuclei | # normal nuclei | # abnormal nuclei | Nuclear morphology classifier (score) | Age at biosy | Fibroblast passage nr. | Sex | Phenotype |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 18 | No LMNA variant | NA | -- | 7.50 | 200 | 185 | 15 | Normal (0.00) | 38 | 3 | Female | Partial lipodystrophy | ||
| 44 | c.1968 + 26A>G | NA | 1 | 8.67 | 300 | 274 | 26 | Normal (0.00) | 52 | 2 | Male | DCM | ||
| 29 | c.892C > T+EMD c.110-112del | p.(Arg298Cys) + EMD p.(Lys37del) | Coil 2 | [30] | 2 (5AR)d 2 (5 X-linked) | 5.00 | 200 | 184 | 16 | Normal (0.00) | 41 | 7 | Female | ACD |
| 35 | c.1619T > C | p.(Met540Thr) | Tail | [3, 31, 38] | 3 (5 AR)e | 9.00 | 600 | 546 | 54 | Normal (0.01) | 32 | 5 | Female | none |
| 27 | c.737A > G | p.(Gln246Arg) | Coil 2 | NA | 3 | 4.00 | 200 | 192 | 8 | Normal (0.00) | 48 | 3 | Male | LVNC |
| 36 | c.161C > T | p.(Thr54Met) | Coil 1a | NA | 3 | 7.00 | 300 | 279 | 21 | Normal (0.00) | 62 | 3 | Male | HCM |
| 38 | c.1718C > T | p.(Ser573Leu) | Tail | [43, 44] | 3 | 4.67 | 300 | 286 | 14 | Normal (0.00) | 45 | 4 | Male | LQT |
| 40 | c.1786_1800del | p.(Asp596_Ala600del) | Tail | NA | 3 | 5.00 | 300 | 285 | 15 | Normal (0.00) | 17 | 3 | Male | NCCM |
| 41 | c.1879C > T | p.(Arg627Cys) | Tail | [45] | 3 | 6.50 | 600 | 561 | 39 | Normal (0.00) | 54 | 4 | Male | none |
| 42 | c.1930C > T | p.(Arg644Cys) | Tail | [46–48] | 3 | 2.67 | 300 | 292 | 8 | Normal (0.00) | 59 | 5 | Male | LGMD, CM |
| 43 | c.1930C > T | p.(Arg644Cys) | Tail | [46–48] | 3 | 3.33 | 300 | 290 | 10 | Normal (0.00) | 33 | 2 | Male | VF, HCM |
| 30 | c.949G > A | p.(Glu317Lys) | Coil 2 | [27, 28, 49] | 4 | 7.33 | 300 | 278 | 22 | Normal (0.00) | 60 | 3 | Female | DCM |
| 25 | c.568C > T | p.(Arg190Trp) | Coil 1b | [22, 27, 50] | 5 | 1.67 | 300 | 295 | 5 | Normal (0.00) | 34 | 3 | Male | DCM |
| 45 | c.514_1995del | Haploinsuffiency | [12] | 5 | 7.00 | 300 | 279 | 21 | Normal (0.00) | 50 | 3 | Male | AF | |
| 37 | c.1634G > A | p.(Arg545His) | Tail | [51–53] | 3 | 9.33 | 300 | 272 | 28 | Unclassified (0.51) | 42 | 3 | Female | LVNC |
| 19 | c.208G > A | p.(Val70Ile) | Coil 1a | NA | 3 | 29.00 | 300 | 213 | 87 | Laminopathy (1.00) | 42 | 3 | Male | DCM |
| 20 | c.236C > A | p.(Ala79Asp) | Coil 1b | NA | 3 | 10.33 | 300 | 269 | 31 | Laminopathy (1.00) | 59 | 3 | Male | DCM |
| 32 | c.1201C > T | p.(Arg401Cys) | Tail | [40, 49, 54, 55] | 3 | 9.67 | 300 | 271 | 29 | Laminopathy (0.99) | 78 | 4 | Male | DCM |
| 39 | c.173G > T | p.(Gly58Val) | Coil 1a | NA | 3 | 12.00 | 300 | 264 | 36 | Laminopathy (1.00) | 52 | 4 | Female | ARVC |
| 21 | c.247delinsCC | p.(Ala83Profs*11) | Coil1b | NA | 4 | 25.33 | 300 | 224 | 76 | Laminopathy (1.00) | 46 | 5 | Female | DCM |
| 22 | c.313_314delinsTT | p.(Glu105Leu) | Coil 1b | NA | 4 | 10.25 | 400 | 359 | 41 | Laminopathy (1.00) | 70 | 3 | Male | DCM |
| 23 | c.313_314delinsTT | p.(Glu105Leu) | Coil 1b | NA | 4 | 18.33 | 300 | 245 | 55 | Laminopathy (1.00) | 46 | 3 | Male | DCM, AF, mitralis insuf. |
| 26 | c.647G > A | p.(Arg216His) | Coil 1b | NA | 4 | 15.67 | 300 | 253 | 47 | Laminopathy (1.00) | 63 | 3 | Male | DCM |
| 28 | c.810G > A | r.766_810del45 | [56] | 4 | 52.33 | 300 | 143 | 157 | Laminopathy (1.00) | 30 | 2 | Female | DCM, muscle weakness | |
| 33 | c.1300G > A | p.(Ala434Thr) | Tail | NA | 4 | 19.33 | 300 | 242 | 58 | Laminopathy (1.00) | 50 | 4 | Male | DCM, VF |
| 34 | c.1592_1594del | p.(Ile531del) | Tail | NA | 4 | 10.67 | 300 | 268 | 32 | Laminopathy (1.00) | 44 | 2 | Female | EDMD |
| 31f | c.[992G > A];[1879C > T] | p.[(Arg331Gln)]; [(Arg627Cys)] | Coil 2 + Tail | [33–35] | 4 3 | 19.33 | 500 | 350 | 150 | Laminopathy (1.00) | 53 | 3 | Male | DCM |
| 24 | c.481G > A | p.(Glu161Lys) | Coil 1b | [22, 50, 57] | 5 | 15.00 | 300 | 255 | 45 | Laminopathy (1.00) | 53 | 3 | Male | DCM, VF |
ACD atrial cardiac disease, AF atrial fibrillation, AR autosomal recessive inheritance, ARVC Arrhythmogenic right ventricle cardiomyopathy, CM cardiomyopathy, DCM dilated cardiomyopathy, EDMD emery dreifuss muscular dystrophy, HCM hypertrophic cardiomyopathy, LVNC left ventricle non-compaction, VF ventricular fibrillation, X-linked X-linked inheritance
aDNA and predicted protein change of identified variant according to LMNA NG_008692.2 NM_170707.3, EMD NG_008677.1 NM_000117.2. All variants were identified heterzygously
bAffected protein domain according to Nextprot structures LMNA isoform A
cInitial classification of genetic variant reported to clinican prior to nuclear morphology analysis, class 1: benign variant, class 2: likely benign variant, class 3: variant of unknown significance, class 4: likely pathogenic variant, class 5: pathogenic variant
dHeterozygous variant and no second LMNA variant demonstrated. This variant has only been demonstrated to cause disease in homozygous state, but no functional effect has been demonstrated in heterozygous state and is regarded as likely benign in the analysis
eHeterozygous variant and no second LMNA variant demonstrated. This variant has been shown to cause disease in compound heterozygous form (ID7), but the functional effect in heterozygous form is unclear and is included als class 3 (VUS) in current analysis
fID31 compound heterozyous carrier of two LMNA variants, these variants were tested in isolation in ID 3 and ID 41
Discussion
The aim of the present study was to assess if aberrant nuclear morphology is indicative of a pathogenic LMNA variant and can be used to aid classification of identified genetic variants in LMNA. We assessed nuclear morphology of 9 laminopathy dermal fibroblast cultures and 8 control fibroblast cultures, as training set, and demonstrated that an increased percentage of abnormal nuclei, irrespective of the type of nuclear malformation (herniation, honeycomb structure or donut shape), is the most discriminating parameter between normal and laminopathy cells. Subsequent assessment of the percentage of abnormal nuclei in validation set of fibroblast cultures containing a (likely) pathogenic LMNA variant (n = 13) or a (likely) benign LMNA variant (n = 3) demonstrated a 100% specificity (CI 29–100%) with respect to determining pathogenicity, as none of the three negative samples were classified as laminopathy. However, the confidence interval is large due to limited availability of cell cultures containing (likely) benign LMNA variants and future inclusion of more negative samples is warranted to reduce this confidence interval. From the 13 samples containing a (likely) pathogenic LMNA variant, 10 samples displayed an excessive percentage of abnormal nuclei and were classified as laminopathy, resulting in a sensitivity of 77% (CI 46–95%). Moreover, the same classification was obtained for two unrelated individuals carrying the LMNA c.313_314delinsTT (p.(Glu105Leu)) variant for two unrelated carriers of the c.1930C>T (p.(Arg644Cys)) variant, demonstrating reproducibility of the developed method. Taken together, we conclude that a laminopathy classification based on significant number of abnormal nuclei in fibroblasts is a quick and reliable method to confirm pathogenicity and aid classification of LMNA variants that predict formation of aberrant lamin A/C protein, while observation of normal nuclear morphology does not rule out pathogenicity.
To our knowledge, few attempts were made to categorize laminopathies based on their nuclear abnormalities, but this is the first and largest study undertaken (n = 45) that uses nuclear morphology assessment of fibroblasts containing an established LMNA variant or no LMNA variant in order to rank newly identified LMNA variants as laminopathy or normal based on the percentage of abnormal nuclei. The percentages of abnormal nuclei observed in the control and laminopathy group in our study are in line with the percentages reported by Decaudain et al., who showed 15–25% of dysmorphic nuclei with herniations in all six patients with a LMNA variant compared to 5% abnormal nuclei in control fibroblasts [20]. In contrast, Muchir et al. observed <1% abnormal nuclei in controls [13], this may be a consequence of various methodological differences, criteria and sensitivity. For instance, the detection of honeycomb-like structures is highly dependent on the quality of the staining and the detection method used. Honeycomb-like structures can be missed due to a weak antibody labeling, as well as by background labeling in the affected nuclei. Moreover, most of these structures are only visible with a high-resolution oil lens, by systemically focusing individual nuclei at a number of horizontal planes. In some of our cases, confocal microscopy was needed to visualize and confirm the gaps in the lamina staining.
We observed abnormal nuclear morphology in fibroblasts containing a (likely) pathogenic variant in the LMNA linker or tail domain, but also for 5 out of 7 (likely) pathogenic variants located in the coil domain of LMNA. In contrast, Muchir et al. only observed a significantly increased percentage of abnormal nuclei in the eight cell-lines from EDMD/LGMD/FPLD patients containing a variant in the LMNA head or tail domain, but in none of the five LMNA coiled-coil domain variants [13]. Our results and those of Decaudain et al. [20] demonstrate that the presence of abnormal nuclei as a consequence of a LMNA pathogenic variant is not limited to certain domains of the lamin A/C protein, even if not all LMNA variants necessarily cause nuclear malformations, which may also be the case for the three false negative results obtained in our validation study. The LMNA c.514_1995del deletion was shown to cause haploinsufficiency [12], supporting a previous hypothesis that only formation of mutated lamin A/C proteins causes nuclear malformations, rather than a reduction in lamin A/C protein quantity, as was also observed in different tissues of mice with reduced lamin A/C expression [21]. Secondly, nuclear morphology analysis in HeLa cells transfected with p.(Arg190Trp) lamin A/C by Bhattacharjee et al. showed nuclei containing aggregates, but no other nuclear abnormalities were described and they concluded that there were no notable changes compared to wild-type [22], which is in line with our classification as normal of lamin A/C (p.(Arg190Trp)) fibroblasts. Of note, nuclear foci and/or aggregates were not taken into account in our study, but have been reported in fibroblasts [13]. Also, foci seem to arise due to overexpression, e.g., after transfection of wild-type lamin A and C [23], or much more prominently after transfection with mutant lamin constructs [8, 24]. For some LMNA variants it has been reported that mutated lamin can form aggregates without interacting with wild-type lamins and thus possibly causing no harm to the nuclear shape nor integrity [25]. In our study, aggregates were only detected in c.1634G>A (p.(Arg545His)) fibroblasts, which also presented with some honeycomb and donut-like structures and a classifier score of 0.51, which we considered as “unclassified”. Future inclusion of nuclear aggregates as parameter may increase sensitivity of the classification tool, but requires further research to establish the occurrence of foci in wild-type cells and a systemic assessment of their significance. Exploring implementation of automated quantification of nuclear morphology in 2D microscopy images, as performed by Core et al. to identify dysmorphic nuclei [26], may also aid our nuclear morphology analysis procedure and classification tool further, but requires optimization as honeycomb structures are difficult to assess using 2D images and likely require 3D analysis methods. Also, automated analysis may pose challenges for implementation of the nuclear morphology analysis in routine genetic laboratories, as the required equipment may not be available. Thirdly, some LMNA pathogenic variants may not cause nuclear abnormalities at all or only cause nuclear abnormalities following stress or present in certain cell-types other than fibroblasts. This may be the case for the likely pathogenic variant c.949G>A (p.(Glu317Lys)), which has been reported in patients with AV-block and DCM [27, 28]. It is unclear if this variant can cause nuclear abnormalities at all, since it has not been functionally analyzed by other groups. Further research is required to assess pathogenicity of this variant and its mode of action.
Taken together, normal nuclear morphology does not rule out pathogenicity of LMNA variants, but detection of excessive abnormal nuclei provides functional evidence of pathogenicity and may warrant reclassification of LMNA VUS as a pathogenic variant. Implementation of this tool in our laboratory enabled reclassification of 4 of the 12 variants from VUS to likely pathogenic variant (33%). Since ~50% of all identified rare missense and splice-site variants in LMNA are being classified as VUS, implementation of this nuclear morphology analysis tool, will have considerable impact on the counseling and follow-up for patients and family members.
Acknowledgements
We would like to thank all persons (A. Bardai, G. Bonne, B. van Engelen, D De Jongh, P. Helderman, R. Hennekam, D. Koolen, C. Marcelis, S. Sallevelt, K. van Kaam, E. Vanhoutte) that provided the fibroblasts included in this study.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Worman HJ. Nuclear lamins and laminopathies. J Pathol. 2012;226:316–25. doi: 10.1002/path.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gonzalo S, Kreienkamp R, Askjaer P. Hutchinson-Gilford Progeria syndrome: a premature aging disease caused by LMNA gene mutations. Ageing Res Rev. 2017;33:18–29. doi: 10.1016/j.arr.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Verstraeten VL, Broers JL, van Steensel MA, et al. Compound heterozygosity for mutations in LMNA causes a progeria syndrome without prelamin A accumulation. Hum Mol Genet. 2006;15:2509–22. doi: 10.1093/hmg/ddl172. [DOI] [PubMed] [Google Scholar]
- 4.Carboni N, Politano L, Floris M, et al. Overlapping syndromes in laminopathies: a meta-analysis of the reported literature. Acta Myol. 2013;32:7–17. [PMC free article] [PubMed] [Google Scholar]
- 5.Brodsky GL, Muntoni F, Miocic S, et al. Lamin A/C gene mutation associated with dilated cardiomyopathy with variable skeletal muscle involvement. Circulation. 2000;101:473–6. doi: 10.1161/01.CIR.101.5.473. [DOI] [PubMed] [Google Scholar]
- 6.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caux F, Dubosclard E, Lascols O, et al. A new clinical condition linked to a novel mutation in lamins A and C with generalized lipoatrophy, insulin-resistant diabetes, disseminated leukomelanodermic papules, liver steatosis, and cardiomyopathy. J Clin Endocrinol Metab. 2003;88:1006–13. doi: 10.1210/jc.2002-021506. [DOI] [PubMed] [Google Scholar]
- 8.Favreau C, Dubosclard E, Ostlund C, et al. Expression of lamin A mutated in the carboxyl-terminal tail generates an aberrant nuclear phenotype similar to that observed in cells from patients with Dunnigan-type partial lipodystrophy and Emery-Dreifuss muscular dystrophy. Exp Cell Res. 2003;282:14–23. doi: 10.1006/excr.2002.5669. [DOI] [PubMed] [Google Scholar]
- 9.Novelli G, Muchir A, Sangiuolo F, et al. Mandibuloacral dysplasia is caused by a mutation in LMNA-encoding lamin A/C. Am J Hum Genet. 2002;71:426–31. doi: 10.1086/341908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vigouroux C, Auclair M, Dubosclard E, et al. Nuclear envelope disorganization in fibroblasts from lipodystrophic patients with heterozygous R482Q/W mutations in the lamin A/C gene. J Cell Sci. 2001;114:4459–68. doi: 10.1242/jcs.114.24.4459. [DOI] [PubMed] [Google Scholar]
- 11.Fidzianska A, Bilinska ZT, Tesson F, et al. Obliteration of cardiomyocyte nuclear architecture in a patient with LMNA gene mutation. J Neurol Sci. 2008;271:91–6. doi: 10.1016/j.jns.2008.03.017. [DOI] [PubMed] [Google Scholar]
- 12.Gupta P, Bilinska ZT, Sylvius N, et al. Genetic and ultrastructural studies in dilated cardiomyopathy patients: a large deletion in the lamin A/C gene is associated with cardiomyocyte nuclear envelope disruption. Basic Res Cardiol. 2010;105:365–77. doi: 10.1007/s00395-010-0085-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muchir A, Medioni J, Laluc M, et al. Nuclear envelope alterations in fibroblasts from patients with muscular dystrophy, cardiomyopathy, and partial lipodystrophy carrying lamin A/C gene mutations. Muscle Nerve. 2004;30:444–50. doi: 10.1002/mus.20122. [DOI] [PubMed] [Google Scholar]
- 14.Houben F, Ramaekers FC, Snoeckx LH, et al. Role of nuclear lamina-cytoskeleton interactions in the maintenance of cellular strength. Biochim Biophys Acta. 2007;1773:675–86. doi: 10.1016/j.bbamcr.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 15.Claes GR, van Tienen FH, Lindsey P, et al. Hypertrophic remodelling in cardiac regulatory myosin light chain (MYL2) founder mutation carriers. Eur Heart J. 2016;37:1815–22. doi: 10.1093/eurheartj/ehv522. [DOI] [PubMed] [Google Scholar]
- 16.Dyer JA, Kill IR, Pugh G, et al. Cell cycle changes in A-type lamin associations detected in human dermal fibroblasts using monoclonal antibodies. Chromosome Res. 1997;5:383–94. doi: 10.1023/A:1018496309156. [DOI] [PubMed] [Google Scholar]
- 17.Akaike H. Information theory and an extension of the maximum likelihood principle. In: Petrov BN, Csàki F, editors. Second International Symposium on Inference Theory: Akadémiai Kiadó, Budapest; 1973; p 267–81.
- 18.Ihaka R, Gentleman R. R: a language for data analysis and graphics. J Comput Graph Stat. 1996;5:299–314. [Google Scholar]
- 19.Ben Yaou R, Toutain A, Arimura T, et al. Multitissular involvement in a family with LMNA and EMD mutations: Role of digenic mechanism? Neurology. 2007;68:1883–94. doi: 10.1212/01.wnl.0000263138.57257.6a. [DOI] [PubMed] [Google Scholar]
- 20.Decaudain A, Vantyghem MC, Guerci B, et al. New metabolic phenotypes in laminopathies: LMNA mutations in patients with severe metabolic syndrome. J Clin Endocrinol Metab. 2007;92:4835–44. doi: 10.1210/jc.2007-0654. [DOI] [PubMed] [Google Scholar]
- 21.Sullivan T, Escalante-Alcalde D, Bhatt H, et al. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J Cell Biol. 1999;147:913–20. doi: 10.1083/jcb.147.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhattacharjee P, Banerjee A, Banerjee A, et al. Structural alterations of Lamin A protein in dilated cardiomyopathy. Biochemistry. 2013;52:4229–41. doi: 10.1021/bi400337t. [DOI] [PubMed] [Google Scholar]
- 23.Broers JL, Machiels BM, van Eys GJ, et al. Dynamics of the nuclear lamina as monitored by GFP-tagged A-type lamins. J Cell Sci. 1999;112(Pt 20):3463–75. doi: 10.1242/jcs.112.20.3463. [DOI] [PubMed] [Google Scholar]
- 24.Broers JL, Kuijpers HJ, Ostlund C, et al. Both lamin A and lamin C mutations cause lamina instability as well as loss of internal nuclear lamin organization. Exp Cell Res. 2005;304:582–92. doi: 10.1016/j.yexcr.2004.11.020. [DOI] [PubMed] [Google Scholar]
- 25.Roblek M, Schuchner S, Huber V, et al. Monoclonal antibodies specific for disease-associated point-mutants: lamin A/C R453W and R482W. PLoS One. 2010;5:e10604. doi: 10.1371/journal.pone.0010604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Core JQ, Mehrabi M, Robinson ZR, et al. Age of heart disease presentation and dysmorphic nuclei in patients with LMNA mutations. PLoS One. 2017;12:e0188256. doi: 10.1371/journal.pone.0188256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arbustini E, Pilotto A, Repetto A, et al. Autosomal dominant dilated cardiomyopathy with atrioventricular block: a lamin A/C defect-related disease. J Am Coll Cardiol. 2002;39:981–90. doi: 10.1016/S0735-1097(02)01724-2. [DOI] [PubMed] [Google Scholar]
- 28.Villa F, Maciag A, Spinelli CC, et al. A G613A missense in the Hutchinson’s progeria lamin A/C gene causes a lone, autosomal dominant atrioventricular block. Immun Ageing. 2014;11:19. doi: 10.1186/s12979-014-0019-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vytopil M, Benedetti S, Ricci E, et al. Mutation analysis of the lamin A/C gene (LMNA) among patients with different cardiomuscular phenotypes. J Med Genet. 2003;40:e132. doi: 10.1136/jmg.40.12.e132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muchir A, van Engelen BG, Lammens M, et al. Nuclear envelope alterations in fibroblasts from LGMD1B patients carrying nonsense Y259X heterozygous or homozygous mutation in lamin A/C gene. Exp Cell Res. 2003;291:352–62. doi: 10.1016/j.yexcr.2003.07.002. [DOI] [PubMed] [Google Scholar]
- 31.De Vos WH, Houben F, Kamps M, et al. Repetitive disruptions of the nuclear envelope invoke temporary loss of cellular compartmentalization in laminopathies. Hum Mol Genet. 2011;20:4175–86. doi: 10.1093/hmg/ddr344. [DOI] [PubMed] [Google Scholar]
- 32.van Spaendonck-Zwarts KY, van Rijsingen IA, van den Berg MP, et al. Genetic analysis in 418 index patients with idiopathic dilated cardiomyopathy: overview of 10 years’ experience. Eur J Heart Fail. 2013;15:628–36. doi: 10.1093/eurjhf/hft013. [DOI] [PubMed] [Google Scholar]
- 33.Moller DV, Pham TT, Gustafsson F, et al. The role of Lamin A/C mutations in Danish patients with idiopathic dilated cardiomyopathy. Eur J Heart Fail. 2009;11:1031–5. doi: 10.1093/eurjhf/hfp134. [DOI] [PubMed] [Google Scholar]
- 34.Gangemi F, Degano M. Disease-associated mutations in the coil 2B domain of human lamin A/C affect structural properties that mediate dimerization and intermediate filament formation. J Struct Biol. 2013;181:17–28. doi: 10.1016/j.jsb.2012.10.016. [DOI] [PubMed] [Google Scholar]
- 35.Hoorntje ET, Bollen IA, Barge-Schaapveld DQ, et al. Lamin A/C-related cardiac disease: late onset with a variable and mild phenotype in a large cohort of patients with the lamin A/C p.(Arg331Gln) founder mutation. Circ Cardiovasc Genet. 2017;10:DOI: 10.1161/CIRCGENETICS.116.001631 1–10. [DOI] [PubMed]
- 36.Verstraeten VL, Caputo S, van Steensel MA, et al. The R439C mutation in LMNA causes lamin oligomerization and susceptibility to oxidative stress. J Cell Mol Med. 2009;13:959–71. doi: 10.1111/j.1582-4934.2009.00690.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shackleton S, Lloyd DJ, Jackson SN, et al. LMNA, encoding lamin A/C, is mutated in partial lipodystrophy. Nat Genet. 2000;24:153–6. doi: 10.1038/72807. [DOI] [PubMed] [Google Scholar]
- 38.Saj M, Dabrowski R, Labib S, et al. Variants of the lamin A/C (LMNA) gene in non-valvular atrial fibrillation patients: a possible pathogenic role of the Thr528Met mutation. Mol Diagn Ther. 2012;16:99–107. doi: 10.1007/BF03256434. [DOI] [PubMed] [Google Scholar]
- 39.Renou L, Stora S, Yaou RB, et al. Heart-hand syndrome of Slovenian type: a new kind of laminopathy. J Med Genet. 2008;45:666–71. doi: 10.1136/jmg.2008.060020. [DOI] [PubMed] [Google Scholar]
- 40.Houben F, De Vos WH, Krapels IP, et al. Cytoplasmic localization of PML particles in laminopathies. Histochem Cell Biol. 2013;139:119–34. doi: 10.1007/s00418-012-1005-5. [DOI] [PubMed] [Google Scholar]
- 41.De Sandre-Giovannoli A, Bernard R, Cau P, et al. Lamin a truncation in Hutchinson-Gilford progeria. Science. 2003;300:2055. doi: 10.1126/science.1084125. [DOI] [PubMed] [Google Scholar]
- 42.Bruston F, Delbarre E, Ostlund C, et al. Loss of a DNA binding site within the tail of prelamin A contributes to altered heterochromatin anchorage by progerin. FEBS Lett. 2010;584:2999–3004. doi: 10.1016/j.febslet.2010.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taylor MR, Fain PR, Sinagra G, et al. Natural history of dilated cardiomyopathy due to lamin A/C gene mutations. J Am Coll Cardiol. 2003;41:771–80. doi: 10.1016/S0735-1097(02)02954-6. [DOI] [PubMed] [Google Scholar]
- 44.Benedetti S, Menditto I, Degano M, et al. Phenotypic clustering of lamin A/C mutations in neuromuscular patients. Neurology. 2007;69:1285–92. doi: 10.1212/01.wnl.0000261254.87181.80. [DOI] [PubMed] [Google Scholar]
- 45.Kumar S, Baldinger SH, Gandjbakhch E, et al. Long-term arrhythmic and nonarrhythmic outcomes of lamin A/C mutation carriers. J Am Coll Cardiol. 2016;68:2299–307. doi: 10.1016/j.jacc.2016.08.058. [DOI] [PubMed] [Google Scholar]
- 46.Genschel J, Bochow B, Kuepferling S, et al. A R644C mutation within lamin A extends the mutations causing dilated cardiomyopathy. Hum Mutat. 2001;17:154. doi: 10.1002/1098-1004(200102)17:2<154::AID-HUMU11>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 47.Cann F, Corbett M, O’Sullivan D, et al. Phenotype-driven molecular autopsy for sudden cardiac death. Clin Genet. 2017;91:22–9. doi: 10.1111/cge.12778. [DOI] [PubMed] [Google Scholar]
- 48.Nouhravesh N, Ahlberg G, Ghouse J, et al. Analyses of more than 60,000 exomes questions the role of numerous genes previously associated with dilated cardiomyopathy. Mol Genet Genom Med. 2016;4:617–23. doi: 10.1002/mgg3.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walsh R, Thomson KL, Ware JS, et al. Reassessment of Mendelian gene pathogenicity using 7855 cardiomyopathy cases and 60,706 reference samples. Genet Med. 2017;19:192–203. doi: 10.1038/gim.2016.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Banerjee A, Rathee V, Krishnaswamy R, et al. Viscoelastic behavior of human lamin A proteins in the context of dilated cardiomyopathy. PLoS One. 2013;8:e83410. doi: 10.1371/journal.pone.0083410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Rijsingen IA, Nannenberg EA, Arbustini E, et al. Gender-specific differences in major cardiac events and mortality in lamin A/C mutation carriers. Eur J Heart Fail. 2013;15:376–84. doi: 10.1093/eurjhf/hfs191. [DOI] [PubMed] [Google Scholar]
- 52.Olfson E, Cottrell CE, Davidson NO, et al. Identification of medically actionable secondary findings in the 1000 genomes. PLoS One. 2015;10:e0135193. doi: 10.1371/journal.pone.0135193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chan D, McIntyre AD, Hegele RA, et al. Familial partial lipodystrophy presenting as metabolic syndrome. J Clin Lipidol. 2016;10:1488–91. doi: 10.1016/j.jacl.2016.08.012. [DOI] [PubMed] [Google Scholar]
- 54.Hanisch F, Neudecker S, Wehnert M, et al. [Hauptmann-Thannhauser muscular dystrophy and differential diagnosis of myopathies associated with contractures] Nervenarzt. 2002;73:1004–11. doi: 10.1007/s00115-002-1388-y. [DOI] [PubMed] [Google Scholar]
- 55.Yang L, Munck M, Swaminathan K, et al. Mutations in LMNA modulate the lamin A—Nesprin-2 interaction and cause LINC complex alterations. PLoS One. 2013;8:e71850. doi: 10.1371/journal.pone.0071850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scharner J, Brown CA, Bower M, et al. Novel LMNA mutations in patients with Emery-Dreifuss muscular dystrophy and functional characterization of four LMNA mutations. Hum Mutat. 2011;32:152–67. doi: 10.1002/humu.21361. [DOI] [PubMed] [Google Scholar]
- 57.Sebillon P, Bouchier C, Bidot LD, et al. Expanding the phenotype of LMNA mutations in dilated cardiomyopathy and functional consequences of these mutations. J Med Genet. 2003;40:560–7. doi: 10.1136/jmg.40.8.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stenson PD, Ball EV, Mort M, et al. The Human Gene Mutation Database (HGMD) and its exploitation in the fields of personalized genomics and molecular evolution. Curr Protoc Bioinform. 2012;Chapter 1: Unit113. [DOI] [PubMed]