Abstract
According to the current dogma, cholesteryl ester transfer protein (CETP) decreases high-density lipoprotein (HDL)-cholesterol (C) and increases low-density lipoprotein (LDL)-C. However, detailed insight into the effects of CETP on lipoprotein subclasses is lacking. Therefore, we used a Mendelian randomization approach based on a genetic score for serum CETP concentration (rs247616, rs12720922 and rs1968905) to estimate causal effects per unit (µg/mL) increase in CETP on 159 standardized metabolic biomarkers, primarily lipoprotein subclasses. Metabolic biomarkers were measured by nuclear magnetic resonance (NMR) in 5672 participants of the Netherlands Epidemiology of Obesity (NEO) study. Higher CETP concentrations were associated with less large HDL (largest effect XL-HDL-C, P = 6 × 10–22) and more small VLDL components (largest effect S-VLDL cholesteryl esters, P = 6 × 10–6). No causal effects were observed with LDL subclasses. All these effects were replicated in an independent cohort from European ancestry (MAGNETIC NMR GWAS; n ~20,000). Additionally, we assessed observational associations between ELISA-measured CETP concentration and metabolic measures. In contrast to results from Mendelian randomization, observationally, CETP concentration predominantly associated with more VLDL, IDL and LDL components. Our results show that CETP is an important causal determinant of HDL and VLDL concentration and composition, which may imply that the CETP inhibitor anacetrapib decreased cardiovascular disease risk through specific reduction of small VLDL rather than LDL. The contrast between genetic and observational associations might be explained by a high capacity of VLDL, IDL and LDL subclasses to carry CETP, thereby concealing causal effects on HDL.
Subject terms: Genetic variation, Disease genetics, Cardiovascular diseases
Introduction
Cholesteryl ester transfer protein (CETP) is able to transfer cholesteryl esters from high-density lipoproteins (HDL) to apolipoprotein B (ApoB) containing triglyceride-rich lipoproteins, mainly very-low-density lipoproteins (VLDL). In exchange, triglycerides are transferred from VLDL to triglyceride-poor particles, which are both HDL and low-density lipoprotein (LDL) [1]. As such, CETP facilitates an atherogenic lipoprotein profile, as has been extensively studied in both humans and in mice transgenic for human CETP [2, 3].
Despite the promising results of preclinical studies [4, 5], the clinical trials with the initial CETP inhibitors, torcetrapib, dalcetrapib and evacetrapib, were terminated: torcetrapib had off-target effects on blood pressure and caused an increase in cardiovascular events [6], and both dalcetrapib and evacetrapib lacked efficacy in reducing cardiovascular events on top of statin therapy [7, 8]. All these CETP inhibitors caused a large increase in HDL-cholesterol (C), accompanied by a modest or no decrease in non-HDL-C. Although a high HDL-C concentration was previously proposed to decrease the risk of cardiovascular disease (CVD) based on observational studies [9], Mendelian randomization showed that higher HDL-C concentrations do not lower the risk of myocardial infarction [10–13]. Although HDL functionality may still influence CVD risk, this implies that at least the association between HDL-C and CVD is not causal. This may provide one of the possible explanations for the lack of efficacy of the three initial CETP inhibitors [6–8].
Interestingly, the fourth clinical trial with the CETP inhibitor anacetrapib (REVEAL) did meet its primary endpoint by showing a 9% relative risk reduction in major coronary events [14]. The reduction in coronary events by anacetrapib, albeit limited, was attributed to a reduction in LDL-C and a corresponding reduction in ApoB-containing lipoprotein particles [14, 15]. However, the specific lipoprotein subclasses affected by CETP have not been determined yet. Specific lipoprotein fractions have previously been associated with increased CVD risk [16], and more in-depth insight in the causal effects of CETP on the circulating lipoprotein profile may therefore assist in understanding CETP inhibitor trial outcomes.
We recently performed a genome-wide association study (GWAS) on serum CETP concentration in the Netherlands Epidemiology of Obesity (NEO) study, and showed that CETP concentration has a strong genetic component [17]. Notably, three independent single-nucleotide polymorphisms (SNPs), all mapped to the CETP gene, together explained 16.4% of the total variation in serum CETP concentration, and were associated with a non-statistically significant odds ratio of 1.08 (0.94–1.23) for coronary artery disease risk [17]. With the use of these SNPs as genetic instruments in Mendelian randomization, the causal effects of circulating CETP on lipoprotein subclasses can be determined to provide more insight into CETP biology. Mendelian randomization assumes random assortment of alleles across a population, and is increasingly used to ascertain causality between genetic exposures and outcomes in the absence of confounding [18, 19]. Furthermore, potential reverse causation can be ruled out given that an outcome cannot influence genetic information.
In the present study, we thus aimed to assess the causal effects of CETP concentration on 159 circulating metabolic measures, primarily lipoprotein subclasses, using a Mendelian randomization approach in a cohort of the Dutch general population [20]. We also compared the causal effect estimates with observational associations between serum CETP concentration and these measures of lipid metabolism.
Materials and methods
Study design and populations
The present study is embedded in the NEO study, a population-based prospective cohort study of men and women aged 45 to 65 years. For the present study, we excluded participants with missing data on serum CETP concentration, metabolic profiling or genotype. Therefore, the present study population consists of 5672 individuals. The NEO study was approved by the medical ethics committee of the Leiden University Medical Center (LUMC), and all participants gave their written informed consent. The study conformed to the principles outlined in the Declaration of Helsinki. Detailed information about the study design and data collection has been described elsewhere [21].
Genotyping and imputation
DNA was isolated from venous blood samples. Genotyping was performed in participants from European ancestry, using the Illumina HumanCoreExome-24 BeadChip (Illumina Inc., San Diego, California, USA). Subsequently, genotypes were imputed to the 1000 Genome Project reference panel (v3 2011) [22] using IMPUTE (v2.2) software [23]. From the whole-genome data, we extracted the three independent genetic variants that have been previously identified in relationship to CETP concentration in the NEO study population, notably rs247616 (chr16:g.56989590C>T; directly genotyped; coding allele (C) frequency 0.67), rs12720922 (chr16:g.57000885G>A; imputation quality 0.98; coding allele frequency (A) 0.17) and rs1968905 (chr16:g.57010948T>G; imputation quality 0.85; coding allele frequency (G) 0.82) [17]. The CETP-increasing alleles are rs247616-C, rs12720922-A and rs1968905-G. Based on these three polymorphisms, we calculated a weighted genetic score per participant. The genetic score was constructed as the sum of the number of CETP-increasing alleles weighted by their effect size on CETP concentration, as previously described [17].
Serum CETP concentration and routine-lipid profile
After centrifugation, aliquots of serum were stored at –80 °C. From 11 April until 16 July 2014, CETP concentrations were measured with enzyme-linked immune sorbent assay (ELISA) kits according to the manufacturer’s instructions (DAIICHI CETP ELISA, Alpco, Salem, USA; coefficient of variation (CV) 11.7%) in serum that had undergone one previous freeze–thaw cycle. Fasting serum total cholesterol and triglycerides concentrations were measured with enzymatic colorimetric assays (Roche Modular P800 Analyzer, Roche Diagnostics, Mannheim, Germany) and fasting serum HDL-C concentrations with third-generation homogenous HDL-C methods (Roche Modular P800 Analyzer, Roche Diagnostics, Mannheim, Germany). Fasting LDL-C concentrations were calculated using the Friedewald equation [24].
NMR-based metabolic biomarker profiling
A high-throughput proton nuclear magnetic resonance (NMR) metabolomics platform [20] (Nightingale Health Ltd., Helsinki, Finland) was used to quantify 159 lipid and metabolite measures. The NMR spectroscopy was conducted at the Medical Research Council Integrative Epidemiology Unit (MRC IEU) at the University of Bristol, Bristol, UK, and processed by Nightingale’s biomarker quantification algorithms (version 2014). This method provides quantification of lipoprotein subclass profiling with lipid concentrations within 14 lipoprotein subclasses. The 14 subclass sizes were defined as follows: extremely large VLDL with particle diameters from 75 nm upwards and a possible contribution of chylomicrons, five VLDL subclasses (average particle diameters of 64.0 nm, 53.6 nm, 44.5 nm, 36.8 nm and 31.3 nm), IDL (28.6 nm), three LDL subclasses (25.5 nm, 23.0 nm and 18.7 nm) and four HDL subclasses (14.3 nm, 12.1 nm, 10.9 nm and 8.7 nm). Within the lipoprotein subclasses the following components were quantified: total cholesterol, total lipids, phospholipids, free cholesterol, cholesteryl esters and triglycerides. The mean size for VLDL, LDL and HDL particles was calculated by weighting the corresponding subclass diameters with their particle concentrations. Furthermore, 58 metabolic measures were determined that belong to classes of apolipoproteins, cholesterol, fatty acids, glycerides, phospholipids, amino acids, fluid balance, glycolysis-related metabolites, inflammation and ketone bodies. Details of the experimentation and applications of the NMR metabolomics platform have been described previously [20], as well as CVs for the metabolic biomarkers [25]. A full list of the measured biomarkers is included in Supplementary Table 1.
Statistical analyses
For all analyses, metabolic measures were natural-log transformed to obtain normal distributions. For comparison of the strengths of the associations between the different metabolic measures, standardized z-scores were composed. Consequently, the outcome variable of all analyses was the natural-log transformed SD difference in metabolic measure. When the concentration of a metabolic measure for an individual was below the detection limit, a value of half of the minimum concentration of that metabolic measure in the total population was imputed.
First, we performed a Mendelian randomization analysis to determine the causal associations between serum CETP concentration and the 159 metabolic measures. The genetic score was used as determinant in a linear regression analysis to assess the effect of a one unit (µg/mL) increase in serum CETP concentration on the metabolic measures. Second, the observational associations between serum CETP concentration and the 159 metabolic measures were determined with linear regression analyses. Participants with a serum CETP concentration beyond four SD from the mean were excluded (n = 1). All linear regression analyses were adjusted for age and sex. Since we previously observed that serum CETP concentration is not associated with possible confounding factors such as measures of body fat [26], markers of liver inflammation and function [27] and hepatic triglyceride content (unpublished data), we did not adjust for any additional factors apart from age and sex. Beta coefficients, SE and P values from linear regression analyses were reported. In addition, a linear regression analysis adjusted for age and sex was performed to determine the associations of serum CETP concentration with routinely measured HDL-C concentration and calculated Friedewald LDL-C concentration.
A power analysis was conducted to assess the minimally required effect size in the NEO study population. For this, we used the online calculator available on: http://cnsgenomics.com/shiny/mRnd/. [26] Considering the number of independent metabolites (alpha = 0.00134) and a power of 0.80, the power calculation showed that we were able to observe associations with an effect size of 0.132 SD in the NEO study population.
The metabolic biomarkers used for the present study are correlated with each other, and therefore conventional correction for multiple testing (e.g., Bonferroni) is too stringent. To obtain the number of independent metabolic biomarkers, we used the method as described by Li and Ji [27], which takes the correlation between the different metabolic biomarkers into account. Based on this method, we found 37 independent metabolic markers. For this reason, associations were considered to be statistically significant in case the P value was below 0.00134 (i.e., 0.05/37).
An important assumption of Mendelian randomization is that the genotype is only associated with the outcome via the exposure. This assumption can be violated when pleiotropy is present, which means that the genotype may be associated with the outcome via phenotypes other than the exposure. To gain insight into the possible pleiotropic effects of the individual variants, we performed an instrumental variable analysis for each of the three genetic variants separately. This analysis was performed by dividing the per-allele effect of an individual SNP for each metabolite by the per-allele effect of this SNP on CETP concentration. The per-allele effects of the genetic instruments on CETP concentration were 0.32 µg/mL for rs247616, 0.35 µg/mL for rs12720922 and 0.12 µg/mL for rs1968905, as we showed previously [17]. These ratios define the effects of the SNPs on the metabolic measure per one unit (1 µg/mL) increase in CETP concentration, thereby allowing for direct comparison of the effects of the three different SNPs on the lipoprotein profile. In case no pleiotropy is present, it is expected that ratios are comparable between the different SNPs, whereas observed differences in effects between the SNPs indicate possible pleiotropic effects. In addition, we checked whether the genetic variants were associated with potential confounders of the associations between CETP concentration and metabolite measures, i.e., alanine transaminase, aspartate transaminase, systolic blood pressure, diastolic blood pressure, body mass index (BMI), body fat percentage and C-reactive protein.
All results were based on analyses weighted towards the reference BMI distribution of the general Dutch population, and therefore apply to a population-based study without oversampling of individuals with overweight or obesity (see Supplementary Materials and Methods). Analyses were performed using STATA Statistical Software version 12.0 (Statacorp, College Station, Texas, USA) and R version 3.4.0 (The R Project, https://www.r-project.org/). Figures were designed with Python version 2.7.6 (Python Software Foundation, https://www.python.org/).
Replication
We aimed to replicate the findings from the Mendelian randomization analyses in the NEO study in an independent population. For that purpose, we used publically available summary statistics from the MAGNETIC NMR GWAS dataset [25], which comprises the additive (per-allele) beta coefficients with accompanying standard errors of the associations between genome-wide SNPs and 123 metabolic measures, of which 111 overlapped with the 159 metabolic measures that were quantified in the NEO study. This GWAS meta-analysis included data of ~20,000 individuals from 14 datasets that were derived from cohorts of European ancestry. The 123 metabolic measures were quantified by a prior version of the same high-throughput proton NMR metabolomics platform [20] (Nightingale Health Ltd., Helsinki, Finland) as used in the NEO study. In these metabolomics data, 22 principal components were identified [25], and therefore associations were considered to be statistically significant in case the P value was below 0.00227 (i.e., 0.05/22).
For the Mendelian randomization analyses, we summarized the genetically determined effect of 1 µg/mL CETP using the same genetic instruments (i.e., rs247616, rs12720922 and rs1968905) as in the analysis of the NEO study population to estimate the causal effect of CETP concentration on the metabolic measures. We weighted this combined effect estimate of the CETP SNPs on the metabolic measures by the inverse of the variance for each individual additive (per-allele) effect on the metabolic measures, and incorporated the individuals additive effects of the genetic instruments on CETP concentration. Additionally, we determined the correlation (R2) between the effect estimates from the NEO study and the MAGNETIC NMR GWAS dataset. A strong correlation indicates high consistency in the overall association profile of higher CETP concentrations with the lipid and metabolite measures.
Results
Population characteristics
Characteristics of the total study population are summarized in Table 1. The mean (SD) age was 56 (6) years. Mean (SD) concentration of CETP was 2.47 (0.65) µg/mL, of LDL-C 3.56 (0.96) mmol/L and of and HDL-C 1.57 (0.46) mmol/L.
Table 1.
Characteristics of the total study population from the Netherlands Epidemiology of Obesity (NEO) study
| Characteristics | |
|---|---|
| Age (year) | 56 (6) |
| Women (%) | 56 |
| Educational level (% higha) | 47 |
| Body mass index (kg/m2) | 26.3 (4.4) |
| Tobacco smoking (% current and former smokers) | 62 |
| Cardiovascular disease (%) | 5 |
| Lipid-lowering drug users (%) | 11 |
| Fasting serum concentrations | |
| CETP (µg/mL) | 2.47 (0.65) |
| Total cholesterol (mmol/L) | 5.70 (1.05) |
| LDL-cholesterol (mmol/L) | 3.56 (0.96) |
| HDL-cholesterol (mmol/L) | 1.57 (0.46) |
| Triglycerides (mmol/L) | 1.23 (0.85) |
N = 5672 (missing data: n = 4 for ethnicity, n = 56 for educational level, n = 4 for smoking, n = 21 for cardiovascular disease, n = 8 for total cholesterol concentration, n = 10 for LDL-cholesterol concentration, n = 9 for HDL-cholesterol concentration and n = 9 for triglyceride concentration). Results are based on analyses weighted towards the reference BMI distribution of the general Dutch population, and presented as mean (SD) or percentage
CETP cholesteryl ester transfer protein, HDL high-density lipoprotein, LDL low-density lipoprotein
aHigh educational level: higher secondary education (according to Dutch educational system), higher vocational education, university, PhD
Causally, CETP is negatively associated with large HDL components and positively associated with small VLDL components
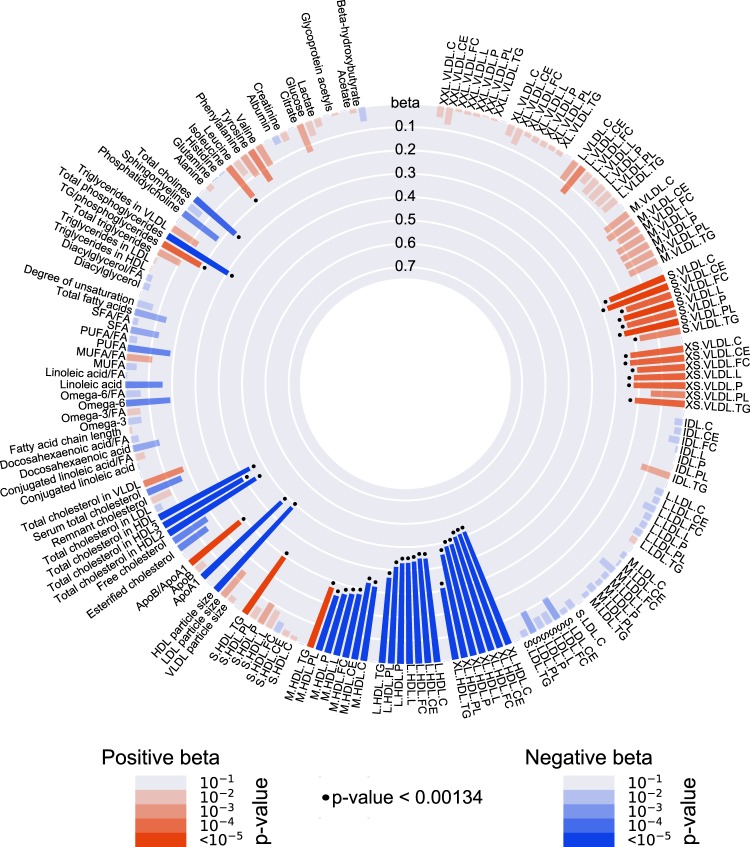
Figure 1 and Supplementary Table 2 show the results from the Mendelian randomization analyses of CETP and 159 circulating metabolic measures. The association with the CETP genetic score was statistically significant for 46 metabolic measures (P < 0.00134).
Fig. 1.
Causal associations between CETP concentration and 159 circulating metabolic measures, which were assessed with a Mendelian randomization approach based on a CETP genetic score, in the Netherlands Epidemiology of Obesity (NEO) study (n = 5672). The genetic score for Mendelian randomization is based on the CETP SNPs rs247616, rs12720922 and rs1968905, as previously determined with a genome-wide association study on serum CETP concentration [17]. Bar heights represent the magnitude of the beta-coefficient from linear regression, which is expressed as the SD difference in metabolic measure per 1 µg/mL increase in CETP concentration. Red bars indicate positive betas and blue bars indicate negative betas. The transparency of the bars indicates the level of statistical significance. A P value < 0.00134 is regarded statistically significant, as represented by the black dots. Full names and descriptions of metabolic measures are listed in Supplementary Table 1. Results are based on analyses weighted towards the reference BMI distribution of the general Dutch population
CETP concentration most strongly affected very large, large and medium HDL subclasses. With a 1 µg/mL increase in CETP, all components of these lipoprotein subclasses decreased. The only exception was the triglyceride content in medium HDL, for which the effect size was positive (effect (SE): 0.272 (0.055), P = 9 × 10–7). In line with this, triglycerides in small HDL also showed a positive beta (effect (SE): 0.293 (0.055), P = 1 × 10–9). In accordance with the decrease in larger HDL subclasses with higher CETP concentration, higher CETP was also associated with a smaller HDL diameter and less ApoA1. Overall, the largest effects were found for cholesterol in very large HDL (effect (SE): –0.517 (0.053), P = 6 × 10–22). When comparing the lipoprotein components within the very large, large and medium HDL subclasses, the cholesterol components, i.e., cholesteryl esters and free cholesterol, consistently showed the largest effect sizes.
Remarkably, CETP concentration did not associate with any of the LDL subclass components, while a higher CETP concentration associated with more small and very small VLDL. The largest increasing effect was found for cholesteryl esters in small VLDL (effect (SE): 0.276 (0.061), P = 6 × 10–6). There were no pronounced differences in the effect sizes between the various components within VLDL subclasses.
The results from the independent replication dataset can be found in Supplementary Figure 1 and Supplementary Table 2. The pattern of lipidomic associations was consistent in the replication dataset (Fig. 2), also when evaluated for the three SNPs of the genetic score separately (Supplementary Figures 2–4). To get insight into the possible pleiotropic effects, the results of the instrumental variable analyses are shown in Supplementary Figure 5. Notably, effects of the rs1968905 variant on HDL metabolites deviated from those of the two other SNPs, which may indicate pleiotropy. The genetic score was not associated with possible confounders of the association between CETP concentration and metabolic measures (Supplementary Table 3).
Fig. 2.
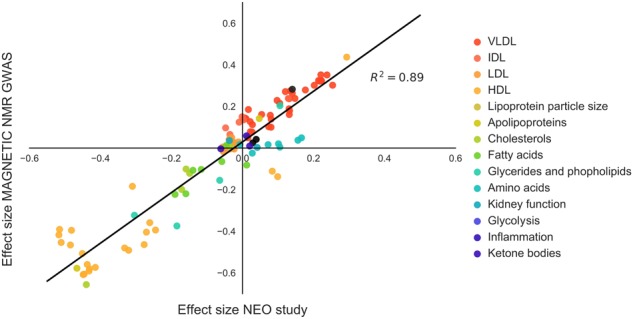
The effects sizes of the causal associations between CETP concentration and metabolic measures in the Netherlands Epidemiology of Obesity (NEO) study are strongly replicated in independent European populations (MAGNETIC NMR GWAS dataset), as shown by the high correlation between the beta-coefficients from both cohorts. Beta-coefficient from linear regression are expressed as the SD difference in metabolic measure per 1 µg/mL increase in CETP concentration. NEO study results are based on analyses weighted towards the reference BMI distribution of the general Dutch population
Observationally, CETP concentration is predominantly positively associated with very small VLDL, IDL and LDL components
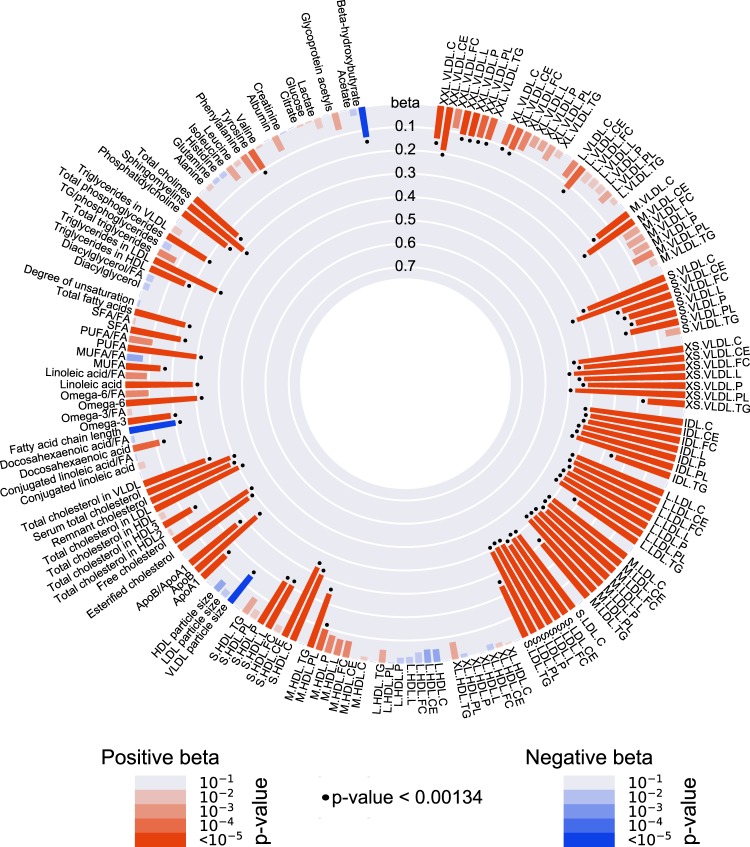
Figure 3 and Supplementary Table 4 show the observational associations between measured serum CETP concentration and the 159 circulating metabolic measures. A higher CETP concentration was positively associated with all VLDL, IDL and LDL subclasses, of which very small VLDL, IDL and all three LDL subclasses showed the strongest associations. This is in line with the associations between serum CETP concentration and the routine-lipid measurements (age- and sex-adjusted), since we observed a large effect on Friedewald LDL-C concentration per unit increase in CETP concentration (effect size 0.46 mmol/L; 95% confidence interval (CI) 0.40–0.52), compared to the relatively small effect on HDL-C concentration (effect size –0.05 mmol/L; 95% CI –0.07 to –0.02). Crude associations are shown in Supplementary Figure 6.
Fig. 3.
Observational associations between serum CETP concentration and 159 circulating metabolic measures, in the Netherlands Epidemiology of Obesity (NEO) study (n = 5672). Bar heights represent the magnitude of the beta-coefficient from linear regression, which is expressed as the SD difference in metabolic measure per 1 µg/mL increase in serum CETP. Red bars indicate positive betas and blue bars indicate negative betas. The transparency of the bars indicates the level of statistical significance. A P value < 0.00134 is regarded statistically significant, as represented by the black dots. Full names and descriptions of metabolic measures are listed in Supplementary Table 1. Results are based on analyses weighted towards the reference BMI distribution of the general Dutch population
The largest effect sizes for observational associations were found for free cholesterol in very small VLDL (effect (SE): 0.476 (0.030), P = 2 × 10–55) and phospholipids in very small VLDL (effect (SE): 0.472 (0.030), P = 2 × 10–56). When comparing the lipoprotein components (i.e., cholesteryl esters, free cholesterol, phospholipids and triglycerides) within the very small VLDL, IDL and LDL subclasses, the shell components, i.e., phospholipids and free cholesterol, quite consistently (apart from small LDL) showed the largest effect sizes, whereas the effect for the core component triglycerides was, consistently, the smallest. Concerning the HDL subclasses, higher CETP concentration was associated with a higher concentration of small HDL particles, whereas no associations were found with concentrations of very large, large and medium HDL particles.
Overall, observational and genetic associations were hardly consistent. Only for VLDL effect directions were similar (i.e., positive) between genetic and observational associations.
Discussion
In this study, we determined the causal effects of serum CETP on the circulating lipoprotein profile, and contrasted the results to observational associations. We used three recently identified CETP SNPs [17] (i.e., rs247616, rs12720922 and rs1968905) that together explain 16.4% of the total variation in serum CETP concentration as genetic instruments in Mendelian randomization to enable inferences of causality on 159 circulating metabolic measures. Higher circulating CETP concentrations were causally most strongly associated with lower concentrations of very large, large and medium-sized HDL components, a smaller overall HDL diameter, less ApoA1 and more small VLDL components, while there was no association with LDL components. In contrast, observationally, measured serum CETP concentration predominantly associated with more VLDL, IDL and LDL components.
Based on the principle of Mendelian randomization, the association between the CETP genetic score and lipoprotein subclasses can be interpreted as causal, thereby providing insight into human CETP biology [28, 29]. The results from our Mendelian randomization analyses are fully consistent with the mechanism by which CETP transfers cholesteryl esters from HDL towards ApoB-containing triglyceride-rich lipoproteins, mainly VLDL, and mediates the transfer of triglycerides from VLDL towards triglyceride-poor particles, such as HDL [1, 30, 31]. CETP accelerates the clearance of HDL from blood by enrichment of HDL with triglycerides, which makes this HDL a preferred substrate for hepatic triglyceride lipase [32]. Catabolism of triglyceride-rich HDL by hepatic triglyceride lipase leads to the formation of very small remnant HDL that is cleared by the liver and kidneys [32–37]. Indeed, in the present study, the CETP genetic score was associated with a smaller HDL diameter, and with a higher triglyceride content specifically in the medium and small HDL particles. Also, our results show that the CETP mainly affects the HDL pool, which is in accordance with the lipoprotein profile observed in CETP-deficient individuals, who have markedly increased amounts of large HDL particles [38–40], while effects on LDL and VLDL subclasses are less pronounced [40]. Our results are also in line with previously performed genetic association studies that identified CETP loci as strong determinants of HDL measures [41–43], without having effects on LDL-C [42].
Importantly, since effects of the rs1968905 variant on HDL metabolites deviated from those of the two other SNPs, it is plausible that pleiotropy plays in role in the effects of the rs1968905 variant on HDL metabolites, as we suggested before [17]. Thus, possibly the rs1968905 variant is associated with the HDL metabolites via phenotypes other than CETP concentration. Therefore, the HDL-related results for this variant specifically should be interpreted with caution, when making statements about direct effects of CETP.
Our results may also shed light on the mechanisms that underlie the effectiveness of CETP inhibition [28, 29]. Therefore, it is highly interesting to discuss the present study results in light of the results from the REVEAL trial with the CETP inhibitor anacetrapib, which showed a reduction in major coronary events [44]. Our findings implicate that CETP inhibition causes a relatively large increase in HDL components, which is predominantly caused by an increase in large and medium-sized HDL particles, in addition to a more modest reduction in VLDL components, which is mainly caused by a decrease in small and extra small VLDL particles. Of note, LDL concentration and composition were not affected by CETP, which was unexpected based on the current dogma that CETP increases LDL-C. However, it should be realized that LDL-C is generally not measured directly but calculated from the Friedewald formula [24], which may well misclassify cholesterol contained within small VLDL subclasses as LDL-C. It was previously reported that small and very small VLDL particle concentrations (reflecting triglyceride-rich lipoprotein remnants) and cholesterol in VLDL (reflecting remnant cholesterol) are among the lipoprotein components that most strongly associate with an increased CVD risk [16, 45, 46]. Thus, although we cannot rule out that CETP inhibition may beneficially affect HDL functionality and reduce CVD, the positive outcome of the latest CETP inhibition trial with anacetrapib [14] may well be explained by a reduction of small VLDL particles.
We compared the genetic associations with non-causal observational associations between serum CETP concentration measured with ELISA and all 159 metabolic measures. Observationally, associations were markedly different, as CETP concentration was predominantly positively associated with very small VLDL, IDL and LDL components, whereas no negative associations with HDL components were observed. This indicates that confounding factors conceal the true causal association. Although we showed previously that CETP concentration is strongly genetically determined [17], still the majority of the total variation in CETP concentration between individuals is caused by unknown factors. It should be realized that, just like apolipoproteins, CETP is an amphiphilic protein that binds to lipid surfaces of lipoproteins. It is thus conceivable that the capacity of lipoproteins to carry CETP is one of the factors that may explain the marked differences between the genetic and observational associations.
We propose that the contrast between genetic and observational associations may be explained by the higher capacity of VLDL, IDL and LDL subclasses to carry CETP, thereby concealing causal associations with HDL. The distribution of CETP over circulating lipoproteins has been extensively studied in the past, however, with diverging conclusions. The earliest studies proposed that CETP is predominantly bound to HDL particles [47–49]. However, thereafter, it was shown that CETP also avidly binds to LDL and VLDL, although the binding to these particles might be more labile [50]. It should be noted that the distribution of CETP over the various lipoprotein subclasses has commonly been determined after separating lipoproteins with ultracentrifugation, thereby presumably disrupting the binding within the least stable CETP-lipoprotein complexes. In addition, storage of plasma samples may also alter the distribution of CETP over lipoproteins. Indeed, marked effects of storage on the lipoprotein distribution have been observed for ApoC3 and ApoE following fast protein liquid chromatography [51]. Results from previous studies may therefore not accurately represent the distribution of CETP over lipoproteins within the circulation. Remarkably, in the present study, observational serum CETP concentration showed the strongest associations with the shell components (i.e., phospholipids and free cholesterol) of very small VLDL, IDL and LDL subclasses. This may indicate that CETP is mostly carried on the surface of these particles in the circulation, although future studies are needed to substantiate this hypothesis.
The main strength of the present study is the use of a strong genetic instrument in Mendelian randomization to draw conclusions on the causal effects of CETP concentration on the circulating lipoprotein profile. In addition, sufficient statistical power for the analyses was provided, as we had genetic data and NMR-metabolomic profiles available of 5672 individuals, which we were able to replicate in an independent dataset (n ~20,000). However, the study populations were from European ancestry and results may therefore not be generalizable to other populations. Interestingly, a recent study in a Chinese population, which used a different CETP genetic score (i.e., rs3764261, rs1800775, rs708272, rs9939224 and rs2303790), showed comparable results [52]. This indicates consistency of the results among different ancestries.
Based on Mendelian randomization, we conclude that CETP is an important determinant of HDL concentration and composition, without affecting LDL concentration and composition. Our finding challenges the current dogma that CETP increases LDL-C, mainly based on indirect LDL-C estimation using the Friedewald formula [24]. Instead, by directly assessing the lipoprotein profile by NMR, we now show that CETP specifically increases small VLDL fractions that likely represent VLDL remnants. Therefore, we speculate that the CETP inhibitor anacetrapib attenuated cardiovascular disease risk through specific reduction of remnant cholesterol rather than LDL-C.
Electronic supplementary material
Acknowledgements
We express our gratitude to all individuals who participate in the NEO study. We are grateful to all participating general practitioners for inviting eligible participants. We furthermore thank Pat van Beelen and all research nurses for collecting the data, Petra Noordijk and her team for laboratory management and DNA isolation, and Ingeborg de Jonge for all data management of the NEO study. We sincerely thank Chris van der Bent for performing the serum CETP concentration measurements. The genotyping in the NEO study was supported by the Centre National de Génotypage (Paris, France), headed by Jean-Francois Deleuze. This work was supported by the participating Departments, the Division and the Board of Directors of the Leiden University Medical Center, and by the Leiden University, Research Profile Area ‘Vascular and Regenerative Medicine; a grant from the Board of Directors of the Leiden University Medical Center to LLB; the Dutch Science Organization (ZonMW-VENI Grant 91617027 to YW); the European Commission funded project HUMAN (Health-2013-INNOVATION-1-602757 to DvH); the Dutch Science Organization (ZonMW-VENI Grant 91614023 to DOM-K); the Academy of Finland (312476 and 312477 to PW); the Novo Nordisk Foundation (15998 to PW); and the Netherlands CardioVascular Research Initiative: the Dutch Heart Foundation, Dutch Federation of University Medical Centers, the Netherlands Organisation for Health Research and Development and the Royal Netherlands Academy of Sciences for the GENIUS project ‘Generating the best evidence-based pharmaceutical targets for atherosclerosis’ (CVON2011-9).
Compliance with ethical standards
Conflict of interest
SS and PW are employees of Nightingale Health Ltd, a company offering NMR-based metabolite profiling. PW is a shareholder of Nightingale Health Ltd. All other authors declare that they have no conflict of interest.
Electronic supplementary material
The online version of this article (10.1038/s41431-018-0301-5) contains supplementary material, which is available to authorized users.
References
- 1.Tall AR. Plasma cholesteryl ester transfer protein. J Lipid Res. 1993;34:1255–74. [PubMed] [Google Scholar]
- 2.Chapman MJ, Le Goff W, Guerin M, Kontush A, et al. Cholesteryl ester transfer protein: at the heart of the action of lipid-modulating therapy with statins, fibrates, niacin, and cholesteryl ester transfer protein inhibitors. Eur Heart J. 2010;31:149–64. doi: 10.1093/eurheartj/ehp399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Westerterp M, van der Hoogt CC, de Haan W, Offerman EH, Dallinga-Thie GM, Jukema JW, et al. Cholesteryl ester transfer protein decreases high-density lipoprotein and severely aggravates atherosclerosis in APOE*3-Leiden mice. Arterioscler Thromb Vasc Biol. 2006;26:2552–9. doi: 10.1161/01.ATV.0000243925.65265.3c. [DOI] [PubMed] [Google Scholar]
- 4.Morehouse LA, Sugarman ED, Bourassa PA, Sand TM, Zimetti F, Geo F, et al. Inhibition of CETP activity by torcetrapib reduces susceptibility to diet-induced atherosclerosis in New Zealand White rabbits. J Lipid Res. 2007;48:1263–72. doi: 10.1194/jlr.M600332-JLR200. [DOI] [PubMed] [Google Scholar]
- 5.Okamoto H, Yonemori F, Wakitani K, Minowa T, Maeda K, Shinkai H, et al. A cholesteryl ester transfer protein inhibitor attenuates atherosclerosis in rabbits. Nature. 2000;406:203–7. doi: 10.1038/35018119. [DOI] [PubMed] [Google Scholar]
- 6.Barter PJ, Caulfield M, Eriksson M, Grundy SM, Kastelein JJ, Komajda M, et al. Effects of torcetrapib in patients at high risk for coronary events. N Engl J Med. 2007;357:2109–22. doi: 10.1056/NEJMoa0706628. [DOI] [PubMed] [Google Scholar]
- 7.Schwartz GG, Olsson AG, Abt M, Ballantyne CM, Barter PJ, Brumm J, et al. Effects of dalcetrapib in patients with a recent acute coronary syndrome. N Engl J Med. 2012;367:2089–99. doi: 10.1056/NEJMoa1206797. [DOI] [PubMed] [Google Scholar]
- 8.Lincoff AM, Nicholls SJ, Riesmeyer JS, Barter PJ, Brewer HB, Fox KAA, et al. Evacetrapib and cardiovascular outcomes in high-risk vascular disease. N Engl J Med. 2017;376:1933–42. doi: 10.1056/NEJMoa1609581. [DOI] [PubMed] [Google Scholar]
- 9.Gordon DJ, Probstfield JL, Garrison RJ, Neaton JD, Castelli WP, Knoke JD, et al. High-density lipoprotein cholesterol and cardiovascular disease. Four prospective American studies. Circulation. 1989;79:8–15. doi: 10.1161/01.CIR.79.1.8. [DOI] [PubMed] [Google Scholar]
- 10.Voight BF, Peloso GM, Orho-Melander M, Frikke-Schmidt R, Barbalic M, Jensen MK, et al. Plasma HDL cholesterol and risk of myocardial infarction: a mendelian randomisation study. Lancet. 2012;380:572–80. doi: 10.1016/S0140-6736(12)60312-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frikke-Schmidt R, Nordestgaard BG, Stene MC, Sethi AA, Remaley AT, Schnohr P, et al. Association of loss-of-function mutations in the ABCA1 gene with high-density lipoprotein cholesterol levels and risk of ischemic heart disease. JAMA. 2008;299:2524–32. doi: 10.1001/jama.299.21.2524. [DOI] [PubMed] [Google Scholar]
- 12.Johannsen TH, Kamstrup PR, Andersen RV, Jensen JB, Sillesen H, Tybjaerg-Hansen A, et al. Hepatic lipase, genetically elevated high-density lipoprotein, and risk of ischemic cardiovascular disease. J Clin Endocrinol Metab. 2009;94:1264–73. doi: 10.1210/jc.2008-1342. [DOI] [PubMed] [Google Scholar]
- 13.Haase CL, Tybjaerg-Hansen A, Grande P, Frikke-Schmidt R, et al. Genetically elevated apolipoprotein A-I, high-density lipoprotein cholesterol levels, and risk of ischemic heart disease. J Clin Endocrinol Metab. 2010;95:E500–510. doi: 10.1210/jc.2010-0450. [DOI] [PubMed] [Google Scholar]
- 14.Bowman L, Hopewell JC, Chen F, Wallendszus K, Stevens W, Collins R, et al. Effects of anacetrapib in patients with atherosclerotic vascular disease. N Engl J Med. 2017;377:1217–27. doi: 10.1056/NEJMoa1706444. [DOI] [PubMed] [Google Scholar]
- 15.Ference BA, Kastelein JJP, Ginsberg HN, Chapman MJ, Nicholls SJ, Ray KK, et al. Association of genetic variants related to CETP inhibitors and statins with lipoprotein levels and cardiovascular risk. JAMA. 2017;318:947–56. doi: 10.1001/jama.2017.11467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Würtz P, Havulinna AS, Soininen P, Tynkkynen T, Prieto-Merino D, Tillin T, et al. Metabolite profiling and cardiovascular event risk: a prospective study of 3 population-based cohorts. Circulation. 2015;131:774–85. doi: 10.1161/CIRCULATIONAHA.114.013116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blauw LL, Li-Gao R, Noordam R, de Mutsert R, Trompet S, Berbée JFP, et al. CETP (cholesteryl ester transfer protein) concentration: a genome-wide association study followed by Mendelian randomization on coronary artery disease. Circ Genom Precis Med. 2018;11:e002034. doi: 10.1161/CIRCGEN.117.002034. [DOI] [PubMed] [Google Scholar]
- 18.Davey Smith G, Ebrahim S. What can mendelian randomisation tell us about modifiable behavioural and environmental exposures. BMJ. 2005;330:1076–9. doi: 10.1136/bmj.330.7499.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith GD, Ebrahim S. ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. 2003;32:1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- 20.Soininen P, Kangas AJ, Würtz P, Suna T, Ala-Korpela M. Quantitative serum nuclear magnetic resonance metabolomics in cardiovascular epidemiology and genetics. Circ Cardiovasc Genet. 2015;8:192–206. doi: 10.1161/CIRCGENETICS.114.000216. [DOI] [PubMed] [Google Scholar]
- 21.de Mutsert R, den Heijer M, Rabelink TJ, Smit JW, Romijn JA, Jukema JW, et al. The Netherlands Epidemiology of Obesity (NEO) study: study design and data collection. Eur J Epidemiol. 2013;28:513–23. doi: 10.1007/s10654-013-9801-3. [DOI] [PubMed] [Google Scholar]
- 22.Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–13. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 24.Friedewald WT, Levy RI, Fredrickson DS. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin Chem. 1972;18:499–502. [PubMed] [Google Scholar]
- 25.Kettunen J, Demirkan A, Würtz P, Draisma HH, Haller T, Rawal R, et al. Genome-wide study for circulating metabolites identifies 62 loci and reveals novel systemic effects of LPA. Nat Commun. 2016;7:11122. doi: 10.1038/ncomms11122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brion MJ, Shakhbazov K, Visscher PM. Calculating statistical power in Mendelian randomization studies. Int J Epidemiol. 2013;42:1497–501. doi: 10.1093/ije/dyt179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J, Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity. 2005;95:221–7. doi: 10.1038/sj.hdy.6800717. [DOI] [PubMed] [Google Scholar]
- 28.Thanassoulis G, O’Donnell CJ. Mendelian randomization: nature’s randomized trial in the post-genome era. JAMA. 2009;301:2386–8. doi: 10.1001/jama.2009.812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ference BA: How to use Mendelian randomization to anticipate the results of randomized trials. Eur Heart J. 2018;39:360–2. [DOI] [PubMed]
- 30.Qiu X, Mistry A, Ammirati MJ, Chrunyk BA, Clark RW, Cong Y, et al. Crystal structure of cholesteryl ester transfer protein reveals a long tunnel and four bound lipid molecules. Nat Struct Mol Biol. 2007;14:106–13. doi: 10.1038/nsmb1197. [DOI] [PubMed] [Google Scholar]
- 31.Postmus I, Warren HR, Trompet S, Arsenault BJ, Avery CL, Bis JC, et al. Meta-analysis of genome-wide association studies of HDL cholesterol response to statins. J Med Genet. 2016;53:835–45. doi: 10.1136/jmedgenet-2016-103966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goldberg IJ. Lipoprotein lipase and lipolysis: central roles in lipoprotein metabolism and atherogenesis. J Lipid Res. 1996;37:693–707. [PubMed] [Google Scholar]
- 33.Xiao C, Watanabe T, Zhang Y, Trigatti B, Szeto L, Connelly PW, et al. Enhanced cellular uptake of remnant high-density lipoprotein particles: a mechanism for high-density lipoprotein lowering in insulin resistance and hypertriglyceridemia. Circ Res. 2008;103:159–66. doi: 10.1161/CIRCRESAHA.108.178756. [DOI] [PubMed] [Google Scholar]
- 34.Charles MA, Kane JP. New molecular insights into CETP structure and function: a review. J Lipid Res. 2012;53:1451–8. doi: 10.1194/jlr.R027011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guendouzi K, Collet X, Perret B, Chap H, Barbaras R. Remnant high density lipoprotein2 particles produced by hepatic lipase display high-affinity binding and increased endocytosis into a human hepatoma cell line (HEPG2) Biochemistry. 1998;37:14974–80. doi: 10.1021/bi9810508. [DOI] [PubMed] [Google Scholar]
- 36.Newnham HH, Barter PJ. Synergistic effects of lipid transfers and hepatic lipase in the formation of very small high-density lipoproteins during incubation of human plasma. Biochim Biophys Acta. 1990;1044:57–64. doi: 10.1016/0005-2760(90)90218-M. [DOI] [PubMed] [Google Scholar]
- 37.Rye KA, Clay MA, Barter PJ. Remodelling of high density lipoproteins by plasma factors. Atherosclerosis. 1999;145:227–38. doi: 10.1016/S0021-9150(99)00150-1. [DOI] [PubMed] [Google Scholar]
- 38.Brown ML, Inazu A, Hesler CB, Agellon LB, Mann C, Whitlock ME, et al. Molecular basis of lipid transfer protein deficiency in a family with increased high-density lipoproteins. Nature. 1989;342:448–51. doi: 10.1038/342448a0. [DOI] [PubMed] [Google Scholar]
- 39.Arai T, Tsukada T, Murase T, Matsumoto K. Particle size analysis of high density lipoproteins in patients with genetic cholesteryl ester transfer protein deficiency. Clin Chim Acta. 2000;301:103–17. doi: 10.1016/S0009-8981(00)00328-4. [DOI] [PubMed] [Google Scholar]
- 40.Koizumi J, Inazu A, Yagi K, Koizumi I, Uno Y, Kajinami K, et al. Serum lipoprotein lipid concentration and composition in homozygous and heterozygous patients with cholesteryl ester transfer protein deficiency. Atherosclerosis. 1991;90:189–96. doi: 10.1016/0021-9150(91)90114-I. [DOI] [PubMed] [Google Scholar]
- 41.Tukiainen T, Kettunen J, Kangas AJ, Lyytikäinen LP, Soininen P, Sarin AP, et al. Detailed metabolic and genetic characterization reveals new associations for 30 known lipid loci. Hum Mol Genet. 2012;21:1444–55. doi: 10.1093/hmg/ddr581. [DOI] [PubMed] [Google Scholar]
- 42.Shim H, Chasman DI, Smith JD, Mora S, Ridker PM, Nickerson DA, et al. A multivariate genome-wide association analysis of 10 LDL subfractions, and their response to statin treatment, in 1868 Caucasians. PLoS One. 2015;10:e0120758. doi: 10.1371/journal.pone.0120758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chasman DI, Pare G, Mora S, Hopewell JC, Peloso G, Clarke R, et al. Forty-three loci associated with plasma lipoprotein size, concentration, and cholesterol content in genome-wide analysis. PLoS Genet. 2009;5:e1000730. doi: 10.1371/journal.pgen.1000730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eyvazian VA, Frishman WH. Evacetrapib: another CETP inhibitor for dyslipidemia with no clinical benefit. Cardiol Rev. 2017;25:43–52. doi: 10.1097/CRD.0000000000000137. [DOI] [PubMed] [Google Scholar]
- 45.Varbo A, Benn M, Tybjærg-Hansen A, Jørgensen AB, Frikke-Schmidt R, Nordestgaard BG, et al. Remnant cholesterol as a causal risk factor for ischemic heart disease. J Am Coll Cardiol. 2013;61:427–36. doi: 10.1016/j.jacc.2012.08.1026. [DOI] [PubMed] [Google Scholar]
- 46.Varbo A, Benn M, Nordestgaard BG. Remnant cholesterol as a cause of ischemic heart disease: Evidence, definition, measurement, atherogenicity, high risk patients, and present and future treatment. Pharmacol Ther. 2014;141:358–67. doi: 10.1016/j.pharmthera.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 47.Pattnaik NM, Zilversmit DB. Interaction of cholesteryl ester exchange protein with human plasma lipoproteins and phospholipid vesicles. J Biol Chem. 1979;254:2782–6. [PubMed] [Google Scholar]
- 48.Groener JE, Van Rozen AJ, Erkelens DW. Cholesteryl ester transfer activity. Localization and role in distribution of cholesteryl ester among lipoproteins in man. Atherosclerosis. 1984;50:261–71. doi: 10.1016/0021-9150(84)90074-1. [DOI] [PubMed] [Google Scholar]
- 49.Barter PJ, Hopkins GJ, Gorjatschko L, Jones ME. A unified model of esterified cholesterol exchanges between human plasma lipoproteins. Atherosclerosis. 1982;44:27–40. doi: 10.1016/0021-9150(82)90050-8. [DOI] [PubMed] [Google Scholar]
- 50.Morton RE. Binding of plasma-derived lipid transfer protein to lipoprotein substrates. The role of binding in the lipid transfer process. J Biol Chem. 1985;260:12593–9. [PubMed] [Google Scholar]
- 51.Cohn JS, Rodriguez C, Jacques H, Tremblay M, Davignon J, et al. Storage of human plasma samples leads to alterations in the lipoprotein distribution of apoC-III and apoE. J Lipid Res. 2004;45:1572–9. doi: 10.1194/jlr.D300041-JLR200. [DOI] [PubMed] [Google Scholar]
- 52.Millwood IY, Bennett DA, Holmes MV, Boxall R, Guo Y, Bian Z, et al. Association of CETP gene variants with risk for vascular and nonvascular diseases among Chinese adults. JAMA Cardiol. 2018;3:34–43. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.