Abstract
Aberrant left-right patterning in the developing human embryo can lead to a broad spectrum of congenital malformations. The causes of most laterality defects are not known, with variants in established genes accounting for <20% of cases. We sought to characterize the genetic spectrum of these conditions by performing whole-exome sequencing of 323 unrelated laterality cases. We investigated the role of rare, predicted-damaging variation in 1726 putative laterality candidate genes derived from model organisms, pathway analyses, and human phenotypes. We also evaluated the contribution of homo/hemizygous exon deletions and gene-based burden of rare variation. A total of 28 candidate variants (26 rare predicted-damaging variants and 2 hemizygous deletions) were identified, including variants in genes known to cause heterotaxy and primary ciliary dyskinesia (ACVR2B, NODAL, ZIC3, DNAI1, DNAH5, HYDIN, MMP21), and genes without a human phenotype association, but with prior evidence for a role in embryonic laterality or cardiac development. Sanger validation of the latter variants in probands and their parents revealed no de novo variants, but apparent transmitted heterozygous (ROCK2, ISL1, SMAD2), and hemizygous (RAI2, RIPPLY1) variant patterns. Collectively, these variants account for 7.1% of our study subjects. We also observe evidence for an excess burden of rare, predicted loss-of-function variation in PXDNL and BMS1- two genes relevant to the broader laterality phenotype. These findings highlight potential new genes in the development of laterality defects, and suggest extensive locus heterogeneity and complex genetic models in this class of birth defects.
Subject terms: Congenital heart defects, Medical genomics
Introduction
In humans, the normal patterning of internal organs on the left or right side of the body is a complex, yet highly orchestrated, developmental process. Consequently, disordered left-right (LR) patterning can lead to a broad spectrum of laterality defects, including situs inversus totalis (SIT), in which there is complete transposition of visceral organs with maintenance of organ concordance, and heterotaxy (HTX) in which at least one organ is discordant along the left-right axis [1]. SIT is rarely associated with congenital organ malformations, whereas heterotaxy is highly associated with a host of congenital malformations that include isomerisms (in which normally asymmetric structures appear duplicated on both left and right e.g., bilateral bilobed lung), segmental reversals of sidedness (e.g., dextrocardia), and failures of embryonic structures to regress (e.g., persistent left superior vena cava) or complete loss of structures (e.g., asplenia). The conventional anatomic classification of HTX has been based on splenic and atrial/bronchial situs—namely asplenia-type/right atrial isomerism and polysplenia-type/left atrial isomerism. Although there are characteristic lesions within each of these classifications, their considerable anatomic and mechanistic overlap justifies their combination in epidemiological and genetic studies [2].
Heart formation in the early embryo requires proper LR axis signaling [3], and therefore, it is not surprising that cardiovascular malformations occur in approximately 80% of individuals with heterotaxy. The resulting spectrum of lesions is extensive and includes anomalies of pulmonary and systemic venous return, atrioventricular septal defects, ventricular inversion, and hypoplasia, transposition of the great arteries (TGA), double outlet right ventricle (DORV), and heart rhythm disorders [4]. In fact, abnormal LR patterning may contribute to as much as 3–7% of all congenital heart defects, and, with the associated multi-organ involvement, the medical and surgical management of such cases is particularly challenging [5]. The mortality for HTX thus remains appreciably high [6].
The etiologies of congenital laterality defects are complex and thought to include both environmental and genetic factors. Contributing environmental factors are poorly understood, but have been consistently observed [7]. From a genetic standpoint, laterality defects are occasionally observed in patients with chromosome abnormalities and genomic disorders [6, 8], and rare families have been reported with apparent segregation of HTX or related heart defects as autosomal dominant, autosomal recessive or X-linked traits [9–12]. There is thus some support for highly-penetrant rare variants, i.e., monogenic or Mendelian inheritance, causing laterality defects. At the same time, it has been noted that the same gene can cause the divergent phenotypes of SIT and HTX, and that polysplenia- or asplenia-types of HTX can be observed in families segregating the same pathogenic allele in the same disease gene, as well as in isolated cases with distinct alleles in the same gene [9, 10, 13]. In this context, the complexity of the heart defects and associated birth defects observed among patients with presumed protein-damaging variants in ZIC3 and CFC1 is most illustrative; there is little genotype-phenotype correlation among patients with ZIC3 variants, and the spectrum of heart defects is extensive, including defects not traditionally associated with laterality phenotypes and complex extra-cardiac malformations [10, 13]. Similarly, CFC1 variants have been associated with a variety of isolated congenital heart defects in addition to laterality abnormalities [9]. These observations demonstrate that there is substantial overlap in the genetic etiology of congenital heart defects, regardless of their phenotypic classification.
The known genes contributing to laterality defects account for only 15–20% of cases, and are largely associated with NODAL/TGFβ signaling (NODAL, CFC1, ACVR2B, LEFTYB, GDF1, TGFBR2, FOXH1), SHH signaling (ZIC3, LZTFL1) and monocilia functions (NPHP2, NPHP3, NPHP4, PKD2, TTC8) (see Supplementary Table S1). This connection to ciliary function is further reinforced by the observation that ~50% of patients with primary ciliary dyskinesia (PCD) defects have SIT and ~10% present with heterotaxy [8, 14]. Other gene variants with well-studied connections to early cardiac development (NKX2-5, CRELD1, MMP21, PKD1L1) and genes with relatively limited functional annotation (BCL9L, SHROOM3, MEGF8) have also been implicated in laterality defects. A critical review of these reports demonstrates that, although significant recurrence in first-degree relatives [15] and increased frequency in consanguineous populations [16] support a single gene view of laterality defects, putative pathogenic variants are often transmitted from unaffected parents, suggesting additional factors affect disease penetrance. Thus, these primary epidemiological observations are compatible with both Mendelian and oligogenic or complex inheritance mechanisms.
To explore the role of genetic variation in laterality defects, we performed singleton whole-exome sequencing (WES) of 323 unrelated cases, and investigated rare predicted-damaging variation, homo/hemizygous exon deletions, and gene-based burden of rare variation in a set of biologically-implicated cardiovascular genes defined a priori.
Results
High-quality candidate variants identified in the laterality cohort (Methods) were prioritized using three criteria: (1) extremely low allele frequency compared to population-based databases (minor allele <0.05% in ARIC, 1000 Genomes Project, ESP, ExAC, gnomAD—see Methods), (2) prediction of a deleterious functional effect including loss-of-function (LOF – e.g., frameshift-, stopgain-, and splice site- variants within 2 base pairs of intron-exon boundaries) or damaging nonsynonymous variation (Methods), and (3) a priori evidence for the gene playing a role in laterality or cardiac development (Methods; Supplementary Table S3). A total of 24 single nucleotide variants (SNVs) or small insertions/deletions met these criteria, representing 15 distinct genes (Table 1). The cases carrying these candidate SNVs represented ~7% of our total laterality cohort. Of these genes, nine have been previously implicated in human laterality disorders, and six represent novel candidate genes. We further investigated the clinical presentation of variant-carrying patients as well as their associated inheritance patterns.
Table 1.
Candidate variants in laterality defect cases identified by whole exome sequencing
| Inheritance | Model | Gene | Chr | hg19pos | Ref | Alt | Fnc. | Pred. change | AF | ID | Supporting evidence | Diagnosis | Variant class. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Known HTX & PCD genes | |||||||||||||
| Inherited | Dom | ACVR2B | 3 | 38523761 | C | T | NS | p.(R383C) | - | LAT1543 | MIM613751 | Venous anomaly | VUS |
| Dom | ACVR2B | 3 | 38519870 | C | G | NS | p.(P176R) | 8.67E−05 | LAT1333 | MIM613751 | Conotruncal | VUS | |
| Dom | NODAL | 10 | 72195225 | CGGTGCCT | - | FS | p.(R236fs)^ | - | LAT1763 | MIM270100 | Conotruncal | LP | |
| Dom | NODAL | 10 | 72195740 | G | T | SA | c.[892–1G >T] | - | LAT0022 | MIM270100 | Complex | LP | |
| Unknown | Dom | NODAL | 10 | 72195536 | C | T | NS | p.(E133K) | 4.06E−06 | LAT1696 | MIM270100 | Complex | VUS |
| Dom | NODAL | 10 | 72195225 | CGGTGCCT | - | FS | p.(R236fs)^ | - | LAT0191 | MIM270100 | Complex | LP | |
| Homozygous | Rec | DNAI1 | 9 | 34497158 | C | T | NS | p.(R292W) | 1.63E−03 | LAT1012 | MIM24400 (PCD) | Situs anomaly | LP |
| Rec | DNAH5 | 5 | 13719144 | A | G | NS | p.(I4116V) | 7.02E−04 | LAT1007 | MIM603335 (PCD) | Situs anomaly | LP | |
| Rec | MMP21 | 10 | 127462454 | C | T | NS | p.(E215K) | 8.53E−05 | LAT1451 | MIM608416 | Situs anomaly | P | |
| Rec | MMP21 | 10 | 127462540 | C | A | NS | p.(S186I) | - | LAT1441 | MIM608416 | Situs anomaly | LP | |
| Rec | PKD1L1 | 7 | 47886558 | C | G | NS | p.(C1691S) | - | LAT1180 | MGI, LIT [22] | Situs anomaly | LP | |
| Compound Heterozygous | Rec | HYDIN | 16 | 70884416 | T | C | NS | p.(K4196E) | 2.89E−04 | LAT1042 | MIM610812 (PCD) | Conotruncal | VUS |
| Rec | HYDIN | 16 | 70954829 | G | A | NS | p.(P2484S) | 3.31E−04 | LAT1042 | VUS | |||
| Hemizygous | X-linked | ZIC3 | X | 136649710 | - | G | FS | p.(V287fs) | - | LAT0180 | MIM306955 | Situs anomaly | P |
| X-linked | ZIC3 | X | 136649605 | C | A | SG | p.(S252*) | - | LAT1177 | MIM306955 | Situs anomaly | P | |
| X-linked | ZIC3 | X | 136649607 | T | A | NS | p.C253S | - | LAT0107 | MIM306955 | Ware, et. al., 2004[10] | P | |
| X-linked | ZIC3 | X | 136649595 | C | T | SG | p.(Q249*) | - | LAT0129 | MIM306955 | Ware, et. al., 2004 [10] | P | |
| X-linked | ZIC3 | X | 136648978 | C | A | SG | p.(S43*) | - | LAT0138 | MIM306955 | Ware, et. al., 2004 [10] | P | |
| X-linked | ZIC3 | X | 136649441 | CCCATACCGCCCAGTGG | - | FS | p.(198_203del) | - | LAT0264 | MIM306955 | Situs anomaly | P | |
| Novel candidate genes | |||||||||||||
| Inherited | Dom | ROCK2 | 2 | 11332802 | G | A | SG | p.(Q1242*) | - | LAT1349 | CVMhu, MGI, ZFIN, TGFB, PITX2 | Situs anomaly | - |
| Dom | ISL1 | 5 | 50687107 | G | A | SA | - | - | LAT1079 | MGI, ZFIN, LIT [24] | Complex | - | |
| Dom | SMAD2 | 18 | 45422938 | A | - | FS | p.(S64fs) | - | LAT1612 | MGI, TGFB | Anomalous venous return | - | |
| Compound heterozygous | Rec | ZFYVE16 | 5 | 79734302 | G | A | NS | p.(D600N) | 8.16E−06 | LAT0264 | TGFB | Situs anomaly | - |
| Rec | ZFYVE16 | 5 | 79752344 | G | C | NS | p.(G1252A) | - | LAT0264 | TGFB | Situs anomaly | - | |
| Hemizygous | X-linked | RAI2 | X | 17819419 | C | T | NS | p.(V188I) | 1.27E−05 | LAT0308 | MGI, LIT [24] | Situs anomaly | - |
| Unknown | Dom | SUPT16H | 14 | 21834689 | C | T | SA | - | - | LAT1183 | HHE [24], PITX2 | Situs anomaly Conotruncal | - |
Supporting evidence for known and candidate genes are tabulated under ‘Supporting Evidence’ and correspond to Supplementary Table S3. Cases with one- or neither parent available for segregation analyses were classified under “unknown” inheritance
ASI abdominal situs inversus, ZFIN zebrafish network, HH hedgehog signaling (KEGG), NOTCH NOTCH related (KEGG), PITX2 downstream target of PITX2 transcription factor, MGI overlapping phenotype in mouse, TGFB KEGG definition of TGFB pathway, HHE high heart expression[24], CVMhu previously implicated in human CVM, LIT recent literature report of cardiovascular genomics, PCD primary ciliary dyskinesia, NS nonsynonymous, SG stopgained, FS frameshift, SA splice acceptor; SD splice donor, Variant Class variant classification for known disease gene variants using AMP/ACMG annotations, VUS variant of uncertain significance, LP likely pathogenic, P pathogenic, AF Allele Frequency in gnomAD Browser. Additional variant details are given in Supplementary Table S8.
^Complex variant that includes a deletion at position 72195215 (AGTGAT > A)
Known laterality genes
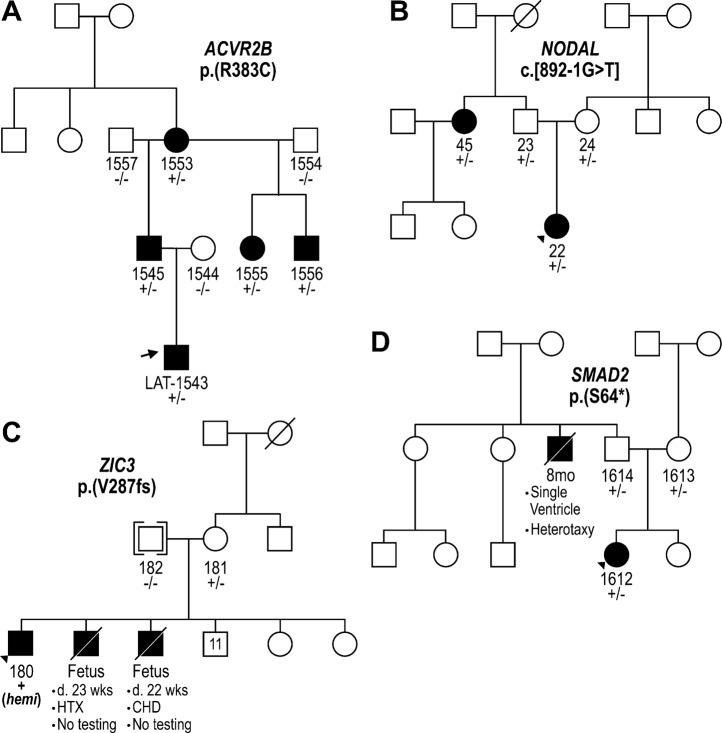
Six cases had rare LOF or nonsynonymous damaging SNVs in two known dominant laterality genes—NODAL and ACVR2B. The p.(R383C) missense variant observed in ACVR2B was inherited from an affected parent, and further analysis of this pedigree demonstrated clear segregation of the variant with five affected individuals from three generations (Fig. 1a). Conversely, all four NODAL variants were inherited from apparently unaffected parents; this is consistent with incomplete penetrance of these pathogenic variants, which is well-described for pathogenic variants in this gene [17]. For example, in the one available NODAL multiplex pedigree, a splice variant in patient LAT0022 was shown to be inherited from an apparently unaffected father and was shared with an affected paternal aunt (Fig. 1b). Somewhat surprisingly, three of the four predicted-damaging SNVs in NODAL were predicted loss-of-function variants—an uncommon observation in this gene (pLI = 0.95 [18]). We also observed compound heterozygous, rare, predicted-damaging SNVs in five genes previously implicated in either PCD—DNAI1, DNAH5, DNAH11, and HYDIN (1 case each), or HTX - MMP21 (2 cases), PKD1L1. PCD was not a part of the study entry criteria, and none of the cases harboring variants in PCD genes were known to have PCD. A genetic and phenotypic overlap between PCD and laterality has been previously postulated [19] and our data support this. Homozygous and compound heterozygous variants in MMP21 have been reported as the cause of HTX in a total of 14 cases (from 12 families) [20, 21]. Our recent report of homozygous recessive PKD1L1 variants as a cause of HTX included both the case reported here and another case with a homozygous splicing variant in PKD1L1 [22]. Amongst known laterality genes, the largest contribution of predicted-damaging SNVs was in ZIC3—we observed five LOF SNVs (5 cases) and one predicted-damaging missense variant (1 case), including pathogenic variants in three cases that we have previously reported [10]. Consistent with X-linked disease inheritance, ZIC3 candidate variants were all found in the hemizygous state in male offspring and were inherited from heterozygous mothers, with evidence of more extensive X-linked segregation of the phenotype in available pedigrees (Fig. 1c and ref. [10]).
Fig. 1.
Multiplex families with laterality defects and CHD. Families segregating variants in ACVR2B (a, dominant), NODAL (b, dominant), ZIC3 (c, X-linked), and SMAD2 (d, dominant) are shown. Shaded individuals are affected with a laterality defect. Presence (+) or absence (−) of variant is indicated for tested individuals. Accompanying numbers indicate LAT IDS (see Supplementary Table S2). Phenotypes are provided for affected individuals for whom a DNA sample was not available
Novel human laterality candidate genes
Finding likely pathogenic variants in known laterality genes served as a benchmark for our a priori filtering approach. Next, we focused on candidate laterality variation in genes not previously implicated in human disease. De novo variants have been shown to be of particular relevance in CHD [23–26]; however, we did not observe any of the candidate variants in our cohort to occur de novo. We observed rare heterozygous singleton LOF SNVs in six candidate genes with strong supporting evidence for a role in cardiac development and a low tolerance for loss-of function variation (pLI) in gnomAD—SMAD2, ROCK2, ISL1, and SUPT16H (Table 1). Where parents and family members were available to assess segregation, these variants, with the exception of SUPT16H but like those in NODAL, all showed evidence of dominant inheritance with incomplete penetrance—all were transmitted from parents without a reported history of significant cardiac disease. De novo damaging variants in SMAD2 were previously reported in two cases from a large genetic study of CHD [23, 24], which, on review, both had phenotypes consistent with laterality. In our cohort, the SMAD2 LOF variant was inherited from an unaffected father whose brother passed away with a diagnosis of HTX prior to genetic testing (Fig. 1d).
We did not systematically perform echocardiograms or additional molecular (e.g., mosaicism) investigations on carrier parents; therefore, reportedly ‘unaffected’ parents could still have less severe, undetected, laterality phenotypes. Given the known incomplete penetrance of dominant inherited variants in these disorders [25], the predicted molecular effect of the identified variants on the protein, and robust prior biological support, we chose to maintain these presumed partially penetrant dominant genes on our candidate list.
We observed biallelic, rare, predicted-damaging variants occurring in trans in one gene not previously associated with either PCD or laterality defects in humans- ZFYVE16 (Table 1). ZFYVE16 encodes a FYVE zinc finger domain protein; related proteins in this family have been implicated in TGF-β and BMP signaling through Smads [27], as well as in ventral folding of the developing mouse embryo, including heart development [28]. Notably, these ZFYVE16 variants were observed in the same male individual with a hemizygous frameshift variant in ZIC3 (LAT0264—see Table 1). In addition to Dextrocardia, this individual was also noted to have an absent stomach; an unusual feature suggestive of a more severe phenotype outside of the true laterality spectrum.
We also assessed evidence for X-linked inheritance models among our candidate genes. We noted a male proband with a hemizygous, maternally-inherited RAI2 variant and SIT in combination with asplenia as well as other cardiac and vascular abnormalities (Table 1; Supplementary Table S5). RAI2 (retinoic-acid induced 2) encodes a protein (RAI2) of unclear function that is thought to regulate transcription. RAI2 is expressed in fetal heart, brain, and kidney [29], and copy number loss of chromosome Xp22, encompassing RAI2, has been noted in two families with a syndromic diagnosis that included congenital heart lesions [30]. Another retinoic-acid induced gene—RAI1 —is the putative phenotypic driver in Smith–Magenis syndrome (MIM# 182290), a neurodevelopmental syndrome that includes cardiac defects and is caused by haploinsufficiency of 17p11.2 due to deletion CNV or LOF SNV in RAI1.
Homozygous and Hemizygous exon deletions in human laterality disorders
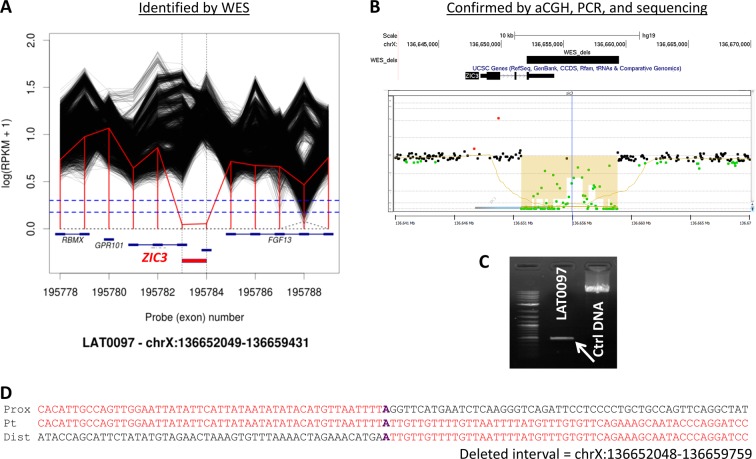
Next, we assessed structural genomic variation in our cohort; specifically, small and rare hemizygous and homozygous deletions detectable from comparison of whole exome sequence reads [31]. We observed a total of 14 high-confidence events (Z < −1.5 (autosomes), Z < −1.0 (X chromosome); maximum 3 events per person and 3 overlapping event calls) (Supplementary Table S6). One of these events overlapped our original a priori list - a hemizygous single exon deletion of ZIC3 observed in a patient with a common AV canal, asplenia, single ventricle, and atrioventricular valve regurgitation. Subsequent validation and parental testing showed this deletion to be maternally inherited (Fig. 2). We also validated an inherited, hemizygous single exon deletion of RIPPLY1 (Ripply Transcriptional Repressor 1) in a male patient with abdominal situs inversus, asplenia, interrupted inferior vena cava (IIVC), total anomalous pulmonary venous return (TAPVR) and complex CHD (D-TGA, common AV canal, ventricular septal defect, atrial septal defect, pulmonary atresia) (Supplementary Figure F1). Although not on our original a priori list, RIPPLY1 encodes a putative transcriptional repressor that acts in the NOTCH pathway and is paralogous to RIPPLY2, which we have recently described in a case of autosomal recessive Klippel-Feil syndrome with SIT [32]. In zebrafish embryos, Ripply1 interacts with Tbx6 and Mesp-b to regulate somite and vertebral development [33], and t-box genes are known to be important regulators of the establishment of asymmetry mediated by cilia in Kupffer’s vesicle [34], making it a strong candidate gene for laterality defects.
Fig. 2.
Hemizygous partial deletions of ZIC3. a WES read count data (RPKM) plotted for subject LAT0097 (red line) and all other BHCMG subjects (black lines) in the indicated regions of the X chromosome. Near-zero RPKM values in (a) suggest a hemizygous deletion of the final exon of ZIC3; b Confirmation and mapping of ZIC3 CNV by array comparative genomic hybridization using a custom 8 × 60k probe Agilent array (design ID = 064211). This array and all array procedures have been described previously [49]. c To more accurately map the CNV, long-range PCR with primers spanning the predicted breakpoints was employed, resulting in a smaller product compared to wild-type (ctrl DNA); sequencing of the resulting PCR product illustrated the deleted region (d)
Possible multigenic contribution to human laterality disorders
Recent studies have revealed associations between common variants and a variety of cardiovascular traits [35, 36], as well as highlighting the complex multigene/multi-variant interactions in hypoplastic left heart syndrome [37]. Therefore, we chose to expand our interrogation beyond single marker Mendelian analyses to perform gene-based aggregation analyses across the entire exome by comparing the burden of putative loss-of-function and predicted-damaging nonsynonymous variation between laterality cases and ARIC controls (see Methods). This analysis revealed compelling statistical evidence (surpassing our significance threshold of p < 7.06 × 10−6) for two genes—PXDNL and BMS1 (Supplementary Table S7), with little evidence of systematic stratification between the cohorts.
Although neither Peroxidasin-like (PXDNL; p = 1.61 × 10−6) or Ribosomal biogenesis factor (BMS1; p = 7.29 × 10−7) were included on our a priori list, both are good biological candidates in the context of the broader laterality phenotype. PXDNL is a homolog of human peroxidase that is exclusively expressed by cardiomyocytes in the human heart and thought to interact with peroxidase in remodeling of the extracellular matrix after stress exposure [38]. BMS1 is highly conserved across species. The zebrafish homolog, bms1l, is strongly expressed throughout the digestive tract and accessory organs including the liver, and mutant bmsl1 has been associated with hypoplasia of the liver and digestive tract [39]. Thus, BMS1 may be of particular relevance to the ~2/3rds of our cohort with extra-cardiac laterality defects, which included liver abnormalities and intestinal malrotation (Supplementary Tables S2 and S5), as was evident in two of the six LOF variant carriers.
Discussion
Our analysis of rare damaging variation and exonic deletions with suspected and established CHD genes detected 28 compelling monogenic candidate variants (26 SNV/indels, 2 deletion CNV) in 25 of 323 cases, or 7.1% of our starting cohort of unrelated laterality patients. This approach identified pathogenic and likely pathogenic variants in known and putative laterality genes, expanding the spectrum of disease-causing variation within these genes. Though we incorporated evidence from familial segregation patterns of these variants when possible, these variants were not primarily ascertained using family-based methods. This demonstrates that stringent variant filtering strategies can be used to identify candidate variants in rare disease case cohorts without the necessity of performing WES in all family members. Nevertheless, the overall “solve” rate would likely improve using a combination of these approaches.
We anticipate that further genetic studies of this phenotype will confirm many of our candidates and identify new ones, thereby improving the overall molecular diagnostic rate for laterality defects. For instance, our approach was able to confirm SMAD2 (dominant) as a laterality CHD gene alongside two reported de novo cases identified from large CHD cohorts [23, 24], and was instrumental in identifying and confirming PKD1L1 (recessive) [22]. We thus posit RIPPLY1 (X-linked hemizygous) and RAI2 (two X-linked hemizygous cases) as particularly compelling laterality candidates emerging from our analyses.
The majority of cases remained without an identifiable genetic etiology. In part, this reflects the limitations of our somewhat conservative analytical framework—we restricted our Mendelian SNV analyses to high-confidence biological candidates and imposed a high threshold for pathogenic variant classification among identified variants. This enabled us to bring our analytical framework in line with recent recommendations for inferring pathogenic and likely-pathogenic genomic variants [40]—the variants in newly described candidate genes presented here would be considered to have (at least) moderate evidence of pathogenicity. Nevertheless, our stringent criteria and cohort approach could miss potential pathology-contributing variants in individual patients and families. Future surveys that incorporate data from the entire exome/genome, utilize segregation in larger pedigrees, and prioritize functional validation of novel variation will be of high value in further expanding the genetic spectrum of these defects.
The variants and candidate genes we identified in individual subjects suggest that alleles contributing to laterality-related traits largely segregate with either recessive or X-linked inheritance, although we did observe suggestive examples of dominant and complex/polygenic modes of inheritance. Almost all of the dominant heterozygous LOF variants we observed—both in known and new candidate genes—were inherited from apparently unaffected parents. With new candidate genes, it is difficult to know a priori whether haploinsufficiency is sufficient to cause disease; however, this phenomenon is consistently observed in known laterality-causing genes such as NODAL, and was congruent with the disease segregation of SMAD2 LOF variants in extended families. One potential mechanism contributing to this observation is parental mosaicism. We did not perform WES on parents, and although validation traces were not highly suggestive of mosaicism, a role for low-level gonadal- or germline- mosaicism cannot be completely ruled out. Another consideration is that the phenotype of dominantly-inherited laterality genes may involve more complex mechanisms of non-penetrance [37] and variable expressivity [41], such that ‘unaffected’ carrier parents might have milder phenotypes that do not readily come to clinical attention. It may also be that multiple ‘hits’ are required to disturb the complex processes involved in early embryonic left-right patterning, such that the effect of a single heterozygous ‘hit’ is most relevant in the context of epistatic modifiers around the genome [42]. Similarly, multi-locus variation may provide an adverse variant burden in a pathway, system or interactome [43]. The latter two speculations are bolstered by the associations between the burden of rare variation in BCL1 and PDXNL and laterality defects. A similar argument can be made for rare compound heterozygous damaging variants in ZFYVE16 occurring in the same individual (LAT0264) carrying a ZIC3 frameshift variant (Table 1). Functional studies of our candidate variants in model organisms together with consideration of more complex multi-locus genetic models may help to confirm or refute these notions.
Expanded assessments of coding-sequence variation, however, may leave our understanding of the etiology of laterality defects incomplete. For instance, LOF variants accounted for all but one of the ZIC3 variants, and included distinct types of LOF variants (LAT0180—frameshift; LAT1177—stopgain; LAT0097—hemizygous deletion). The effects of these variants converge on absence of ZIC3 protein; thus, it may follow that other variants with a similar effect could also cause HTX and congenital heart disease (e.g., miRNA, gene silencer motifs). Screening for these and other categories of LOF-like variation in ZIC3 could be prioritized in future genomic analyses of HTX, in addition to the exome-centric analyses presented here.
This study provides a comprehensive coding-sequence survey for variants at known and putative candidate loci in laterality defects. The results implicate a total of six candidate genes for human laterality defects and reinforce that the genetic architecture of such defects is complex, spanning several single gene inheritance models, potential multi-locus contributions and other putative disease mechanisms. These data provide a basis for future investigation of additional monogenic causes of heterotaxy and related defects in left-right patterning as well as a starting point for discovery of complex genetic mechanisms underlying CHD and other human birth defects.
Methods
Case ascertainment
Cases were recruited through Texas Children’s Hospital (TCH) in Houston, TX. The Institutional Review Board of Baylor College of Medicine approved the study and all participating subjects gave informed consent. When available, parents and affected family members of the index cases were also invited to participate. The details of case ascertainment have been previously published [17]; briefly, patients were eligible if they presented with evidence of disturbed left-right patterning, which included situs abnormalities (SIT, HTX) or an isolated congenital malformation consistent with disturbed left-right patterning such as D-TGA or DORV (summarized in Supplementary Table S2). The clinical diagnoses were confirmed by a detailed review of medical records, including radiologic and cardiac-specific imaging. DNA samples were obtained from blood, saliva, and skin fibroblasts as previously described [17].
Cardiovascular gene set
Given the strong relationship between laterality and CHD, we compiled a list of 1702 human genes with a priori evidence for a role in laterality and/or cardiovascular malformation (CVM) from multiple public resources (Supplementary Table S3). This list was compiled from searches of human disorders including laterality defects and CVM (OMIM, NCBI, literature), relevant biological pathways and interactions (HH, NOTCH, TGFβ, PITX2) as determined by the Kyoto Encyclopedia of Genes and Genomes (KEGG) database, and model organism support including zebrafish heart expression (ZFIN) and abnormal cardiac morphology in mouse models (MGI, MP:0000266).
Population-based allele frequency comparison
In additional to population-based databases of variant minor allele frequencies—Exome Aggregation Consortium (ExAC, v 0.3.1), Genome Aggregation Database (gnomAD, v3.0.1), Exome Variant Server (EVS)—WES data from 5492 European American (EA) individuals from the population-based Atherosclerosis Risk in Communities (ARIC) study—sequenced and annotated using the same pipeline as the laterality cases [44]—were utilized as a variant frequency comparison group. ARIC samples with heart failure, major Q-wave, or LVH by the Cornell definition were excluded from the analyses.
Whole exome sequencing, annotation, and validation
WES was performed on 323 individual cases at the Human Genome Sequencing Center at Baylor College of Medicine through the Baylor-Hopkins Center for Mendelian Genomics initiative using the Illumina HiSeq platform and the Mercury pipeline [45]. ARIC samples were captured using VCRome 2.1 (42 Mb) and HTX cases were captured using HGSC-CORE (52 Mb), and all analyses were restricted to the intersection of the targeted regions of these reagents. Furthermore, we analyzed only high-quality genotypes including single base substitutions with a minimum coverage of 10x, and small insertions/deletions (indels) with >30x coverage. Read mapping to Genome Reference Consortium Human Build 37 (GRCh37) was performed with Burrows-Wheeler alignment [46], and allele calling was performed with the Atlas2 suite [47] (Atlas-SNP, Atlas-Indel) in order to identify high-quality variants annotated for their potential protein-damaging effect. All candidate SNVs from the laterality cohort were validated on an orthogonal platform (Dideoxy -Sanger- sequencing) in cases. In order to assess inheritance patterns, validated variants were then genotyped, using the same methodology (dideoxy -Sanger- sequencing), in parents and available family members.
Variant filtering
The Variant Call File (VCF) contained flagged low-quality variants including SNPs with posterior probability lower than 0.95, total depth of coverage less than 10x, fewer than 3 variant reads, an allelic fraction less than 10%, 99% reads in a single direction, and homozygous reference alleles with < 6x coverage. We increased stringency to remove low-quality indels with a total depth less than 30x and allelic fraction below 30%. Eight cases presented extremely high or low heterozygosity (more than 3 standard deviations from sample mean) and were excluded from burden analyses. Variants were annotated to Refseq gene definitions using ANNOVAR. Conservative loss-of-function (LOF) annotation was performed by selecting only included premature stopgains in the non-terminal exon, essential splice sites used by all gene isoforms, and frameshift indels similarly mapping to all isoforms. Damaging nonsynonymous (DNS) variation was defined as protein-altering substitutions predicted to be damaging by a consensus of at least 3 out of 6 prediction scores downloaded via dbNSFP (SIFT, Polyphen2 HDIV, LRT, Mutation Taster, Mutation Assessor, FATHMM). A PHRED-like scaled C-score (CADD) was also used to assess pathogenicity of variants (LOF and DNS), but was not used to exclude candidate sites. LOF constraint was quantified by first calculating the sum of all LOF alleles in ARIC (gene-wise observed LOF). Next, we simulated all potential nucleotide substitutions in exonic regions to determine the number of total potential LOF sites for each gene and calculated the ratio of observed to potential LOF alleles (OP ratio). OP ratios were used alongside pLI (probability of loss-of-function scores from ExAC/gnomAD), to filter for genes with a very low OP ratio (zero, or lowest 30th percentile) or high pLI score (>0.9) amongst laterality candidates. Variants identified in known laterality genes were also curated for pathogenicity using ACMG/AMP criteria [48], and then annotated as either a variant of uncertain significance (VUS), a likely pathogenic (LP), likely benign (LB), or pathogenic (P) variant (Table 1; Supplementary Table S8). Finally, for large genes in which we observed singleton biallelic variants in trans (e.g., DNAH5, DNAH11 and HYDIN), we also calculated the chance of observing two such variants by taking the maximum frequency of the two observed variants and catalogued all other variants (in gnomAD) in the gene at that frequency or lower. The resulting frequencies were added, assuming independent segregation, to give the proportion of people with rare variants. Assuming random mating, we then estimated the chance that any two variants would be observed in the same person. If this probability of biallelic variants occurring by chance was higher than observed in our cohort, we removed the gene in question (e.g., DNAH11), keeping those that were less than observed.
WES of laterality cases revealed 1,217,653 total variants (1,152,651 single nucleotide substitutions, and 65,002 small indels; Supplementary Table S4) across the entire allele frequency spectrum.
Validation of hemizygous/homozygous copy losses
CNVs were confirmed and mapped by array comparative genomic hybridization using a custom 8 × 60k probe Agilent array (design ID = 064211). This array and all array procedures have been described previously [49]. To more accurately map the CNV and provide substrate for breakpoint sequencing, long-range PCR with primers spanning the predicted breakpoints was employed. PCR reagents and concentrations have been described previously [50]. The thermal cycler was programmed as follows: 94 °C x 1 min; 30 cycles of 94 °C x 30 sec followed by 68 °C x 7 min; 72 °C x 10 min. PCR primers are listed in the Supplemental Methods. Breakpoint PCR products were treated with ExoSAP-IT (Affymetrix) according to the manufacturer’s instructions, then sequenced by Sanger di-deoxynucleotide sequencing (Baylor College of Medicine Sequencing Core, Houston, TX, USA).
Gene burden testing
Firth logistic regression was performed on case-control status using the total number of heterozygous sites per individual as a covariate in order to address potential platform differences between sequencing batches. These analyses were restricted to samples of reported European ancestry (111 HTX cases; 5,752 ARIC participants) and excluded genes in the MHC region of chromosome 6 and multiallelic variants. Only rare DNS/LOF variation that were covered at 10x or better and meeting our variant-filtering criteria in both cohorts were included in our analyses; a p-value of 7 × 10−6—reflecting a Bonferroni correction for 7086 genes harboring DNS in the case cohort – was deemed statistically significant.
Web resources
1000 Genomes, http://browser.1000genomes.org
ExAC Browser, http://exac.broadinstitute.org/
gnomAD Browser, http://gnomad.broadinstitute.org/
dbNSFP, http://varianttools.sourceforge.net/Annotation/DbNSFP
OMIM, http://www.omim.org/
UCSC Genome Browser, http://genome.ucsc.edu
HMZDelfinder, https://github.com/BCM-Lupskilab/HMZDelFinder
Supplementary information
Acknowledgements
This work was supported in part by the US National Human Genome Research Institute/National Heart Blood Lung Institute jointly funded Baylor Hopkins Center for Mendelian Genomics (UM1HG006542) and by National Institutes of Health (NIH) grants to JWB (1U54 HD083092, 5RO1 HD039056, 5RO1 HL090506, 5RO1 HL091771). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. NH is funded by a Clinical Scientist Development Award from the Doris Duke Charitable Foundation (Grant #:2013096).
Compliance with ethical standards
Conflict of interest
JRL holds stock ownership in 23andMe, Inc. and Lasergen, Inc., is a paid consultant for Regeneron Pharmaceuticals and is a co-inventor on multiple United States and European patents related to molecular diagnostics. The Department of Molecular and Human Genetics at Baylor College of Medicine derives revenue from molecular genetic testing offered in the Baylor Genetics Laboratories. JRL is on the Scientific Advisory Board of Baylor Genetics. JWB is a fulltime employee of Illumina Inc, but all work was performed under the listed affiliation. AHL is a fulltime employee of Regeneron Pharmaceuticals, but all analyses were performed under the listed affiliation.
Supplementary information
The online version of this article (10.1038/s41431-018-0307-z) contains supplementary material, which is available to authorized users.
References
- 1.Shiraishi I, Ichikawa H. Human heterotaxy syndrome – from molecular genetics to clinical features, management, and prognosis –. Circ J. 2012;76:2066–75. doi: 10.1253/circj.CJ-12-0957. [DOI] [PubMed] [Google Scholar]
- 2.Lin AE, Krikov S, Riehle-Colarusso T, Frías JL, Belmont J, Anderka M, et al. Laterality defects in the national birth defects prevention study (1998-2007): Birth prevalence and descriptive epidemiology. Am J Med Genet Part A. 2014;164:2581–91. doi: 10.1002/ajmg.a.36695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raya A, Izpisúa Belmonte JC. Left-right asymmetry in the vertebrate embryo: from early information to higher-level integration. Nat Rev Genet. 2006;7:283–93. doi: 10.1038/nrg1830. [DOI] [PubMed] [Google Scholar]
- 4.Jacobs JP, Anderson RH, Weinberg PM, Walters HL, Tchervenkov CI, Del Duca D, et al. The nomenclature, definition and classification of cardiac structures in the setting of heterotaxy. Cardiol Young. 2007;17:1–28. doi: 10.1017/S1047951107001138. [DOI] [PubMed] [Google Scholar]
- 5.Escobar-Diaz MC, Friedman K, Salem Y, Marx GR, Kalish BT, Lafranchi T, et al. Perinatal and infant outcomes of prenatal diagnosis of heterotaxy syndrome (asplenia and polysplenia) Am J Cardiol. 2014;114:612–7. doi: 10.1016/j.amjcard.2014.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anagnostopoulos PV, Pearl JM, Octave C, Cohen M, Gruessner A, Wintering E, et al. Improved current era outcomes in patients with heterotaxy syndromes. Eur J Cardio-thoracic Surg. 2009;35:871–8. doi: 10.1016/j.ejcts.2008.12.018. [DOI] [PubMed] [Google Scholar]
- 7.Kuehl KS, Loffredo C. Risk factors for heart disease associated with abnormal sidedness. Teratology. 2002;66:242–8. doi: 10.1002/tera.10099. [DOI] [PubMed] [Google Scholar]
- 8.M. Ware S, Lynn Jefferies J. New Genetic Insights into Congenital Heart Disease. J Clin Exp Cardiolog 2012;01. [DOI] [PMC free article] [PubMed]
- 9.Bamford RN, Roessler E, Burdine RD, Saplakoğlu U, dela Cruz J, Splitt M, et al. Loss-of-function mutations in the EGF-CFC gene CFC1 are associated with human left-right laterality defects. Nat Genet. 2000;26:365–9. doi: 10.1038/81695. [DOI] [PubMed] [Google Scholar]
- 10.Ware SM, Peng J, Zhu L, Fernbach S, Colicos S, Casey B, et al. Identification and functional analysis of ZIC3 mutations in heterotaxy and related congenital heart defects. Am J Hum Genet. 2004;74:93–105. doi: 10.1086/380998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gebbia M, Ferrero GB, Pilia G, Bassi MT, Aylsworth A, Penman-Splitt M, et al. X-linked situs abnormalities result from mutations in ZIC3. Nat Genet. 1997;17:305–8. doi: 10.1038/ng1197-305. [DOI] [PubMed] [Google Scholar]
- 12.Aylsworth AS. Clinical aspects of defects in the determination of laterality. Am J Med Genet. 2001;101:345–55. doi: 10.1002/ajmg.1219. [DOI] [PubMed] [Google Scholar]
- 13.Cowan J, Tariq M, Ware SM. Genetic and Functional Analyses of ZIC3 Variants in Congenital Heart Disease. Hum Mutat. 2014;35:66–75. doi: 10.1002/humu.22457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knowles MR, Daniels LA, Davis SD, Zariwala MA, Leigh MW. Primary ciliary dyskinesia: Recent advances in diagnostics, genetics, and characterization of clinical disease. Am J Respir Crit Care Med. 2013;188:913–22. doi: 10.1164/rccm.201301-0059CI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Øyen N, Poulsen G, Wohlfahrt J, Boyd HA, Jensen PKA, Melbye M. Recurrence of discordant congenital heart defects in families. Circ Cardiovasc Genet. 2010;3:122–8. doi: 10.1161/CIRCGENETICS.109.890103. [DOI] [PubMed] [Google Scholar]
- 16.Shieh JTC, Bittles AH, Hudgins L. Consanguinity and the risk of congenital heart disease. Am. J. Med. Genet. Part A. 2012;158 A:1236–41. doi: 10.1002/ajmg.a.35272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mohapatra B, Casey B, Li H, Ho-Dawson T, Smith L, Fernbach SD, et al. Identification and functional characterization of NODAL rare variants in heterotaxy and isolated cardiovascular malformations. Hum Mol Genet. 2009;18:861–71. doi: 10.1093/hmg/ddn411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kennedy MP, Omran H, Leigh MW, Dell S, Morgan L, Molina PL, et al. Congenital heart disease and other heterotaxic defects in a large cohort of patients with primary ciliary dyskinesia. Circulation. 2007;115:2814–21. doi: 10.1161/CIRCULATIONAHA.106.649038. [DOI] [PubMed] [Google Scholar]
- 20.Perles Z, Moon S, Ta-Shma A, Yaacov B, Francescatto L, Edvardson S, et al. A human laterality disorder caused by a homozygous deleterious mutation in MMP21. J Med Genet. 2015;52:840–7. doi: 10.1136/jmedgenet-2015-103336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guimier A, Gabriel GC, Bajolle F, Tsang M, Liu H, Noll A, et al. MMP21 is mutated in human heterotaxy and is required for normal left-right asymmetry in vertebrates. Nat Genet. 2015;47:1260–3. doi: 10.1038/ng.3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Francesco Vetrini, Lisa C.A. D’Alessandro, Zeynep C. Akdemir, Alicia Braxton, Mahshid S. Azamian, Mohammad K. Eldomery, et al. Bi-allelic Mutations in PKD1L1 Are Associated with Laterality Defects in Humans. Am J Hum Genet 2016. [DOI] [PMC free article] [PubMed]
- 23.Homsy J, Zaidi S, Shen Y, Ware JS, Samocha KE, Karczewski KJ, et al. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science (80-) 2015;350:1262–6. doi: 10.1126/science.aac9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zaidi S, Choi M, Wakimoto H, Ma L, Jiang J, Overton JD, et al. De novo mutations in histone-modifying genes in congenital heart disease. Nature. 2013;498:220–3. doi: 10.1038/nature12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sifrim A, Hitz M-P, Wilsdon A, Breckpot J, Turki SHAl, Thienpont B, et al. Distinct genetic architectures for syndromic and nonsyndromic congenital heart defects identified by exome sequencing. Nat Genet. 2016;48:1060–5. doi: 10.1038/ng.3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin SC, Homsy J, Zaidi S, Lu Q, Morton S, Depalma SR, et al. Contribution of rare inherited and de novo variants in 2,871 congenital heart disease probands. Nat Genet. 2017;49:1593–601. doi: 10.1038/ng.3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moustakas A, Heldin C-H. From mono- to oligo-Smads: the heart of the matter in TGF-beta signal transduction. Genes Dev. 2002;16:1867–71. doi: 10.1101/gad.1016802. [DOI] [PubMed] [Google Scholar]
- 28.Komada M, Soriano P. Hrs, a FYVE finger protein localized to early endosomes, is implicated in vesicular traffic and required for ventral folding morphogenesis. Genes Dev. 1999;13:1475–85. doi: 10.1101/gad.13.11.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walpole SM, Hiriyana KT, Nicolaou a, Bingham EL, Durham J, Vaudin M, et al. Identification and characterization of the human homologue (RAI2) of a mouse retinoic acid-induced gene in Xp22. Genomics. 1999;55:275–83. doi: 10.1006/geno.1998.5667. [DOI] [PubMed] [Google Scholar]
- 30.Liao H-M, Niu D-M, Chen Y-J, Fang J-S, Chen S-J, Chen C-H. Identification of a microdeletion at Xp22.13 in a Taiwanese family presenting with Nance-Horan syndrome. J Hum Genet. 2011;56:8–11. doi: 10.1038/jhg.2010.121. [DOI] [PubMed] [Google Scholar]
- 31.Gambin T, Akdemir ZC, Yuan B, Gu S, Chiang T, Carvalho CMB, et al. Homozygous and hemizygous CNV detection from exome sequencing data in a Mendelian disease cohort. Nucleic Acids Res. 2017;45:1633–48. doi: 10.1093/nar/gkw1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karaca E, Yuregir OO, Bozdogan ST, Aslan H, Pehlivan D, Jhangiani SN, et al. Rare variants in the notch signaling pathway describe a novel type of autosomal recessive Klippel-Feil syndrome. Am J Med Genet Part A. 2015;167:2795–9. doi: 10.1002/ajmg.a.37263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Windner SE, Doris Ra, Ferguson CM, Nelson aC, Valentin G, Tan H, et al. Tbx6, Mesp-b and Ripply1 regulate the onset of skeletal myogenesis in zebrafish. Development. 2015;142:1159–68. doi: 10.1242/dev.113431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Amack JD, Wang X, Yost HJ. Two T-box genes play independent and cooperative roles to regulate morphogenesis of ciliated Kupffer’s vesicle in zebrafish. Dev Biol. 2007;310:196–210. doi: 10.1016/j.ydbio.2007.05.039. [DOI] [PubMed] [Google Scholar]
- 35.Cordell HJ, Bentham J, Topf A, Zelenika D, Heath S, Mamasoula C, et al. Genome-wide association study of multiple congenital heart disease phenotypes identifies a susceptibility locus for atrial septal defect at chromosome 4p16. Nat Genet. 2013;45:822–4. doi: 10.1038/ng.2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hanchard NA, Swaminathan S, Bucasas K, Furthner D, Fernbach S, Azamian MS, et al. A genome-wide association study of congenital cardiovascular left-sided lesions shows association with a locus on chromosome 20. Hum Mol Genet. 2016;25:2331–41. doi: 10.1093/hmg/ddw071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Timberlake AT, Choi J, Zaidi S, Lu Q, Nelson-Williams C, Brooks ED, et al. Two locus inheritance of non-syndromic midline craniosynostosis via rare SMAD6 and common BMP2 alleles. eLife. 2016;5:1–19. doi: 10.7554/eLife.20125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Péterfi Z, Donkó A, Orient A, Sum A, Prókai A, Molnár B, et al. Peroxidasin is secreted and incorporated into the extracellular matrix of myofibroblasts and fibrotic kidney. Am J Pathol. 2009;175:725–35. doi: 10.2353/ajpath.2009.080693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Y, Luo Y, Hong Y, Peng J, Lo L. Ribosome Biogenesis Factor Bms1-like Is Essential for Liver Development in Zebrafish. J Genet Genomics. 2012;39:451–62. doi: 10.1016/j.jgg.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 40.Jarvik GP, Browning BL. Consideration of Cosegregation in the Pathogenicity Classification of Genomic Variants. Am J Hum Genet. 2016;98:1077–81. doi: 10.1016/j.ajhg.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gonzaga-Jauregui C, Harel T, Gambin T, Kousi M, Griffin LB, Francescatto L, et al. Exome Sequence Analysis Suggests that Genetic Burden Contributes to Phenotypic Variability and Complex Neuropathy. Cell Rep. 2015;12:1169–83. doi: 10.1016/j.celrep.2015.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu X, Yagi H, Saeed S, Bais AS, Gabriel GC, Chen Z, et al. The complex genetics of hypoplastic left heart syndrome. Nat Genet. 2017;49:1152–9. doi: 10.1038/ng.3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Posey JE, Harel T, Liu P, Rosenfeld JA, James RA, Coban Akdemir ZH, et al. Resolution of Disease Phenotypes Resulting from Multilocus Genomic Variation. N Engl J Med. 2017;376:21–31. doi: 10.1056/NEJMoa1516767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.The ARIC Investigators. The Atherosclerosis Risk in Communities (ARIC) Study: design and objectives. Am J Epidemiol. 1989;1294:687–702. [PubMed] [Google Scholar]
- 45.Reid JG, Carroll A, Veeraraghavan N, Dahdouli M, Sundquist A, English A, et al. Launching genomics into the cloud: deployment of Mercury, a next generation sequence analysis pipeline. BMC Bioinformatics. 2014;15:30. doi: 10.1186/1471-2105-15-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–95. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Challis D, Yu J, Evani US, Jackson AR, Paithankar S, Coarfa C, et al. An integrative variant analysis suite for whole exome next-generation sequencing data. BMC Bioinformatics. 2012;13:8. doi: 10.1186/1471-2105-13-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–23. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trivellin G, Daly AF, Faucz FR, Yuan B, Rostomyan L, Larco DO, et al. Gigantism and acromegaly due to Xq26 microduplications and GPR101 mutation. N Engl J Med. 2014;371:2363–74. doi: 10.1056/NEJMoa1408028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boone PM, Bacino CA, Shaw CA, Eng PA, Hixson PM, Pursley AN, et al. Detection of clinically relevant exonic copy-number changes by array CGH. Hum Mutat. 2010;31:1326–42. doi: 10.1002/humu.21360. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.