Abstract
Clinical exome sequencing (CES) has become the preferred diagnostic platform for complex pediatric disorders with suspected monogenic etiologies. Despite rapid advancements, the major challenge still resides in identifying the casual variants among the thousands of variants detected during CES testing, and thus establishing a molecular diagnosis. To improve the clinical exome diagnostic efficiency, we developed Phenoxome, a robust phenotype-driven model that adopts a network-based approach to facilitate automated variant prioritization. Phenoxome dissects the phenotypic manifestation of a patient in concert with their genomic profile to filter and then prioritize variants that are likely to affect the function of the gene (potentially pathogenic variants). To validate our method, we have compiled a clinical cohort of 105 positive patient samples that represent a wide range of genetic heterogeneity. Phenoxome identifies the causative variants within the top 5, 10, or 25 candidates in more than 50%, 71%, or 88% of these exomes, respectively. Furthermore, we show that our method is optimized for clinical testing by outperforming the current state-of-art method. We have demonstrated the performance of Phenoxome using a clinical cohort and showed that it enables rapid and accurate interpretation of clinical exomes. Phenoxome is available at https://phenoxome.chop.edu/.
Subject terms: Genetic testing, Computational biology and bioinformatics
Introduction
Individual Mendelian pediatric diseases are rare, yet approximately 8% of the worldwide population is diagnosed with at least one genetic disorder before reaching adulthood [1]. Exome and genome sequencing have rapidly altered the landscape of clinical genetics by enabling researchers and physicians to make novel gene-disease associations [2] and precise molecular diagnoses [3]. However, molecular testing using Clinical Exome Sequencing (CES) remains challenging with only about 30% of patients receiving a definitive diagnosis [4]. Making a diagnosis is often complicated by the sheer volume of variants detected and the presentation of overlapping phenotypic characteristics in affected individuals [5].
A carefully designed analysis workflow is essential for high quality interpretation of CES [6]. Clinical correlation, which includes concurrent assessment of the patient’s phenotypes and genotypes, is central to the overall interpretation [7]. During this step, putative causative genes and variants that may contribute to the clinical findings are identified. Nonetheless, clinical correlation is often time consuming and requires extensive medical and genetic expertize [8].
The use of prior biological and clinical knowledge, such as previously known disease-causing genes and variants, can assist in gene-disease clinical correlation [9]. A number of databases [10–12] that curate gene-disease associations have been developed, along with several computational variant annotation tools, which are now routinely used in CES [13–15]. Detailed phenotypic information greatly enhances the diagnostic power of exome sequencing, prompting an array of phenotype-driven approaches that often employ machine learning methods, including eXtasy [16], Phenomizer [5], PHIVE [17], Phevor [18], PhenIX [19], Phen-Gen [20], SimReg [21], Phenolyzer [22], and PVP [23]. Most of these tools use the vocabulary from the Human Phenotype Ontology (HPO) [24] to describe a patient’s phenotypic abnormalities.
These tools have clearly demonstrated the utility of using gene-curated phenotype data to improve disease gene identification. Most of these approaches have been validated on a large number of simulated scenarios, but a limited number of clinical samples. However, in some cases, clinician evaluation of variants has been shown to outperform computational approaches. [25]. Additionally, none of these machine-learning based tools have been validated on a large-scale clinical sequencing cohort. As variant interpretation remains a challenge for clinical laboratories, continuous refinement of computational algorithms is necessary.
Here, we present a computational framework, Phenoxome, to filter and then prioritize candidate variants using population frequency, deleteriousness and clinical relevance of the affected gene (Fig. 1). Phenoxome uses two inputs, (i) a variant call format (VCF) file representing the genotypes of the affected individual, and (ii) a set of symptoms described using HPO terms. Our approach first filters the variants according to rarity, predicted protein effects and other prior knowledge. Following this, Phenoxome generates a personalized gene panel (PGP) derived from the phenotypic manifestations, and each gene in the PGP is scored based on its potential involvement in these phenotypes. Finally, each variant is prioritized based upon a composite score combining the knowledge inferred from both variant level and gene level information. We first evaluate the performance of our method on comprehensive computational simulations of different scenarios. We then demonstrate the effectiveness of Phenoxome using 105 positive clinical exomes from Children’s Hospital of Philadelphia (CHOP). Our approach outperforms the state-of-art method by yielding superior overall rankings of the causative variants of the clinical samples.
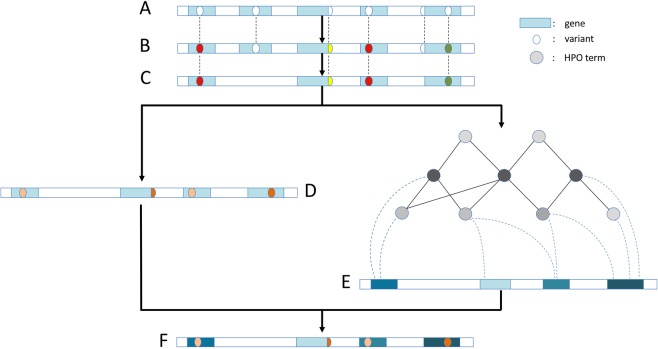
Fig. 1.
Step-wise schema of Phenoxome’s overall workflow. a Raw variants yielded from sequencing the patient’s exome and subsequent bioinformatics analysis. Blue rectangles imply genes and ovals indicate variants. b Variants annotated by Phenoxome using a series of bioinformatics resources. Distinct color schemes indicate different predicted effects on protein products. c Variants retained after filtering procedure depending on HGMD annotations, population allele frequency and functional effects. d Variants deleterious score are derived from the tier strategy where a darker color implies a more disruptive variant. e Genes harboring post-filtered variants are assigned phenotypic relevance scores inferred by their associations with relevant phenotypes in HPO. A darker color implies the gene is more pertinent to the patient’s phenotypic manifestation. f Each of post-filtration variants receives an overall score by integrating both variant deleterious score and the gene’s phenotypic relevance score. Hence a global prioritization of the variants is achieved in the framework
Material and methods
Variant annotation and filtration
Variants are first annotated using SnpEff package v4.214 with hg19 RefSeq database. In addition, the variants are also annotated with the Human Gene Mutation Database (HGMD) v2017.210, and minor allele frequencies from the Genome Aggregation Database (gnomAD) v2.0 [26].
Similar to the clinically-validated strategy implemented at CHOP [27], Phenoxome retains a variant if it meets one of the following criteria:
AF < 1% in gnomAD and classified as disease mutation (DM or DM?) in HGMD
AF < 0.2% in all sub-populations in gnomAD and predicted to alter protein or splice sites (i.e splice acceptor/donor, stop retained/gained, start/stop loss, inframe deletion/insertion, frameshift and missense variants)
A detailed variant filtering scheme is demonstrated in Fig. 2.
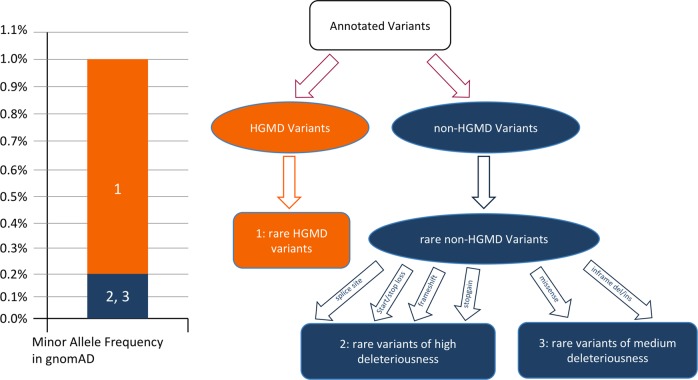
Fig. 2.
Variant filtration strategy and tiers of deleterious score. A HGMD variant of DM or DM? class is retained if the minor allele frequency of the variant is less than 1% in general population in gnomAD database. This variant is binned tier one and is assigned 1.0 as deleterious score (shown in orange 1). A non-HGMD variant is retained if the predicted protein effect is disruptive, and its minor allele frequency is less than 0.2% in general population, as well as five sub-populations (East Asian, Finnish, Non-finnish European, African/African American and South Asian) in gnomAD. If the predicted effects of the variant include change of splice site, start/stop loss, frameshift and stopgain, the variant is binned tier two and is assigned 0.8 as deleterious score (shown in blue 2). The variant is binned tier three otherwise and is assigned 0.6 as deleterious score (shown in blue 3)
Global variant prioritization strategy
In Phenoxome, each variant that passes filtration receives a composite score reflecting its likelihood of affecting the protein’s function and causing disease, and hence a ranking of the variants is achieved based upon the scores. We define a pathogenic variant as a disease causing variant in the context of a patient’s phenotypes. Similar to other approaches [19, 20], the composite score of each variant is constructed using a deleterious score and a phenotypic relevance score, derived from variant level and gene level measurements, respectively. A variant level score usually indicates the disruptiveness of the variant, inferred by characteristics such as rarity, evolutionary conservation and predicted functional impact [28]. A gene level score generally reflects the assessment of the affected gene’s functional involvement in the observed phenotypes. Unlike other approaches that calculate the composite score by averaging the variant score and the gene relevance score [17, 19], our approach assigns greater weight to the phenotypic component while generating the overall score of a variant. This empirical implementation was derived from the clinical observation that most of the rare variants with disruptive protein effects were harbored by genes that shared little or no known disease overlap with the phenotypic manifestation of the affected individual.
Deleterious score
Each of the variant that passes filtration is evaluated and assigned a deleterious score based upon its predicted functional impact and HGMD label. Inspired by clinical protocols classifying variants into different categories [29], we implement a tier system to triage the variants into three different bins. A variant is deemed the most damaging if it is in HGMD with a DM/DM? class (bin 1). The damage level is deemed high if the functional impact of a non-HGMD variant is any of the following: splice site aberration, frameshift, stop gain, start loss or stop loss (bin2). The rest of the variants are deemed medium damaging if the predicted effects include missense, inframe deletion or insertion (bin3). Binned variants are given a deleterious score of 1.0, 0.8, and 0.6 for bins 1, 2, and 3 respectively (Fig. 2).
Phenotypic relevance score
Variants are also assessed on the gene level using HPO. HPO is a computational representation of a wide collection of phenotype abnormalities in human. Each of the phenotypes in the vocabulary is annotated with genes implicated with the clinical symptoms, curated from resources including OMIM [11] and Orphanet [30]. Because of its strictly controlled and standardized vocabulary, hierarchical structure and well-defined phenotype-gene relationships, HPO has become an ideal resource for clinical phenotyping [31].
Phenotypic terms in HPO are organized in a directed acyclic graph where they are associated by “is a” relationships. An “is a” relationship indicates that one phenotype is a subclass of another phenotype that is a more generic parent term [32]. For instance, Abnormality of the atrial septum “is an” Abnormality of the cardiac setpa which “is an” Abnormal heart morphology. The design of Phenoxome takes the advantage of the hierarchical structure of HPO and assembles a Personalized Gene Panel (PGP) for each patient, where each gene of the PGP is potentially associated with the input phenotypes. Our approach starts from each of the provided phenotypes, and then traverses down the ontology tree to retrieve all of its direct and indirect subclass nodes/phenotypes until a terminal node is encountered (Fig. S1). The nature of “is a” associations guarantees that all of the children nodes are essentially subclasses of the primary phenotypes by the definition of the hierarchy. In addition, to account for imprecision in selecting the primary phenotypes in clinical scenarios, the algorithm also visits the immediate parent nodes of the input phenotypes. The original terms describing the phenotypes of the patient are considered primary, while the terms retrieved during the extension process are termed secondary. Following this, PGP is compiled to collect all of the genes associated with any of the primary or secondary phenotypes. These genes are reported to have caused corresponding symptoms and therefore are potentially relevant to the patient’s phenotypes.
Once Phenoxome identifies the primary and secondary terms, and a sub-graph of the HPO tree containing all of the nodes and their parent-child relationships is also generated, we then employ a network-based approach, PageRank with Priors [33], to prioritize each phenotype in the sub-graph. The algorithm evaluates the significance of each node of a graph with a clear-defined transition matrix by imitating a random walker surfing the graph. Starting from a root node, the surfer selects an outgoing edge from the current node randomly to jump to the next node in each iteration. The algorithm converges when the significance scores of the nodes become steady. In a directed acyclic graph, this process is similar to the ontology propagation described by Singleton et al. [18]. However, with a set of priors (root set), the random surfer opts to jump back to any of the node in the root set regardless of its current location with a predefined probability in each iteration. The iterative stationary probability equation of a node n is given by
| 1 |
where is the back probability. The first component of the equation summarizes the likelihood of arriving at this node from all of its neighboring nodes while the second component indicates transporting back to the root set. In Eq. 1, when the node is in the root set and 0 if otherwise. is the size of the root set. The stationary distribution after the convergence of the algorithm represents the probability of the random surfer landing on each node at any given moment.
In order to implement the algorithm in the context of the sub-graph of the HPO tree, we set the primary HPO terms as the priors and the back probability β to be 0.5 as it was within the range that was suggested to yield optimal performance by previous studies [34], meaning there is a 50% chance of the random surfer returning to the primary terms in each step. It is intuitive to see several benefits with this implementation for the clinical utility. The primary phenotypes are ranked highly because of the back probability; the secondary phenotypes that are close to the primary phenotypes are ranked highly because they are easily accessible from the root set; and the secondary phenotypes with more “is a” relationships are ranked highly because they are more likely to be visited during the random walk. Each gene in PGP may be associated with multiple primary and secondary phenotypes, thus a variant receives a phenotypic score that is the sum of all of the phenotypes’ scores the affected gene is associated with. In this way, variants harbored by genes associated with more significant phenotypes are ranked higher.
Integrated variant pathogenicity score
As discussed earlier, an overall score is assigned to each of the candidate variants. The first component is the gene-phenotypic score and the second is the deleterious score. Each of the components are normalized to the range of [0, 1] across all the variants. Following this, weight factor α is employed to combines the two components together in the final significance score:
| 2 |
where α is intuitively set to 0.8 to ensure the global implementation is phenotype-driven. The final scores of all variants are then normalized to the range of [0, 1] and hence a prioritization of the variants is achieved. The configuration of α was re-evaluated after initial benchmarking, which is discussed in the Supplementary Materials and Methods.
Clinical sample cohort
Acquiring validated large-scale clinical cohorts for differential clinical diagnostics studies is challenging and limited access to these resources has forced most of the abovementioned computational tools to perform their validation largely or solely using in silico patient profiles. We have collected a large cohort of clinical patients (n = 105) where each individual patient received a positive molecular diagnosis from clinical exome sequencing (CES) analysis. Eighty-five positive samples were from patients diagnosed by the clinical exome test at the Division of Genomic Diagnostics (DGD) at CHOP between 2014 and 2017. Twenty positive exomes were collected through the CHOP Pediatric Sequencing (PediSeq) project, which was a part of the National Human Genome Research Institute (NHGRI) Clinical Sequencing Exploratory Research (CSER) consortium. For this study, we define positive cases as having at least one pathogenic variant in the final clinical laboratory report. The variant interpretation procedure followed ACMG/AMP guidelines [35], and the pathogenic variants were thoroughly evaluated and classified as disease causing with concrete supporting evidence [27]. The phenotypic features of these patients were carefully discussed and documented by physicians upon clinical chart reviews, and the corresponding HPO terms were selected to best represent the symptoms. All of the samples in the cohort were from pediatric patients representing a wide range of complex syndromes. Additional phenotypic characteristics, as well as the sequencing details and subsequent bioinformatics processing of the samples are described in Supplementary Materials and Methods.
Results
Ranking candidate genes using synthetic patient profiles
Since the phenotypic scores of candidate variants are imperative to the overall prioritization and due to the general lack of clinical data, we first assessed the performance of the candidate gene ranking using in silico patients [5, 21]. We focused on 33 monogenic diseases with known causative genes and used a similar strategy discussed by Masino et al. [36]. Three clinical scenarios were considered during the simulations, including “optimal”, “noisy” and “imprecise”. 1000 simulated profiles were generated per scenario for each diseases. The details of the generation of these profiles are discussed in the Supplementary Materials and Methods.
We carried out the candidate gene prioritization approach of Phenoxome on all simulated patient profiles of the three scenarios. For each synthetic patient, our algorithm first generated the PGP from the phenotypes and then prioritized the genes in PGP using the phenotypic relevance scores presented in Methods. In all of the simulated cases, the causative genes were constantly captured by the PGP across the 33 diseases of the above scenarios. In the “optimal” scenario, the causative gene was ranked in the first place for 98.5% of the simulations. Introducing the “noise” terms did not have any substantial impact on the rankings. In the “noisy” scenario, the causative gene was ranked first for 94.4% of the cases. Consistent with previous studies, a deteriorated performance of Phenoxome in the “imprecise” scenarios was observed where the causative gene was ranked first in only 3.7% of the cases. However, the target gene was ranked within top 10% of the PGPs in 89.8% of the cases. The overall summary of the performance of Phenoxome in three scenarios is demonstrated in Fig. 3, with corresponding AUC (area under curve) values for the “optimal”, “noisy” and “imprecise” scenarios of 0.995, 0.991, and 0.952, respectively [5, 36].
Fig. 3.
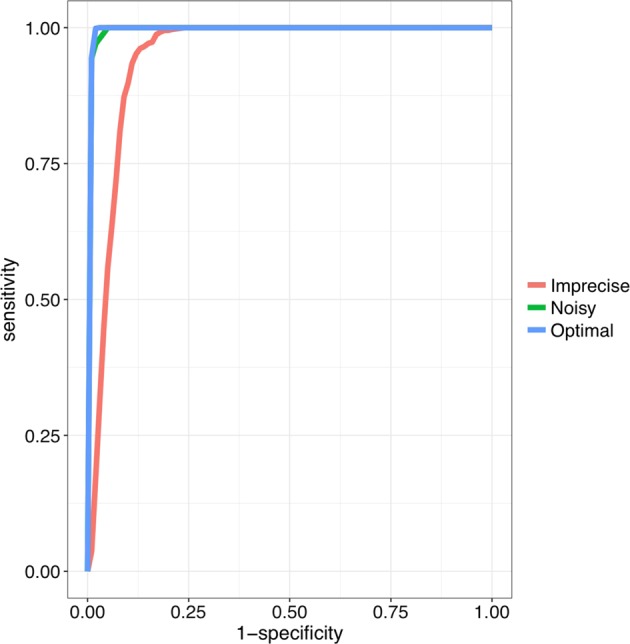
Benchmarking Phenoxome on simulated patient profiles. Blue curve is the ROC of Phenoxome’s performance on the “optimal” scenarios based on score ranks; green and red curves indicate the “noisy” and “imprecise” scenarios, respectively. Each of the plots was generated from 33,000 simulated cases. Sensitivity was defined as the frequency of “target” genes that are ranked above a particular threshold position, and specificity as the percentage of genes ranked below the threshold. For instance, a sensitivity/specificity value of 70/90 indicates that the disease gene (the “target”) is ranked among the best-scoring 10% of genes in 70% of the prioritizations
Performance on clinical samples
We then used the clinical cohort to validate Phenoxome performance. Paired VCF files and HPO terms of each clinical sample were utilized as described in Methods. These VCF files were generated by the original version of the clinical bioinformatics pipeline at the time the case was analyzed. Phenoxome generated a list of ranked variants independent of the original clinical evaluation. The numbers of variants in the original input VCF files ranged from 37,150 to 258,968. Since these samples were collected during a period of four years, various enhancements were made to the clinical bioinformatics pipeline, such as better use of laboratory-specific sequence quality metrics for variant filtration. These enhancements led to less variants in more recent version of the pipeline. The clinically reported pathogenic variants were consistently retained in the final ranked lists and their ranks were recorded for each of the patient. If more than one pathogenic variant was reported, the best rank of these variants was used in the benchmarking for the case.
The median rank of the pathogenic variants in the 105 patients was 5 with a standard deviation of 23. Specifically, 92 (88%) pathogenic variants were ranked in the top 25, 74 (71%) in the top 10, 53 (51%) within the top 5, while 17 pathogenic variants (16%) were ranked in the first place.
Causal genes harboring the pathogenic variants were captured in the PGP in 95 out of 105 cases (associated with at least one primary or secondary phenotype). The median rank of the pathogenic variants was 4 among those 95 cases. Three primary and/or secondary phenotypes were annotated to the causal gene on average among these cases. Causal genes were associated with at least one primary phenotype in 78 out of 95 cases while they were associated with only secondary phenotypes in the other 17 cases. No statistical differences between the two groups were observed regarding the ranks of the pathogenic variants. For the 17 cases where the causal genes were associated with only secondary phenotypes, the ranking of causative variants were in the range of 1 to 24 with an average of 6. It is common that the causal gene was associated with a primary phenotype when it was also implicated in secondary phenotypes inferred from other primary phenotypes. In a striking instance where the patient was documented with phenotypes of Chronic mucocutaneous candidiasis, Recurrent fungal infections, Recurrent candida infections and Impaired T cell function, the pathogenic variant was identified in IL12RB1, which was not directly annotated with any of the primary phenotypes but was associated with Onychomycosis which was a sub-class of Recurrent fungal infections. Thus, the causal gene was captured in the PGP and the missense pathogenic variant was ranked in the second position for this patient.
Phenoxome heavily relies on the provided phenotypes and the gene-phenotype associations in prioritizing the variants. Thus, using the most accurate and up-to-date phenotypes is essential to achieving the optimal performance. On the other hand, as phenotypic features of patients evolve over time, as well as new gene-phenotype associations are uncovered, re-analysis using Phenoxome could yield new diagnosis. In our clinical validation cohort, 10 pathogenic variants (marked in orange in Fig. 4) were not in PGP during the initial benchmarking. These variants were retained after filtration but the affected genes were not associated with any primary or secondary phenotypes. The pathogenic variants in these 10 cases were ranked in the range of 5 to 140. With one exception where the pathogenic variant was ranked in top 5, the rest of these variants all scored well below the median rank of the cohort, with an average rank of 59. To investigate the ten cases, re-analysis was performed using the latest build of HPO (build 1249, January 2018). Three out of the ten causative genes were annotated with at least one pertinent phenotype in the re-analysis, resulting substantially better ranks of the pathogenic variants (Table 1). The improvement was due to novel gene-phenotype relationships curated by HPO that were absent in the HPO database version at the time of the initial analysis. These findings highlight the clinical utility of re-analysis of exome data to yield additional diagnosis in a systematic manner [37]. For the remaining seven cases, we noted that precise HPO terms were not provided in the clinical HPO phenotyping information.
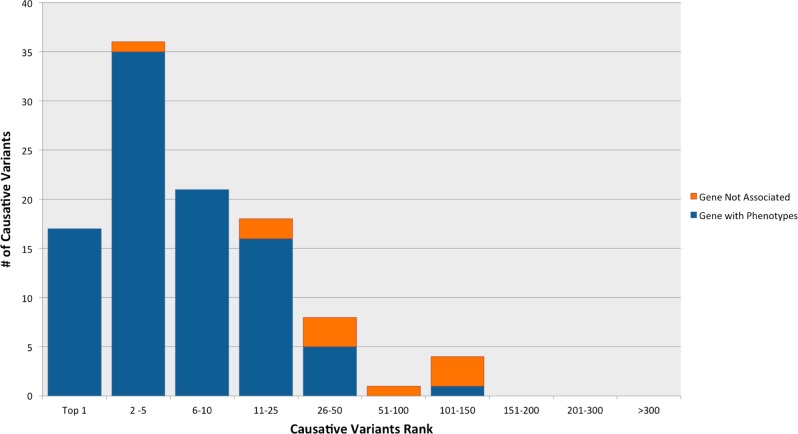
Fig. 4.
Summary of performance of phenoxome on clinical cohort. Blue bars represent the cases where the causative variants/genes are associated with at least one pertinent phenotype. Orange bars represent the cases where the causative variants/genes are not associated with any pertinent phenotype. 53(51%) target variants were ranked in top 5 while 92(88%) target variants were ranked in top 25 among 105 clinical cases
Table 1.
Re-analysis results on 3 clinical samples
| Sample | HPO terms | Initial analysis rank | Re-analysis rank |
|---|---|---|---|
| CHOP-PA-S28 | HP:0000577;HP:0001249;HP:0000717;HP:0000739;HP:0000047;HP:0004322;HP:0001999;HP:0002275;HP:0011098 | 41 | 31 |
| CHOP-PA-S52 | HP:0001263;HP:0001508;HP:0008872;HP:0001385;HP:0004474;HP:0004482;HP:0000280;HP:0010813 | 48 | 8 |
| CHOP-PA-S78 | HP:0000410;HP:0001508;HP:0001263;HP:0001738;HP:0000010;HP:0012330;HP:0100785;HP:0004322;HP:0000076 | 41 | 3 |
Comparison with PhenIX
Unlike Phenoxome, most previously published computational approaches were primarily assessed using simulated patient data (see Table S1). Studies have suggested that the performance of such tools could vary significantly when using actual clinical cases [38, 39]. Specifically, a recent comparative study examined the performance of a wide range of phenotype-driven variant prioritization methods on 21 positive clinical exomes, and determined that PhenIX was the most effective [40]. Thus, we benchmarked the performance of PhenIX on the exomes in our cohort and compared the rank positions of the causative variants with Phenoxome. The analyses of the clinical samples in the cohort using PhenIX is discussed in the Supplementary Materials and Methods.
As the result, the pathogenic variants were ranked in the range of 1 to 497 among 86 patients in the cohort. PhenIX ranked the causative variants in first place in 22 (21%) out of the 86 cases, showing a slight advantage over Phenoxome (17/105). However, PhenIX scored notably fewer causative variants in the top 5 (45, 43%), top 10 (50, 48%), and top 25 (57, 54%) than Phenoxome, respectively (Table S2). Moreover, PhenIX did not report the causative variants in the final ranked list in 19 out of the 105 cases.
Collectively, Phenoxome outperformed PhenIX on the clinical cohort by yielding more robust rankings of the same pathogenic variants (Fig. S2) as suggested by the statistical analysis on the overall rankings of the pathogenic variants (p = 0.0015; Mann–Whitney test).
Discussion
Several academic and commercial variant prioritization tools have been introduced in recent years. These algorithms have demonstrated efficacy and efficiency in a wide variety of scenarios albeit only a handful of them have been validated on a limited set of clinical sequencing data. Here, we presented a clinical laboratory focused computational framework to aid with clinical correlation and variant interpretation. Through a series of benchmarking using both in silico and clinical data, Phenoxome has demonstrated high clinical utility in identifying the causative variants in a wide range of scenarios and outperformed the semantic similarity-based approaches such as PhenIX.
Phenoxome’s advantage over PhenIX is exhibited through the consistently retention of causative variants during the filtration process and by achieving better rankings overall. Specifically, we believe our model outperforms PhenIX in the CES cohort because Phenoxome is more patient-centric by utilizing PGP. In semantic similarity-based models, the phenotypic relevance score of a gene is calculated by comparing the set of phenotypes manifested by the patient and all of the phenotypes associated with the gene, which may lead to what we call “phenotype dilution”. In clinical chart reviews, the symptoms are summarized into several key phenotypes. However, a well-studied gene may be associated with a wide range of diseases that may be unrelated to each other. All of these associated phenotypes contribute to the semantic similarity calculation, which may “dilute” the associated phenotypes specific to this patient. On the other hand, our approach only takes into account the primary and secondary phenotypes of the patient and thus the signal is enhanced for the causal gene, as other irrelevant phenotypes associated with the gene are not considered in the analysis. For instance, in a clinical sample where a patient was documented with Volvulus, Intestinal pseudo-obstruction, Cholestatis and Intestinal malrotation, a missense variant in ACTG2 was classified pathogenic. ACTG2 was associated with a total of 34 different phenotypes in HPO, ranging from Camptodactyly of finger to Sepsis, including Intestinal malrotation. Most of these phenotypes were not observed and unrelated to this patient, as they were “noise” in the similarity metrics and PhenIX prioritized this variant at rank 70. In contrast, Phenoxome did not consider those “noise” phenotypes in its modeling and ranked the causative variant in the second place.
Trio (an affected proband with both parents) clinical exome sequencing has been proven more effective in detecting de novo and compound heterozygous variants compared to a proband-only approach [41]. However, since parents are not always available for CES analysis, variant prioritization in Phenoxome is designed to be independent of mode of inheritance.
We also compared the number of variants requiring manual review in the 85 DGD CES samples retrospectively. On average, Phenoxome reported 240 less variants per case which could save significant review time from the interpretation workflow. Phenoxome not only reduces the number of variants for manual review but also allows the clinical laboratories to sift through the variants using a prioritized list. Specifically, the search for the pathogenic variants can stop after identifying those that explain all of the patient’s phenotypes [35], and hence enabling rapid interpretation of clinical exomes.
Phenoxome is different from attempts to optimize the input phenotypes such as Bauer et al. [42]. Instead, it generates a patient phenotypic profile from the HPO terms provided. Furthermore, Phenoxome does not make inferences from non-human genomic data, unlike several other tools [18, 20]. By utilizing only well-established evidence of human disease and associated genes, it is designed for clinical sequencing applications and is not optimized for novel gene discovery. This strategy offers clinical robustness that precludes non-human genomic data, which often does not benefit clinical diagnostics [40]. While Phenoxome is able to effectively reduce raw variants to a more manageable prioritized list, clinical input and further curation are still essential for evaluating the pathogenic status of the variants.
The performance benchmarking highlights the importance of validating computational algorithms using real-world clinical data sets. Furthermore, our validation results also indicate that selecting the most accurate phenotypes to describe the symptom manifestations of a patient is crucial for CES interpretation and ultimately an accurate diagnostic. We have shown several scenarios that could have contributed to those cases where the causal genes were not in the PGP. One plausible explanation is the phenotype-gene annotations are absent from HPO, due to the lag between publication and annotation database updates. It is also possible that a phenotype abnormality, which could potentially direct Phenoxome to the causal gene, might have been missed or unobserved during the clinical chart review. Nonetheless, with its robust algorithm and regular database updates, Phenoxome provides a platform that enables clinicians and laboratories to re-analyze undiagnosed cases.
Supplementary information
Supplementary materials and methods revised
Acknowledgements
Clinical sequencing and analyses were supported in part by the Division of Genomic Diagnostics and the Individualized Medical Genetics Center at CHOP. The PediSeq project at CHOP was supported by NHGRI Clinical Sequencing Exploratory Research (CSER) consortium (grant U01HG006546). The authors thank Michael Hammond and Leonard Hu for their assistance in supporting the web application of this work.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Ahmad N. Abou Tayoun, Email: ahmad.tayoun@ajch.ae.
Mahdi Sarmady, Phone: +1 267426 1373, Email: sarmadym@chop.edu.
Supplementary information
The online version of this article (10.1038/s41431-018-0328-7) contains supplementary material, which is available to authorized users.
References
- 1.Baird PA, Anderson T, Newcombe H, Lowry R. Genetic disorders in children and young adults: a population study. Am J Hum Genet. 1988;42:677. [PMC free article] [PubMed] [Google Scholar]
- 2.Bamshad MJ, Ng SB, Bigham AW, et al. Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet. 2011;12:745–55. doi: 10.1038/nrg3031. [DOI] [PubMed] [Google Scholar]
- 3.Yang Y, Muzny DM, Reid JG, et al. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. New Engl J Med. 2013;369:1502–11. doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee H, Deignan JL, Dorrani N, et al. Clinical exome sequencing for genetic identification of rare Mendelian disorders. JAMA. 2014;312:1880–7. doi: 10.1001/jama.2014.14604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kohler S, Schulz MH, Krawitz P, et al. Clinical diagnostics in human genetics with semantic similarity searches in ontologies. Am J Hum Genet. 2009;85:457–64. doi: 10.1016/j.ajhg.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seaby EG, Pengelly RJ, Ennis S. Exome sequencing explained: a practical guide to its clinical application. Brief Funct Genom. 2015;15:374–84. doi: 10.1093/bfgp/elv054. [DOI] [PubMed] [Google Scholar]
- 7.Segal MM. Genome interpretation: clinical correlation is recommended. Appl Transl Genom. 2015;6:26–27. doi: 10.1016/j.atg.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quintáns B, Ordóñez-Ugalde A, Cacheiro P, Carracedo A, Sobrido M. Medical genomics: the intricate path from genetic variant identification to clinical interpretation. Appl Transl Genom. 2014;3:60–67. doi: 10.1016/j.atg.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gudivada RC, Qu XA, Chen J, Jegga AG, Neumann EK, Aronow BJ. Identifying disease-causal genes using Semantic Web-based representation of integrated genomic and phenomic knowledge. J Biomed Inform. 2008;41:717–29. doi: 10.1016/j.jbi.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 10.Stenson PD, Mort M, Ball EV, Evans K, Hayden M, Heywood S, et al. The Human Gene Mutation Database: towards a comprehensive repository of inherited mutation data for medical research, genetic diagnosis and next-generation sequencing studies. Hum Genet. 2017;136:1–13. [DOI] [PMC free article] [PubMed]
- 11.McKusick VA. Online Mendelian inheritance in man, OMIM. http://www ncbi nlm nih gov/omim/. 2006. [DOI] [PubMed]
- 12.Aymé S. [Orphanet, an information site on rare diseases]. Soins; la revue de référence infirmière 2003; 46. [PubMed]
- 13.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164–e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cingolani P, Platts A, Wang LL, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strainw1118; iso-2; iso-3. Fly. 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, Cunningham F. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26:2069–70. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sifrim A, Popovic D, Tranchevent LC, et al. eXtasy: variant prioritization by genomic data fusion. Nat Methods. 2013;10:1083–4. doi: 10.1038/nmeth.2656. [DOI] [PubMed] [Google Scholar]
- 17.Robinson PN, Kohler S, Oellrich A, et al. Improved exome prioritization of disease genes through cross-species phenotype comparison. Genome Res. 2014;24:340–8. doi: 10.1101/gr.160325.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singleton MV, Guthery SL, Voelkerding KV, et al. Phevor combines multiple biomedical ontologies for accurate identification of disease-causing alleles in single individuals and small nuclear families. Am J Hum Genet. 2014;94:599–610. doi: 10.1016/j.ajhg.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zemojtel T, Kohler S, Mackenroth L, et al. Effective diagnosis of genetic disease by computational phenotype analysis of the disease-associated genome. Sci Transl Med. 2014;6:252ra123. doi: 10.1126/scitranslmed.3009262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Javed A, Agrawal S, Ng PC. Phen-Gen: combining phenotype and genotype to analyze rare disorders. Nat Methods. 2014;11:935–7. doi: 10.1038/nmeth.3046. [DOI] [PubMed] [Google Scholar]
- 21.Greene D, BioResource N, Richardson S, Turro E. Phenotype similarity regression for identifying the genetic determinants of rare diseases. Am J Human Genet. 2016;98:490–9. doi: 10.1016/j.ajhg.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang H, Robinson PN, Wang K. Phenolyzer: phenotype-based prioritization of candidate genes for human diseases. Nat Methods. 2015;12:841–3. doi: 10.1038/nmeth.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boudellioua I, Razali RBM, Kulmanov M, et al. Semantic prioritization of novel causative genomic variants. PLoS Comput Biol. 2017;13:e1005500. doi: 10.1371/journal.pcbi.1005500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Köhler S, Vasilevsky NA, Engelstad M, et al. The human phenotype ontology in 2017. Nucleic Acids Res. 2017;45:D865–D876. doi: 10.1093/nar/gkw1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stark Z, Dashnow H, Lunke S, et al. A clinically driven variant prioritization framework outperforms purely computational approaches for the diagnostic analysis of singleton WES data. Eur J Hum Genet. 2017;25:1268–72. doi: 10.1038/ejhg.2017.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gibson KM, Nesbitt A, Cao K, Yu Z, Denenberg E, DeChene E, et al. Novel findings with reassessment of exome data: implications for validation testing and interpretation of genomic data. Genet Med. 20.3 (2018): 329. [DOI] [PubMed]
- 28.Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berg JS, Khoury MJ, Evans JP. Deploying whole genome sequencing in clinical practice and public health: meeting the challenge one bin at a time. Genet Med. 2011;13:499–504. doi: 10.1097/GIM.0b013e318220aaba. [DOI] [PubMed] [Google Scholar]
- 30.Weinreich SS, Mangon R, Sikkens J, Teeuw M, Cornel M. Orphanet: a European database for rare diseases. Ned Tijdschr Geneeskd. 2008;152:518–9. [PubMed] [Google Scholar]
- 31.Girdea M, Dumitriu S, Fiume M, et al. PhenoTips: patient phenotyping software for clinical and research use. Hum Mutat. 2013;34:1057–65. doi: 10.1002/humu.22347. [DOI] [PubMed] [Google Scholar]
- 32.Robinson PN, Köhler S, Bauer S, Seelow D, Horn D, Mundlos S. The Human Phenotype Ontology: a tool for annotating and analyzing human hereditary disease. Am J Human Genet. 2008;83:610–5. doi: 10.1016/j.ajhg.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.White S, and P Smyth, Algorithms for estimating relative importance in networks. Proceedings of the Proceedings of the ninth ACM SIGKDD international conference on Knowledge discovery and data mining. ACM 2003 Washington DC, USA, pp. 266–75.
- 34.Chen J, Aronow BJ, Jegga AG. Disease candidate gene identification and prioritization using protein interaction networks. BMC Bioinforma. 2009;10:73. doi: 10.1186/1471-2105-10-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Masino AJ, Dechene ET, Dulik MC, et al. Clinical phenotype-based gene prioritization: an initial study using semantic similarity and the human phenotype ontology. BMC Bioinforma. 2014;15:248. doi: 10.1186/1471-2105-15-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wenger AM, Guturu H, Bernstein JA, Bejerano G. Systematic reanalysis of clinical exome data yields additional diagnoses: implications for providers. Genet Med. 2017;19:209. doi: 10.1038/gim.2016.88. [DOI] [PubMed] [Google Scholar]
- 38.Bone WP, Washington NL, Buske OJ, et al. Computational evaluation of exome sequence data using human and model organism phenotypes improves diagnostic efficiency. Genet Med. 2016;18:608. doi: 10.1038/gim.2015.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eilbeck K, Quinlan A, Yandell M. Settling the score: variant prioritization and Mendelian disease. Nat Rev Genet. 2017;18:599. doi: 10.1038/nrg.2017.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pengelly RJ, Alom T, Zhang Z, Hunt D, Ennis S, Collins A. Evaluating phenotype-driven approaches for genetic diagnoses from exomes in a clinical setting. Sci Rep. 2017;7:13509. doi: 10.1038/s41598-017-13841-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sawyer S, Hartley T, Dyment D, et al. Utility of whole‐exome sequencing for those near the end of the diagnostic odyssey: time to address gaps in care. Clin Genet. 2016;89:275–84. doi: 10.1111/cge.12654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bauer S, Köhler S, Schulz MH, Robinson PN. Bayesian ontology querying for accurate and noise-tolerant semantic searches. Bioinformatics. 2012;28:2502–8. doi: 10.1093/bioinformatics/bts471. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary materials and methods revised