Abstract
Background
Osteosarcoma was locally aggressive and frequently metastasizes to the lung. However, the etiology of osteosarcoma was unknown. Thus, exploring the mechanisms behind the occurrence of osteosarcoma was important for its prediction and prevention. To investigate the usefulness of mammalian Eps15 homology domain 1 (EHD1) as a prognostic marker for osteosarcoma, the expression of EHD1 in 57 osteosarcoma patients was measured using immunohistochemistry techniques and correlated with the clinicopathological features of patients.
Methods
Correlations of EHD1 expression levels with clinicopathological features of patients were assessed using the Pearson χ2 test for categorical variables and the Student t test for continuous variables. Cumulative disease-free survival (DFS) curves and overall survival (OS) curves were plotted using the Kaplan–Meier method, and the relationship between each of the variables and survival was assessed by log-rank tests using univariate analysis. Subsequently, the parameters were tested using the multivariate Cox proportional hazards model, which was used to identify independent variables for predicting survival. EHD1 expression [P = 0.020; HR, 5.582; 95% confidence intervals (CI), 1.314–23.72] was an independent prognostic indicator of DFS in osteosarcoma patients; tumor size and EHD1 expression of osteosarcomas were independent prognostic indicators of OS in osteosarcoma patients.
Results
EHD1 protein expression was a positive expression in examined tumor tissues. The median OS time of patients with high expression of EHD1 was 46.8 months (95% CI, 29.8–63.8 months), and the median OS time of patients with low expression of EHD1 was 58.8 months (95% CI, 31.6–86.0 months). The prognosis for patients with low expression of EHD1 in osteosarcomas was significantly better than that for patients with high expression of EHD1 (log-rank test, P = 0.019).
Conclusion
The expression of EHD1 was negatively correlated with DFS and OS of osteosarcoma patients; therefore, the expression of EHD1 is a prognostic marker for prediction and prevention of osteosarcomas.
Keywords: Osteosarcoma, Eps15 homology domain 1, Disease-free survival, Overall survival
Introduction
Osteosarcoma, a malignant tumor that frequently occurs in the distal femur, the proximal tibia, and the proximal humerus [1], is locally aggressive and frequently metastasizes to the lung [2]. A combination of surgery and chemotherapy results in the long-term survival of approximately 60–70% of osteosarcoma patients [3]. However, the etiology of osteosarcoma is unknown. Because of this unknown etiology, prevention and early diagnosis of osteosarcoma is difficult. Thus, exploring the mechanisms behind the occurrence of osteosarcoma is important for its prediction and prevention.
Mammalian Eps15 homology domain 1 (EHD1) is located in the chromosomal band 11q13 and is associated with lung, breast, head, neck, and small-cell lung cancer [4]. EHD1 plays an important role in the control of various cellular events by regulating various proteins [5, 6], including the β1 integrins [7]. The β1 integrins can bind to the extracellular matrix and stimulate the signaling pathways that influence the proliferation, apoptosis, cell spreading, migration, invasion, and metastasis of tumor cells [4]. In addition, high levels of EHD1 expression were associated with poor response to treatment in patients with cutaneous T cell lymphomas [8]. The studies were also reported that EHD1 was the best-studied member of the four highly homologous mammalian proteins (EHD1–4) which regulated the endocytic recycling of membrane and associated cell surface receptors [9, 10]. These studies suggest that EHD1 plays a role in cancer invasion and metastasis.
The present study aimed to investigate the usefulness of EHD1 as a prognostic marker for osteosarcoma. The expression of EHD1 in tumor and normal tissues collected from 57 osteosarcoma patients was measured using immunohistochemistry techniques, and correlations with the clinicopathological features of patients were sought.
Methods
Patients
To assess the prognostic capacity of EHD1 for osteosarcomas, formalin-fixed, paraffin-embedded (FFPE) SCLC tumor tissues were collected from 57 osteosarcoma patients who underwent surgery between January 2011 and September 2015. Inclusion criteria were patients recorded to have osteosarcoma by pathology reports and included cases from stage I to stage III. All cases, representing a spectrum of osteosarcomas, were retrieved from patients attending Harbin Medical University Cancer Hospital, Harbin, China. Exclusion criteria were having stage IV of disease, or a history of other cancers. For each patient, each tissue specimen type was resected during a single surgical procedure. Primary cancers were evaluated in accordance with the seventh edition of the American Joint Committee on Cancer (AJCC) staging system (TNM). All patients were followed up until their death or the study end date (January 21, 2014). The median follow-up time for survivors was 46.8 months (range 34.9–58.7 months). The study was approved by the Ethical Committee of Harbin Medical University. Informed consent was obtained from all patients. All investigators involved in the study, apart from the study statistician, were blinded to patient outcomes throughout all laboratory analyses.
Immunohistochemistry
The expression of EHD1 in FFPE sections was analyzed by immunohistochemistry. The tissue sections were first dried at 70 °C for 3 h. After deparaffinization and hydration, sections were washed in phosphate-buffered saline (PBS, 3 × 3 min). Endogenous peroxidase was quenched with 3% H2O2 for 15 min. After first being washed with distilled water, sections were washed in PBS (3 × 5 min). Antigen retrieval was performed in citrate buffer (pH 6.0). Each section was treated with 300–500 ml EHD1 rabbit polyclonal antibody solution (Abcam, Cambridge, UK, ab75886, diluted at 1:200) overnight at 4 °C. The sections were incubated with peroxidase-conjugated streptavidin for 30 min, and the reaction products were visualized with diaminobenzidine as a chromogen and counterstained with commercial hematoxylin. The percentage of positive cells was determined by counting 500 cells in five randomly selected fields per section. IHC staining was scored based on intensity as follows: 0 (no staining), 1 (weak staining = light yellow), 2 (moderate staining = yellow brown), and 3 (strong staining = brown). The percentage (0–100%) of the extent of reactivity was scored as follows: 0 (no positive tumor cells), 1 (fewer than 10% positive tumor cells), 2 (10–50% positive tumor cells), and 3 (greater than 50% positive tumor cells). Next, the cytoplasmic expression score was obtained by multiplying the intensity and reactivity rate values. Scores of < 4 were classified as low expression, and the remainders were classified as high expression. Two blinded, independent observers interpreted all slides.
Statistical analysis
All analyses were performed using statistical software (SPSS 19.0 for Windows; SPSS, Inc., Chicago, IL, USA). Differences were considered statistically significant when P < 0.05. Correlation of EHD1 expression levels with clinicopathological features of patients was assessed using the Pearson χ2 test for categorical variables and the Student t test for continuous variables. Disease-free survival (DFS) was calculated from the date of surgery resection to the date of last follow-up or relapse. Cumulative DFS curves and overall survival (OS) curves were plotted using the Kaplan–Meier method, and the relationship between each of the variables and survival was assessed by a log-rank test using univariate analysis. Covariates with P ≤ 0.15 in the univariate analyses were included in multivariate analyses. The parameters were then tested using the multivariate Cox proportional hazards model, which was performed to identify independent variables for predicting survival. Risk ratios and their 95% confidence intervals (CIs) were recorded for each factor.
Results
EHD1 protein expression in tumor tissues
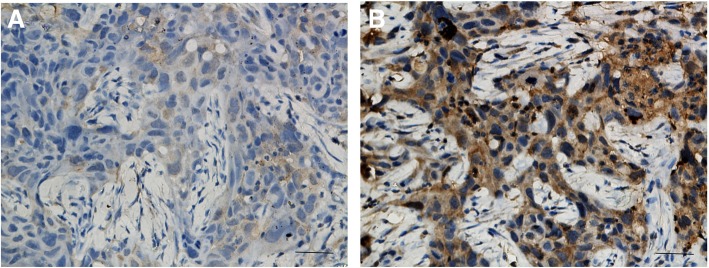
EHD1 protein expression was a positive expression in examined tumor tissues (Fig. 1). The cytoplasmic staining patterns observed for EHD1 were consistent with data from our previous studies.
Fig. 1.
Immunohistochemical staining of EHD1 in FFPE tissue samples (× 40). a EHD1 low expression. b EHD1 high expression
Association of EHD1 protein expression and clinicopathological features
Correlation of EHD1 expression with the clinicopathological features of patients, including age, gender, tumor histology, tumor size, and AJCC stage in SCLC patients was assessed (Table 1). High expression of EHD1 in tumor tissues was positively correlated only with the disease type (P = 0.025). No such significant correlations between EHD1 and other clinicopathological features were found in this study.
Table 1.
The correlation between clinicopathological features and EHD1 expression
| EHD1 expression | |||
|---|---|---|---|
| Low (n = 28) | High (n = 29) | P | |
| Gender | 0.707 | ||
| Male | 17 (60.7%) | 19 (65.5%) | |
| Female | 11 (39.3%) | 10 (34.5%) | |
| Age | 23.0 ± 16.0 | 24.5 ± 14.8 | 0.705 |
| Disease type | 0.025* | ||
| Chondroblastomas | 8 (28.6%) | 1 (3.4%) | |
| Other | 20 (71.4%) | 28 (96.6%) | |
| Tumor size | 0.872 | ||
| ≥ 5 cm | 17 (60.7%) | 17 (58.6%) | |
| < 5 cm | 11 (39.3%) | 12 (41.4%) | |
| Stage | 0.206 | ||
| I | 0 (0%) | 2 (6.9%) | |
| II | 27 (96.4%) | 24 (82.8%) | |
| III | 1 (3.6%) | 3 (10.3%) | |
[0 (negative) ≤ score ≤ 1+] and [2+ ≤ score ≤ 3+] represent low negative and strong positive staining of EHD1, respectively. * P < 0.05
Univariate and multivariate Cox regression analysis of potential prognostic indicators of DFS in osteosarcoma patients
No significant correlations between clinicopathological features and DFS, based on univariate Cox regression models, were found in this study. Multivariate Cox proportional hazards model analysis of the same set of patient data showed that EHD1 expression (P = 0.020; HR, 5.582; 95% CI, 1.314–23.72) was independent of prognostic indicators for DFS in osteosarcoma patients (Table 2).
Table 2.
EHD1 expression in tumor tissues as an independent prognostic factor for DFS in osteosarcoma patients
| Univariatea | Multivariateb | |||
|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | |
| Gender (male vs female) | 1.363 (0.505–3.676) | 0.541 | 1.581 (0.549–4.552) | 0.396 |
| Age | 0.974 (0.939–1.01) | 0.157 | 0.985 (0.912–1.065) | 0.704 |
| Disease type (chondroblastomas vs other) | 1.468 (0.421–5.122) | 0.547 | 4.336 (0.76–24.738) | 0.099 |
| Tumor size (≥ 5 cm vs < 5 cm) | 1.306 (0.504–3.383) | 0.582 | 1.881 (0.616–5.748) | 0.267 |
| EHD1 expression (high vs low) | 2.771 (0.981–7.824) | 0.054 | 5.582 (1.314–23.72) | 0.020 |
CI confidence interval, HR hazard ratio, DFS disease-free survival
aVariables were adopted for their prognostic significance (P < 0.05) in univariate analysis using forward, stepwise selection (forward likelihood ratio)
bA Cox proportional hazards regression model was used for multivariate analysis
Univariate and multivariate Cox regression analysis of potential prognostic indicators of OS in osteosarcoma patients
Univariate Cox regression model analysis showed that age (P = 0.015; HR, 0.945; 95% CI, 0.903–0.989), tumor size (P = 0.042; HR, 3.155; 95% CI, 1.045–9.525), and EHD1 expression (P = 0.026; HR, 3.202; 95% CI, 1.149–8.92) were independent prognostic indicators of OS in osteosarcoma patients (Table 3). Analysis of the same set of patient data using a multivariate Cox proportional hazards model showed that tumor size (P = 0.008; HR, 5.854; 95% CI, 1.587–21.597) and EHD1 expression (P = 0.007; HR, 6.372; 95% CI, 1.645–24.676) were independent prognostic indicators of OS in osteosarcoma patients (Table 3).
Table 3.
EHD1 expression in tumor tissues as an independent prognostic factor for OS in osteosarcoma patients
| Univariatea | Multivariateb | |||
|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | |
| Gender (male vs female) | 0.927 (0.368–2.335) | 0.872 | 1.323 (0.468–3.743) | 0.597 |
| Age | 0.945 (0.903–0.989) | 0.015 | 0.988 (0.887–1.101) | 0.833 |
| Disease type (chondroblastomas vs other) | 0.876 (0.201–3.829) | 0.861 | 2.147 (0.346–13.314) | 0.412 |
| Tumor size (≥ 5 cm vs < 5 cm) | 3.155 (1.045–9.525) | 0.042 | 5.854 (1.587–21.597) | 0.008 |
| EHD1 expression (high vs low) | 3.202 (1.149–8.92) | 0.026 | 6.372 (1.645–24.676) | 0.007 |
CI confidence interval, HR hazard ratio, DFS disease-free survival
aVariables were adopted for their prognostic significance (P < 0.05) in univariate analysis using forward, stepwise selection (forward likelihood ratio)
bA Cox proportional hazards regression model was used for multivariate analysis
Kaplan–Meier survival curve analysis of DFS in osteosarcoma patients
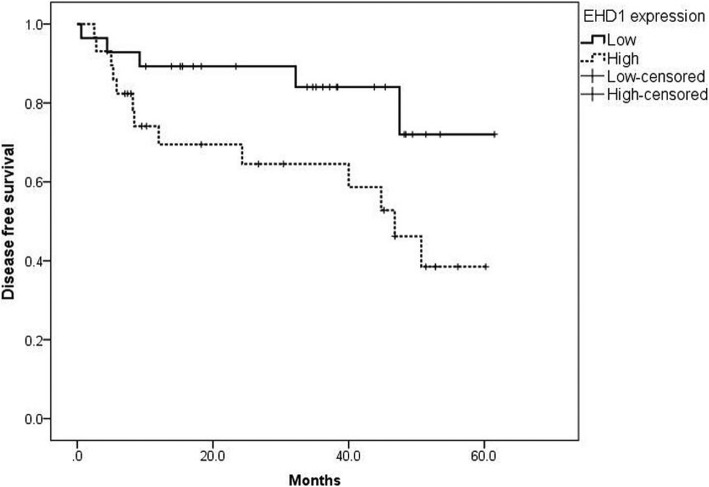
Kaplan–Meier survival curves stratified for EHD1 expression are shown in Fig. 2. The median DFS time in EHD1 high-expression patients was 46.8 months (95% CI, 34.9–58.7 months). The prognosis for patients who had low expression of EHD1 in osteosarcomas was significantly better (log-rank test, P = 0.045) compared with patients who had high expression of EHD1.
Fig. 2.
Kaplan–Meier analysis for disease-free survival (DFS) based on EHD1 expression status in osteosarcoma patients
Kaplan–Meier survival curve analysis of OS in osteosarcoma patients
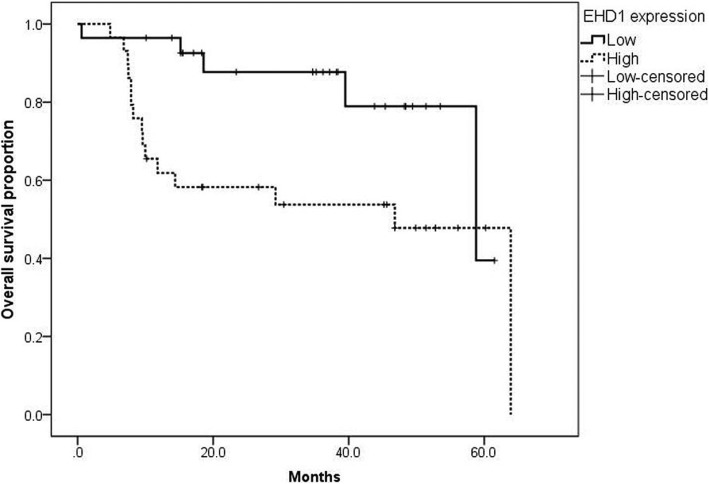
Kaplan–Meier survival curves stratified for EHD1 expression are shown in Fig. 3. The median OS time for patients with high expression of EHD1 was 46.8 months (95% CI, 29.8–63.8 months), and the median OS time for patients with low expression of EHD1 was 58.8 months (95% CI, 31.6–86.0 months). The prognosis for patients with low expression of EHD1 in osteosarcomas was significantly better (log-rank test, P = 0.019) compared with patients with high expression of EHD1.
Fig. 3.
Kaplan–Meier analysis for overall survival (OS) based on EHD1 expression status in osteosarcoma patients
Discussion
Our results revealed that EHD1 expression was an independent prognostic indicator for osteosarcomas, which was similar to results found in previous studies reporting the prognostic value of EHD1 expression in both non-small-cell and small-cell lung cancer [4]. High levels of EHD1 expression have also been associated with poor response to treatment in patients with cutaneous T cell lymphomas [8]. EHD1 is the best-studied member of the four highly homologous mammalian proteins (EHD1–4) which regulate endocytic recycling of membrane and associated cell surface receptors [9, 10]. EHD proteins appear to be involved in critical nodes in the endocytic sorting/recycling process [4]. In other reports, the reports suggested that EHD1 participated in regulating the rate of recycling endocytic compartments traveling to the cell surface via trafficking as part of the slow recycling pathway [11, 12], and the decreased expression of EHD1 enhanced the metastatic ability of well-differentiated pancreatic endocrine neoplasms [13, 14]. EHD1 can be found in exosome secreted by prostate cancer cells [15, 16]. Increased EHD1 expression in lesions associated with cutaneous T cell lymphomas indicated a poor response to treatment [8, 17]. Considering all of these observations, the expression of EHD1 could be considered as a prognostic marker for osteosarcoma prediction and prevention.
In addition, tumor size and EHD1 expression were found to be independent prognostic indicators for osteosarcomas. The median DFS time in patients who showed high expression of EHD1 was 46.8 months, and the prognosis for patients with low expression of EHD1 in osteosarcomas was significantly better than those with high expression of EHD1. The median OS time in patients with high expression of EHD1 was 46.8 months, and the median OS time in patients with low expression of EHD1 was 58.8 months. The prognosis for patients with low expression of EHD1 in osteosarcomas was significantly better than those with high expression of EHD1. A previous study showed that EHD1 expression is associated with a poor response to chemotherapy in cutaneous T cell lymphoma patients, which is consistent with our results [4]. The expression of EHD1 was negatively correlated with DFS and OS in osteosarcoma patients, and thus, the expression of EHD1 could be considered as a prognostic marker for osteosarcoma prediction and prevention. Notwithstanding this study’s limitation, in that it had such a small sample size, the identification of the prognostic potential of EHD1 expression warrants further study in larger cohorts to validate the effects found in osteosarcoma patients.
Conclusion
EHD1 expression was found to be an independent prognostic indicator of OS in osteosarcoma patients. The prognosis for patients with low expression of EHD1 in osteosarcomas was significantly better than patients with high expression of EHD1. The expression of EHD1 was negatively correlated with DFS and OS of osteosarcoma patients, and thus, the expression of EHD1 could be considered as a prognostic marker for the prediction and prevention of osteosarcoma.
Acknowledgements
none
Funding
This study was funded by Science and Technology Research Project of Education Department of Heilongjiang Province (Grant number: 12521328); Scientific Research Project of Heilongjiang Administration of Traditional Chinese Medicine (Grant number: ZHY12-W033); and the Haiyan Foundation of Harbin Medical University Cancer Hospital (Grant number: JJ2008-12).
Availability of data and materials
The raw data were collected and analyzed by the Authors, and are not ready to share their data because the data have not been published.
Authors’ contributions
HWY, GFQ, and YXW participated in the design of this study, and they all performed the statistical analysis. WM and JJB carried out the study and collected important background information. CYS and MY drafted the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Hongwei Yu, Email: hydyhw@126.com.
Guofan Qu, Email: guofanqu@126.com.
Yuxue Wang, Email: wangyuxue888@sina.com.
Wei Mai, Email: bakui210@126.com.
Jun Jie Bao, Email: 13936447803@139.com.
Chunyu Song, Email: 575790473@qq.com.
Meng Yao, Phone: +86-0451-86662961, Email: yaomenghrb@163.com.
References
- 1.Ottaviani G, Jaffe N. The epidemiology of osteosarcoma. Cancer Treat Res. 2009;152:3–13. doi: 10.1007/978-1-4419-0284-9_1. [DOI] [PubMed] [Google Scholar]
- 2.Briccoli A, Rocca M, Salone M, Guzzardella GA, Balladelli A, Bacci G. High grade osteosarcoma of the extremities metastatic to the lung: long-term results in 323 patients treated combining surgery and chemotherapy, 1985-2005. Surg Oncol. 2010;19(4):193–199. doi: 10.1016/j.suronc.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 3.Yamamoto N, Tsuchiya H. Chemotherapy for osteosarcoma - where does it come from? What is it? Where is it going? Expert Opin Pharmacother. 2013;14(16):2183–2193. doi: 10.1517/14656566.2013.827171. [DOI] [PubMed] [Google Scholar]
- 4.Reinecke J, Caplan S. Endocytosis and the Src family of non-receptor tyrosine kinases. Biomol Concepts. 2014;5(2):143–155. doi: 10.1515/bmc-2014-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sato M, Sato K, Liou W, Pant S, Harada A, Grant BD. Regulation of endocytic recycling by C. elegans Rab35 and its regulator RME-4, a coated-pit protein. EMBO J. 2014;27(8):1183–1196. doi: 10.1038/emboj.2008.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Naslavsky N, Caplan S. EHD proteins: key conductors of endocytic transport. Trends Cell Biol. 2011;21(2):122–131. doi: 10.1016/j.tcb.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jović M, Naslavsky N, Rapaport D, Horowitz M, Caplan S. EHD1 regulates β1 integrin endosomal transport: effects on focal adhesions, cell spreading and migration. J Cell Sci. 2007;120:802–814. doi: 10.1242/jcs.03383. [DOI] [PubMed] [Google Scholar]
- 8.Shin J, Monti S, Aires DJ, Duvic M, Golub T, Jones DA, et al. Lesional gene expression profiling in cutaneous T-cell lymphoma reveals natural clusters associated with disease outcome. Blood. 2007;110(8):3015. doi: 10.1182/blood-2006-12-061507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Naslavsky N, Caplan S. C-terminal EH-domain-containing proteins: consensus for a role in endocytic trafficking, EH? J Cell Sci. 2005;118(Pt 18):4093–4101. doi: 10.1242/jcs.02595. [DOI] [PubMed] [Google Scholar]
- 10.Caplan S, Naslavsky N, Hartnell LM, Lodge R, Polishchuk RS, Donaldson JG, et al. A tubular EHD1-containing compartment involved in the recycling of major histocompatibility complex class I molecules to the plasma membrane. EMBO J. 2014;21(11):2557–2567. doi: 10.1093/emboj/21.11.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng X, Xu M, Fang Q. Role of AMPKα in skeletal muscle glycometabolism regulation and adaptation in relation to sepsis. Biomed Res Int. 2014;2014(3):390760. doi: 10.1155/2014/390760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rainey MA, George M, Ying G, Akakura R, Burgess DJ, Siefker E, et al. The endocytic recycling regulator EHD1 is essential for spermatogenesis and male fertility in mice. BMC Dev Biol. 2010;10(1):37. doi: 10.1186/1471-213X-10-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fincham VJ, Frame MC. The catalytic activity of Src is dispensable for translocation to focal adhesions but controls the turnover of these structures during cell motility. EMBO J. 2014;17(1):81–92. doi: 10.1093/emboj/17.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hansel DE, Rahman A, House M, Ashfaq R, Berg K, Yeo CJ, et al. Met proto-oncogene and insulin-like growth factor binding protein 3 overexpression correlates with metastatic ability in well-differentiated pancreatic endocrine neoplasms. Clin Cancer Res. 2004;10(1):6152–6158. doi: 10.1158/1078-0432.CCR-04-0285. [DOI] [PubMed] [Google Scholar]
- 15.Wheeler DL, Iida M, Dunn EF. The role of Src in solid tumors. Oncologist. 2009;14(7):667. doi: 10.1634/theoncologist.2009-0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jansen FH, Krijgsveld J, Van RA, Gj VDB, Ms VDB, van Weerden WM, et al. Exosomal secretion of cytoplasmic prostate cancer xenograft-derived proteins. Mol Cell Proteomics Mcp. 2009;8(6):1192. doi: 10.1074/mcp.M800443-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heidemann F, Schildt A, Schmid K, Bruns OT, Riecken K, Jung C, et al. Selectins mediate small cell lung cancer systemic metastasis. PLoS One. 2014;9(4):e92327. doi: 10.1371/journal.pone.0092327. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw data were collected and analyzed by the Authors, and are not ready to share their data because the data have not been published.