Abstract
Most organisms experience variable and sometimes suboptimal environments in their lifetime. While stressful environmental conditions are normally viewed as a strong selective force, they can also impact directly on the genetic basis of traits such as through environment-dependent gene action. Here, we used the Drosophila melanogaster Genetic Reference Panel to investigate the impact of developmental temperature on variance components and evolutionary potential of cold tolerance. We reared 166 lines at five temperatures and assessed cold tolerance of adult male flies from each line and environment. We show (1) that the expression of genetic variation for cold tolerance is highly dependent on developmental temperature, (2) that the genetic correlation of cold tolerance between environments decreases as developmental temperatures become more distinct, (3) that the correlation between cold tolerance at individual developmental temperatures and plasticity for cold tolerance differs across developmental temperatures, and even switches sign across the thermal developmental gradient, and (4) that evolvability decrease with increasing developmental temperatures. Our results show that the quantitative genetic basis of low temperature tolerance is environment specific. This conclusion is important for the understanding of evolution in variable thermal environments and for designing experiments aimed at pinpointing candidate genes and performing functional analyses of thermal resistance.
Subject terms: Evolutionary genetics, Evolutionary biology, Quantitative trait
Introduction
Stressful environmental conditions not only act as a strong selective force, but also directly affect the genetic architecture of traits in several ways. Firstly, stressful conditions can influence the nature of environmental variation, VE (Hoffmann and Merilä 1999; Ørsted et al. 2018a). Secondly, these conditions can directly affect expression of additive genetic variation, VA (e.g., Czesak et al. 2006; Wilson et al. 2006). Both changes in VE and VA will influence heritability estimates (Gebhardt-Henrich and van Noordwijk 1991; Charmantier and Garant 2005; Wilson et al. 2006; Visscher et al. 2008; Dingemanse et al. 2009; Kristensen et al. 2015). Thirdly, environment-dependent gene action may exist, where the function of genes or gene networks, rather than overall genetic variation, change under stressful conditions (Telonis-Scott et al. 2009). This can lead to a situation where genes influencing a trait in one environment may not have much impact on that trait in a different environment (Sgrò and Hoffmann 2004), which in turn will alter genetic correlations between environments (de Jong 1990; Stearns et al. 1991; Vieira et al. 2000; Stinchcombe et al. 2010). The impact of these environmental effects on genetic variation can be important for evolutionary trajectories (Hoffmann and Merilä 1999; Hercus and Hoffmann 2000), but these mechanisms are rarely studied, as they are typically regarded as a nuisance in quantitative genetic models (but see Wilson et al. 2006; Husby et al. 2011; Wood and Brodie 2016).
Environmental conditions are likely to have particularly large effects on the genetic architecture of ecological traits critical for responding to variable environmental conditions. For this class of traits, the environment can influence VA, VE and gene action (Sgrò and Hoffmann 2004; Kristensen et al. 2015), affecting the extent to which organisms respond to selection and hence persist in the long run. Tolerance to stressful temperatures in ectotherms, especially in Drosophila, provides examples of traits whose genetic architecture is strongly affected by environmental variation, such as the impact of developmental temperature on the resistance to cold extremes (Schou et al. 2017; Ørsted et al. 2017).
The ways in which the environment influences trait expression across genotypes is often characterised through genotype-by-environment (GxE) interactions, where the direction and magnitude of GxE can depend on genotype (Rohde et al. 2017; Ørsted et al. 2018a) and/or as genetic correlations of a trait measured in several environments (Vieira et al. 2000; Sgrò and Hoffmann 2004; Agrawal and Stinchcombe 2009). Genotypic responses to environmental variation are usually expressed as differences in phenotypic plasticity, which is commonly quantified with linear reaction norms, i.e., the slope of trait values under continuous variation in an environmental variable (Valladares et al. 2006). However, while environmental effects on gene action have typically been investigated by comparing two environments (Telonis-Scott et al. 2009; Ellers and Driessen 2011; Levine et al. 2011; Ketola et al. 2012; Gerken et al. 2015), patterns across environmental gradients are not well established. If the slopes of different genotypes along a gradient vary, there will be GxE interactions and genetic correlations will be less than one. Under a continuous environmental gradient, a polygenic trait might be expected to show declining genetic correlations (increasing GxE interactions) as environments become more dissimilar, assuming that there is an increase in variance due to environment-specific gene expression (Via and Lande 1985; Sgrò and Blows 2004; Spichtig and Kawecki 2004). This could lead to a linear or non-linear decrease in genetic correlations with the environmental variable. Surprisingly, this rather simple prediction has rarely been studied experimentally. Knowledge of the effects of environmental similarity on genetic correlations of stress resistance traits can prove valuable when trying to predict adaptive responses in heterogeneous environments (Sheldon et al. 2003; Sgrò and Blows 2004), such as those arising from anthropogenic climate change (Etterson and Shaw 2001; Agrawal and Stinchcombe 2009).
Environment-dependent gene action also raises the question of how plasticity is linked to stress tolerance in a particular environment. Phenotypic plasticity can be costly, because it requires energy and flexibility of the organism on a number of biological levels (Auld et al. 2010; Tonsor et al. 2013; Murren et al. 2015), and might become maladaptive if environments change rapidly and in unpredictable ways (Kristensen et al. 2008). Several studies have found an inverse association between stress resistance in one environment (here defined as “basal” tolerance) and the capacity for plasticity (Stillman 2003; Nyamukondiwa et al. 2011; Gerken et al. 2015; Comte and Olden 2017; Noh et al. 2017), suggesting a trade-off between the ability to cope with current conditions and performance in an altered environment in the future (Murren et al. 2015). However, other studies have not found evidence for such trade-offs (Kellett et al. 2005; Calosi et al. 2008; Franke et al. 2012; Gunderson et al. 2015; Ørsted et al. 2018a). Thus, it remains unclear if plasticity constrains basal thermal tolerance and vice versa. Adding to the ambiguity, some studies have found that trade-offs are species-specific even across related taxa (Nyamukondiwa et al. 2011; Strachan et al. 2011; Comte and Olden 2017), while others have found seasonal variation in trade-offs (Noh et al. 2017), highlighting the influence of environmental conditions on these patterns.
Here, we investigate the effect of developmental temperature on the genetic architecture of cold tolerance in Drosophila melanogaster. The aims were to explore the environmental dependency of quantitative genetic parameters of cold tolerance, and potential trade-offs between cold tolerance in each environment and plasticity in this trait. We used the Drosophila melanogaster Genetic Reference Panel (DGRP; Mackay et al. 2012; Huang et al. 2014), which was established from a natural population of D. melanogaster from Raleigh, USA. The DGRP consists of a collection of lines that has been inbred to an expected inbreeding coefficient of F ~1, resulting in extremely high homozygosity throughout the genome within each line. Thus, by assessing cold tolerance of the same DGRP genotype reared in different thermal environments, knowledge about the genetic aspects of cold tolerance, and the association between cold tolerance at individual temperatures and plasticity, can be obtained. Low temperatures limit the geographical distribution of many species more than warm temperatures (Sunday et al. 2011; Williams et al. 2014), and cold tolerance is therefore a good predictor of present and future distributions of species (Kimura 2004; Overgaard et al. 2011; Araújo et al. 2013). The temperature regimes employed in this study were well within the range of what the cosmopolitan D. melanogaster will experience in its natural habitats (Hoffmann et al. 2003), thus providing ecological relevance to our experimental setup. However, it can be difficult to ascertain which of the thermal environments are optimal and which imposes a level of stress, as this is highly dependent on the biological context and traits investigated (David et al. 1997; Hoffmann 2010).
We reared 166 DGRP lines at five different developmental temperatures and quantified cold tolerance in adults from each developmental temperature. We performed quantitative genetic analyses to estimate genetic and environmental variance components, heritabilities, evolvabilities and phenotypic/genetic correlations of cold tolerance in each developmental environment and for (slope-based) plasticity. This allowed three main questions to be answered. (1) To what extent is cold tolerance phenotypically and genetically correlated between developmental environments, and are these correlations dependent on environmental similarity? (2) Do the developmental temperatures impact on variance components, heritability and evolvability of cold tolerance within developmental temperatures, and on plastic cold tolerance? (3) Is there a trade-off between cold tolerance at individual developmental temperatures and the plasticity of cold tolerance, and is this trade-off dependent on the environment?
Materials and methods
Experimental setup
This study included 166 lines from the D. melanogaster Genetic Reference Panel (DGRP) obtained from Bloomington Drosophila Stock Centre (NIH P40OD018537), maintained at 23 °C, 50% relative humidity and a 12:12 h photoperiod on a standard oatmeal–sugar–agar–yeast medium for two generations before starting the experiment. The experimental procedure follows the setup previously described in Ørsted et al. (2018a). In summary, we reared each DGRP line at five different temperatures: 17, 20, 23, 26 and 29 °C, from eggs to adults in vials containing 7 mL standard medium (for details on medium composition see Ørsted et al. 2018b). Eggs were laid by approximately 20 adult flies in three 12 h periods. Vials were checked daily and eclosed flies were sexed under CO2 anaesthesia. Males were transferred to vials containing fresh medium, and kept at the respective developmental temperature for 48 h. At age 60 ± 12 h, the cold tolerance of around 10 males (for exact numbers see Table S1) per DGRP line per temperature were assessed using the dynamic measure Critical Thermal Minimum (CTmin; Overgaard et al. 2012). In this standardised procedure, individual flies were placed in glass vials in a water bath pre-set to 23 °C. Water in the bath was gradually cooled at a rate of 0.1 °C/min. The temperature at which a fly enters chill coma, i.e., when all movement ceases, was recorded and defines CTmin. In total, 7690 flies were scored for CTmin by the same person to reduce potential scoring bias. We chose males because virginity is not likely to impact on males to the same extent as it does to females (Goenaga et al. 2012), and we were not able to assure that all flies were virgins when assessed.
Phenotypic plasticity of cold tolerance was estimated as the regression coefficient of a linear regression, i.e., slope of CTmin on rearing temperature. Linear regression analysis provided the best fit compared to polynomial regression, as assessed by the coefficient of determination (results not shown). In addition, linear norms-of-reaction are the most commonly used in studies of phenotypic plasticity (Valladares et al. 2006), and especially for the relationship between CTmin and developmental temperature, which is typically linear in Drosophila (Schou et al. 2017; Ørsted et al. 2018a).
Quantitative genetic analyses
Genetic analyses was performed with the average information restricted maximum-likelihood (AI-REML) procedure (Madsen et al. 1994) utilising the R package QGG (http://psoerensen.github.io/qgg/) that links to DMU (http://dmu.agrsci.dk/DMU/). We estimated variance components, genetic correlations and broad and narrow sense heritabilities (H2 and h2, respectively) based on multi-trait linear mixed models. For CTmin we fitted a five-trait multi-trait model including all developmental temperatures, and to estimate genetic correlations between CTmin at individual temperatures and plasticity we fitted a bivariate model. The multi-trait model including i-traits was generalised as:
where, yi was a vector of phenotypic observations (i.e., either temperature specific CTmin or plasticity), Xi and Zi are design matrices linking fixed and random genetic effects to the phenotypes. The bi is a vector of fixed effects (experimental day, water bath, time on day (for CTmin), Wolbachia infection status, and five major chromosomal inversions (only when estimating the additive genetic variance), and ei is a vector of random residuals (). Estimates of the genetic variance components (and therefore H2) were obtained by assuming the DGRP lines to be independent, which we modelled by an identity block matrix (IL) as (co)variance structure of the genetic values, . To estimate the additive genetic variance components (including h2) we estimated the realised relationship among DGRP lines from publicly available single nucleotide polymorphisms (SNPs). Thus h2 was the proportion of phenotypic variance captured by common segregating SNPs within the DGRP. We used standard filtering for the DGRP, i.e., only considering biallelic SNPs with minor allele frequencies ≥0.05, and for which the Phred scaled variant quality was greater than 500, and the genotype call ≥0.8 (Mackay et al. 2012; Huang et al. 2014). This yielded 1,725,755 genetic markers distributed on the six chromosome arms (2L, 2R, 3L, 3R, 4 and X). The notation is similar as above, however, here we included five major polymorphic inversions present in the DGRP system; I2Lt, I2Rns, I3Rp, I3Rk and I3RMo), gi was a vector of random genetic effects, , where G was the additive genomic relationship matrix, and ei was a random residual, . The G matrix was computed as G = WW′/m (VanRaden 2008), where m was the number of genetic markers, and W was a centred and scaled genotype matrix. Each column vector of W, , pi was the frequency of the minor allele at locus i, and ai was the ith column vector of the allele count matrix, A, with the genotypes encoded as 0, 2, referring to the number of the minor allele. The SNP data, including karyotype inversions and Wolbachia infection status, can be obtained from the DGRP2 website (http://dgrp2.gnets.ncsu.edu). To achieve a replicated structure of the plasticity measures, which was needed to obtain accurate estimates of the genetic parameters, we grouped the CTmin data within each DGRP line into three groups of individuals according to day of assay, time of day, and water bath. Based on these replicate groups, we obtained three replicates measures of slope per DGRP line. The slope estimates were robust against re-sampling of random individuals into groups of the same size (for details see Ørsted et al. 2018a).
Phenotypic correlations were computed as Pearson’s correlation using within DGRP line means, and standard errors were estimated as: , where n was the sample size. We also computed correlations between the rank order of the DGRP lines (rr), in which lines are ranked within rearing regimes from 1 to 166.
All variance components were tested under the null hypothesis of being zero using Wald’s test. In addition, we tested if correlations differed from zero, i.e., the estimate deviated more than 1.645 × SE from 0 (p < 0.05). We performed pairwise comparisons of genetic correlations, rg among CTmin in the five rearing temperatures (e.g., rg (17, 20 °C) vs. rg (17, 23 °C)), as well as between correlations between CTmin in each environment and plasticity across environments. These comparisons were based on Fisher’s z transformation. Evolvability (IA) was computed at each developmental temperature as: , where is the mean CTmin across all DGRP lines in a given environment (Houle 1992; Hoffmann et al. 2016). All analyses were done within the R programming environment (R Core Team 2017).
Results
Heritable variation for cold tolerance at the individual rearing temperatures and plasticity
We observed a significant increase in line mean CTmin, i.e., a decrease in cold tolerance, with increasing rearing temperature (Fig. 1, Table 1, F4,804 = 3956; p < 0.001). Average CTmin (±SE) across DGRP lines spanned from 2.81 ± 0.03 °C for flies reared at 17 to 7.80 ± 0.04 °C for flies reared at 29 °C, with considerable variation among DGRP lines within rearing environment (Fig. 2, Table 1 and Table S1). The estimates of H2 and h2 for CTmin were consistent across the developmental temperatures, with estimates in the range of 0.41–0.48 and 0.27–0.34 for H2 and h2, respectively (Table 2). Plasticity of cold tolerance was estimated as the slope of a linear regression of CTmin as a function of developmental temperature. The slope was significantly different from zero in all DGRP lines (t(3–21) > 2.13; p < 0.001). A significant interaction between line and developmental temperature (two-way ANOVA; F(165,477) = 2.22, p < 0.001) indicated a GxE interaction, reflecting differences among DGRP lines in slope ranging from 0.32 to 0.57 °C in CTmin per 1 °C increase in developmental temperature. The heritability estimates for plasticity of CTmin were 0.65 and 0.51 for H2 and h2, respectively. We observed a higher total genetic variance , additive genetic variance and environmental variance of CTmin at 29 °C than at 26 °C (Table 2). At 17, 20 and 23 °C, we observed a decrease in all variance components as compared to 29 °C (Table 2). In contrast, for evolvability (IA; mean corrected ) of CTmin, we observed higher values at lower temperatures, i.e., from 0.67 at 17 to 0.18 at 29 °C (Table 2).
Fig. 1.
Linear regressions of critical thermal minimum (CTmin) across the five developmental temperatures. Each grey line represents one DGRP line (n = 166). The two full black lines represent the DGRP line with the steepest slope and the DGRP line with the flattest slope, with the difference in CTmin from developing at 17–29 °C indicated as ΔCTmin. The two dashed black lines represent the DGRP lines with the highest and lowest average CTmin across the temperatures indicated as . These latter two DGRP lines represent a genotype that performs poorly across environments and a genotype that performs well across environments
Table 1.
Mean ± SE, minimum, and maximum CTmin at the five developmental temperatures (17, 20, 23, 26 and 29 °C) and slope of CTmin across the thermal environments for all DGRP lines (n = 166)
| CTmin 17 °C | CTmin 20 °C | CTmin 23 °C | CTmin 26 °C | CTmin 29 °C | Slope | |
|---|---|---|---|---|---|---|
| Mean ± SE (°C) | 2.81 ± 0.03 | 4.09 ± 0.03 | 5.39 ± 0.03 | 6.92 ± 0.03 | 7.80 ± 0.04 | 0.431 ± 0.003 |
| Min. (°C) | 1.72 | 3.45 | 4.70 | 6.04 | 6.74 | 0.316 |
| Max. (°C) | 4.34 | 5.72 | 6.80 | 8.85 | 9.50 | 0.560 |
Fig. 2.
Correlations of CTmin across developmental thermal environments. Diagonal elements show the DGRP line mean CTmin ( ± SE) sorted by increasing values within rearing regime. Plots above the diagonal show the phenotypic correlations, rp (SE in parenthesis) along with the printed genetic correlations, rg (SE in parenthesis) between CTmin for pairs of environments. Plots below the diagonal show the correlations between the rank order of DGRP lines, rr (SE in parenthesis). In these plots, the DGRP lines are ranked within rearing regimes from 1 to 166. All correlations are Pearson’s product moment correlations. In all plots above and below the diagonal, the grey dots represent one DGRP line. Black lines are the regression lines visualising the correlations. All correlations are significantly different from zero, i.e., the estimate deviate more than 1.645 × SE from 0 (p < 0.05). In the lower right corners of the above diagonal plots, the genetic correlations, rg are compared pairwise within rows, and significantly different correlations are denoted with separate letters, e.g., rg (17, 20 °C) vs. rg (17, 23 °C). In the below diagonal plots, the letters represent significant differences in pairwise comparisons of rank correlations, rr within columns. Some correlations have more letters as they are similar to some but different from other correlations. Comparisons of these correlations are based on Fisher’s z transformation (n = 166)
Table 2.
Estimated variance components (genetic variance,, environmental variance, , and additive genetic variance, ) and broad and narrow sense heritability estimates (H2 and h2, respectively) ± standard errors (SE), as well as evolvability (IA = /(mean CTmin)2 × 100) for CTmin at the five developmental temperatures and for the slope of CTmin across temperatures
| Trait | H 2 | h 2 | I A | |||
|---|---|---|---|---|---|---|
| CTmin 17 °C | 0.1005 ± 0.00101 | 0.1437 ± 0.00042 | 0.0528 ± 0.00055 | 0.41 ± 0.005 | 0.27 ± 0.004 | 0.669 |
| CTmin 20 °C | 0.0973 ± 0.00094 | 0.1078 ± 0.00031 | 0.0505 ± 0.00051 | 0.47 ± 0.005 | 0.32 ± 0.004 | 0.302 |
| CTmin 23 °C | 0.0929 ± 0.00091 | 0.1131 ± 0.00033 | 0.0503 ± 0.00051 | 0.45 ± 0.005 | 0.31 ± 0.004 | 0.173 |
| CTmin 26 °C | 0.1485 ± 0.00142 | 0.1581 ± 0.00046 | 0.0821 ± 0.00082 | 0.48 ± 0.005 | 0.34 ± 0.004 | 0.172 |
| CTmin 29 °C | 0.1943 ± 0.00198 | 0.2703 ± 0.00083 | 0.1116 ± 0.00118 | 0.42 ± 0.006 | 0.29 ± 0.005 | 0.184 |
| Slope | 0.0017 ± 0.00001 | 0.0009 ± 0.000005 | 0.0009 ± 0.00001 | 0.65 ± 0.036 | 0.51 ± 0.041 | 0.519 |
All variance components were significantly different from zero (Wald’s test).
Correlations between CTmin from different developmental temperatures increased with environmental similarity
We found strong positive genetic correlations (rg) between CTmin for DGRP lines at the different rearing temperatures (Fig. 2). The rg values were inversely proportional to the difference between developmental temperatures, i.e., correlations between similar developmental temperatures were larger than between dissimilar developmental temperatures. This pattern was confirmed by statistical pairwise comparisons of rg. The phenotypic correlations (rp) followed the same pattern, decreasing with increasing environmental dissimilarity (Fig. 2). Similarly, the rank order of the DGRP lines was more positively correlated across thermal environments that were more similar to each other (Fig. 2).
Trade-offs between CTmin at individual developmental temperatures and plastic cold tolerance depended on the environment
The phenotypic (rp) correlations between CTmin at the individual temperatures and slope of CTmin across temperatures depended on the environment. At 17 °C, the correlation was significantly negative (Fig. 3), i.e., in this environment genotypes with a high cold tolerance (low CTmin) tended to have a high plasticity of cold tolerance. However, we did not find this association at 20 °C. At higher temperatures, correlations were significantly positive (Fig. 3), thus genotypes with a high cold tolerance (low CTmin) tended to have low plasticity of cold tolerance, suggesting a trade-off between inherent cold tolerance at these developmental temperatures and plasticity. The genetic correlations (rg) followed the same pattern, particularly when involving CTmin scores following development at 17, 26 and 29 °C (Fig. 3).
Fig. 3.
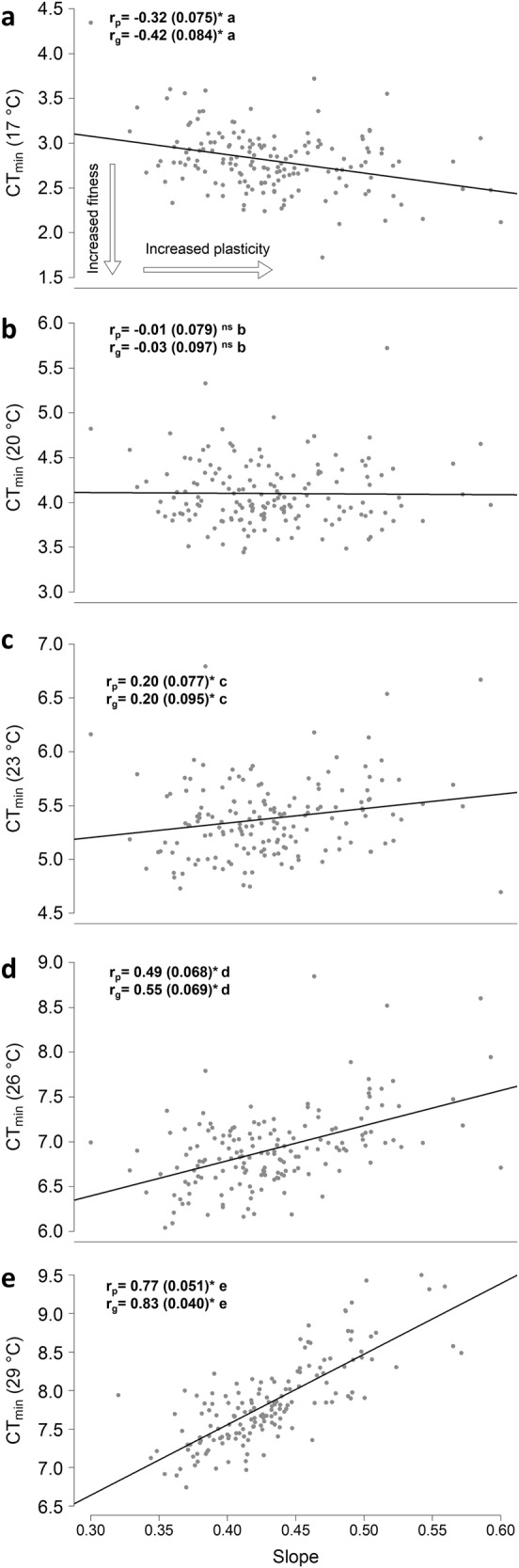
Correlations between CTmin at the individual temperatures and slope of CTmin across temperatures: a 17 °C, b 20 °C, c 23 °C, d 26 °C and e 29 °C. Each dot represents one DGRP line, and the full black lines are the regression lines visualising the phenotypic correlations. In each environment Pearson’s product moment correlation coefficients are shown for the phenotypic correlations, rp (SE in parenthesis) along with the printed genetic correlations, rg (SE in parenthesis) between CTmin. Asterisks designate correlations that are significantly different from 0, i.e., the estimate deviate more than 1.645 × SE from 0 (p < 0.05). Correlations that are not different from 0 are designated “ns”. In pairwise comparisons of the correlations, significantly different correlations are denoted with separate letters after the correlation coefficients, e.g., rg (CTmin 17 °C, slope) vs. rg (CTmin 20 °C, slope). Phenotypic and genetic correlations are compared separately. Comparisons of these correlations are based on Fisher’s z transformation (n = 166). The two arrows in panel a shows the direction of increased fitness (here opposite of increased CTmin) and increased plasticity of CTmin, respectively, for all panels
Discussion
In the present study, we investigated the effects of developmental temperature on quantitative genetic parameters of cold tolerance, which is a key stress tolerance trait. We showed that environmental conditions can directly influence the quantitative genetic basis of cold tolerance by affecting variance components, especially . It is expected that if standing genetic variation is environmentally dependent, selection responses will also be affected (Roff 2002; Bijlsma and Loeschcke 2005; Wilson et al. 2006; Husby et al. 2011; Wood and Brodie 2016). This has been demonstrated in recent studies showing that genetic variation and evolutionary potential of stress resistance, morphological traits and life-history traits depend on the environmental conditions (van Heerwaarden and Sgrò 2011; Bubliy et al. 2012; Kristensen et al. 2015; Bastide et al. 2016; Zhao et al. 2016; Feldman et al. 2017). Such findings can be explained by environment-specific gene expression, i.e., the impact of genes on a trait can depend on prior environmental exposures. This highlights the need to perform experiments and field studies under environmental conditions that are relevant to organisms at the time traits are under selection (Hoffmann and Parsons 1991; Hoffmann and Merilä 1999). This is important not only from an ecological and evolutionary perspective (Bay et al. 2017) but also for animal and plant breeding, in which effects of the environment on the genetic basis of production traits pose a challenge, such as for genomic selection in heterogeneous environments (Kadarmideen et al. 2006; Zhao et al. 2016).
It has long been debated whether some types of environmental conditions, and particularly stressful or benign conditions, affect heritable variation in distinct ways (Hoffmann and Parsons 1991; Møller and Swaddle 1998). Here we showed an increase in of cold resistance with increasing developmental temperature, and heritability estimates being similar across developmental temperatures (Table 2). However, for mean corrected , (i.e., evolvability (IA)), we found higher values at 17 and 20 °C than in the range 23–29 °C, suggesting that the evolutionary capacity for increasing cold tolerance might be more restricted in warm than in cold environments. This is supported by recent Drosophila studies showing that the potential for evolution is lower in relatively warmer conditions (Schou et al. 2014; Kristensen et al. 2015). In nature, individuals reared under cold conditions are more likely to encounter subsequent extreme cold conditions, therefore stronger selection for cold tolerance might be expected in natural populations experiencing low developmental temperatures compared to in hotter environments (Ayrinhac et al. 2004). Thus, in the case of cold tolerance, developmental environments with low temperatures might be considered more optimal than those with high temperatures. However, because we did not have a direct measure of stress in each thermal environment, we cannot confidently determine whether the patterns in and IA are associated with unfavourable environmental conditions (Hoffmann and Merilä 1999; Swindell and Bouzat 2006; Talloen et al. 2009).
The environment can also affect genetic variation via environment-dependent gene action, where genes influencing a certain trait in one environment may not be important for that trait in a different environment. In such cases selection responses can be slowed or even become maladaptive (Steinger et al. 2003). This will often be expressed as altered genetic correlations either between different traits in one environment or between the same trait in a range of environments (Vieira et al. 2000; Sgrò and Hoffmann 2004; Agrawal and Stinchcombe 2009). Here we found positive genetic and phenotypic correlations between cold tolerance across environments, i.e., genotypes with high cold tolerance when developed at one temperature also tended to have high cold tolerance when developed at other temperatures, implying an overlap in the sets of alleles influencing cold tolerance across developmental thermal environments (Via and Lande 1985; Falconer and Mackay 1996; Agrawal and Stinchcombe 2009). However, we also showed that the magnitude of correlations between thermal environments decreased as developmental temperatures diverged. This suggests a shared genetic basis for cold tolerance between rearing environments, which contributes less to genetic variation as environments become more dissimilar. Although evidence for such patterns has rarely been investigated, a few studies have found similar results; Sgrò and Blows (2004) found declining genetic correlations with environmental dissimilarity in three life-history traits in Drosophila serrata, while Stinchcombe et al. (2010) found declining genetic correlations with environmental distance for growth rate in jewelweed.
Our findings are in line with other studies suggesting that the genetic architecture of fitness components is specific to an environment (Bourret and Garant 2015; Rellstab et al. 2017). This represents a challenge when testing for adaptation at specific loci which often seem to be environment and population specific (Zhao et al. 2011; Manenti et al. 2016). Knowledge about the genetic architecture of many complex traits has been rapidly accumulating recently, aided by developments in genomics and quantitative genetics. Typically, such studies identify genes, variance components and heritabilities for a given trait in one environment to predict evolutionary trajectories. In animal and plant breeding, such knowledge may be used to predict selection responses and devise genomic selection procedures. However, our results show that the architecture of an important stress resistance trait is highly dependent on environmental conditions, challenging the notion that there are candidate genes and a particular architecture applicable across a gradient of environmental conditions.
An important issue for the evolution of thermal resistance is whether an organism’s thermotolerance is constrained by plasticity, particularly in response to anthropogenic climate change (Stillman 2003; Chown et al. 2010; Levine et al. 2011; Gunderson et al. 2015; Comte and Olden 2017). Some studies point to a negative association between an organism’s thermal tolerance and plasticity in tolerance (Hoffmann et al. 2003; Kellett et al. 2005; Nyamukondiwa et al. 2011; Gerken et al. 2015; Noh et al. 2017). Here we showed that any association between cold tolerance and plasticity depend on the environment; both the phenotypic and genetic correlations shifted in sign as developmental temperatures shifted from 17 to 29 °C (Fig. 3). Given that the lowest CTmin occurred at the lowest developmental temperature, genotypes with a flat slope would likely have a fitness advantage when exposed to low temperatures, as they would remain the most cold tolerant (i.e., maintain relatively lower CTmin at higher developmental temperatures). However, as indicated by the negative correlation between slope and CTmin at 17 °C, DGRP lines with low CTmin reared at this temperature, typically had a high CTmin (low cold tolerance) when they developed at higher temperatures, indicative of a trade-off (Fig. 3a). At low developmental temperatures, lines with low CTmin and a steep slope could be considered “specialists”; i.e., they performed relatively better in one environment, but relatively poorly across environments. At higher developmental temperatures (23, 26 and 29 °C), we found a positive correlation between CTmin and the plastic response, suggesting that high cold tolerance (low CTmin) at high developmental temperatures was associated with low plasticity (Fig. 3c–e). Thus, genotypes with low CTmin at higher developmental temperatures could also be considered as the “specialists” because they performed relatively better in one environment. This shift in sign of the correlations reflects a reversal in the ordering of genotypes for cold resistance across the thermal gradient, with a tipping point at 20 °C, when there was no correlation between cold tolerance and plasticity.
These genotypic differences in constraints may reflect differential environmental sensitivity and suggests that the fitness effect of plasticity depends heavily on the environment where genotypes are reared. Thus, the fitness landscape of cold tolerance needs to be considered in the context of environmental frequencies, i.e., how likely are individuals reared under one set of thermal conditions to encounter subsequent extreme cold conditions (Ayrinhac et al. 2004)? Our findings support those of Levine et al. (2011) who suggested a “directionality” of plasticity in gene expression depending on the thermal environment commonly encountered in the native range of populations of D. melanogaster. This has implications for understanding developmental processes in variable environments (Stearns et al. 1991), and also for understanding local adaptation in thermal tolerances in populations with a broad geographic distribution along an environmental cline (Ayrinhac et al. 2004; Levine et al. 2011; Cooper et al. 2012; Kellermann et al. 2012; Kristensen et al. 2015). Our results indicate that the quantitative genetic basis of basal cold tolerance and plastic cold tolerance is highly environment specific, which is important for predicting selection responses in natural and variable environments (Sgrò and Hoffmann 2004; Agrawal and Stinchcombe 2009).
Most studies investigating phenotypic and genetic correlations and/or candidate genes between thermal environments focus on only two temperatures (Guerra et al. 1997; Ellers and Driessen 2011; Ketola et al. 2012) or investigate two settings such as field vs. lab conditions (Kristensen et al. 2008; Overgaard et al. 2010), and thus do not provide a comprehensive picture of changing genetic correlations and architectures across environments. A meta-analysis by Charmantier and Garant (2005) specifically called for approaches that consider genetic variability under multiple conditions, and ideally along a continuous environmental gradient such as in the present study. Because correlations within and between traits can change between environments or even shift in sign (Sgrò and Hoffmann 2004; Agrawal and Stinchcombe 2009) as evident in this study, we reiterate this call and caution about genetic constraints and trade-offs deduced from studies that consider only one or two environments.
In conclusion, we showed that the expression of additive genetic variation for cold tolerance was environment dependent, and that evolvability of cold resistance decreased with increasing developmental temperatures. We also showed that the relationship between basal and plastic cold tolerance was affected by the environment, and even switched sign across a gradient of developmental temperatures. We further provided evidence for an evolutionary trade-off between cold tolerance in one environment and cold tolerance across environment. It will be interesting to test for these patterns in relevant ecological settings, such as in populations exposed to strong diurnal and seasonal temperature fluctuations.
Electronic supplementary material
Acknowledgements
DGRP stocks were obtained from the Bloomington Drosophila Stock Centre (NIH P40OD018537). We thank Iben R. Jensen, Helle Blendstrup, Susan M. Hansen, and Iben V. Nielsen for laboratory assistance. This research was financed by the Danish Natural Science Research Council through a Sapere aude stipend to TNK (DFF—4002-00036), the Science Industry Endowment Fund and a grant from the Velux Visiting Professor programme to AAH, and by the Danish Strategic Research Council (GenSAP: Centre for Genomic Selection in Animals and Plants, contract 12-132452) to PS.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
The online version of this article (10.1038/s41437-018-0117-7) contains supplementary material, which is available to authorised users.
References
- Agrawal AF, Stinchcombe JR. How much do genetic covariances alter the rate of adaptation? Proc R Soc B Biol Sci. 2009;276:1183–1191. doi: 10.1098/rspb.2008.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo MB, Ferri-Yáñez F, Bozinovic F, Marquet PA, Valladares F, Chown SL. Heat freezes niche evolution. Ecol Lett. 2013;16:1206–1219. doi: 10.1111/ele.12155. [DOI] [PubMed] [Google Scholar]
- Auld JR, Agrawal AA, Relyea RA. Re-evaluating the costs and limits of adaptive phenotypic plasticity. Proc R Soc B Biol Sci. 2010;277:503–511. doi: 10.1098/rspb.2009.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayrinhac A, Debat V, Gibert P, Kister AG, Legout H, Moreteau B, et al. Cold adaptation in geographical populations of Drosophila melanogaster: phenotypic plasticity is more important than genetic variability. Funct Ecol. 2004;18:700–706. doi: 10.1111/j.0269-8463.2004.00904.x. [DOI] [Google Scholar]
- Bastide H, Lange JD, Lack JB, Yassin A, Pool JE. A variable genetic architecture of melanic evolution in Drosophila melanogaster. Genetics. 2016;204:1307–1319. doi: 10.1534/genetics.116.192492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bay RA, Rose N, Barrett R, Bernatchez L, Ghalambor CK, Lasky JR, et al. Predicting responses to contemporary environmental change using evolutionary response architectures. Am Nat. 2017;189:463–473. doi: 10.1086/691233. [DOI] [PubMed] [Google Scholar]
- Bijlsma R, Loeschcke V. Environmental stress, adaptation and evolution: an overview. J Evol Biol. 2005;18:744–9. doi: 10.1111/j.1420-9101.2005.00962.x. [DOI] [PubMed] [Google Scholar]
- Bourret A, Garant D. candidate gene-environment interactions and their relationships with timing of breeding in a wild bird population. Ecol Evol. 2015;5:3628–3641. doi: 10.1002/ece3.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubliy OA, Kristensen TN, Kellermann V, Loeschcke V. Humidity affects genetic architecture of heat resistance in Drosophila melanogaster. J Evol Biol. 2012;25:1180–1188. doi: 10.1111/j.1420-9101.2012.02506.x. [DOI] [PubMed] [Google Scholar]
- Calosi P, Bilton DT, Spicer JI. Thermal tolerance, acclimatory capacity and vulnerability to global climate change. Biol Lett. 2008;4:99–102. doi: 10.1098/rsbl.2007.0408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charmantier A, Garant D. Environmental quality and evolutionary potential: lessons from wild populations. Proc R Soc B Biol Sci. 2005;272:1415–1425. doi: 10.1098/rspb.2005.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chown SL, Hoffmann AA, Kristensen TN, Angilletta MJ, Stenseth N, Pertoldi C. Adapting to climate change: a perspective from evolutionary physiology. Clim Res. 2010;43:3–15. doi: 10.3354/cr00879. [DOI] [Google Scholar]
- Comte L, Olden JD. Evolutionary and environmental determinants of freshwater fish thermal tolerance and plasticity. Glob Change Biol. 2017;23:728–736. doi: 10.1111/gcb.13427. [DOI] [PubMed] [Google Scholar]
- Cooper BS, Tharp JM, Jernberg II, Angilletta MJ. Developmental plasticity of thermal tolerances in temperate and subtropical populations of Drosophila melanogaster. J Therm Biol. 2012;37:211–216. doi: 10.1016/j.jtherbio.2012.01.001. [DOI] [Google Scholar]
- Czesak ME, Fox CW, Wolf JB. Experimental evolution of phenotypic plasticity: how predictive are cross-environment genetic correlations? Am Nat. 2006;168:323–35. doi: 10.1086/506919. [DOI] [PubMed] [Google Scholar]
- David JR, Gibert P, Gravot E, Petavy G, Morin JP, Karan D, et al. Phenotypic plasticity and developmental temperature in drosophila: analysis and significance of reaction norms of morphometrical traits. J Therm Biol. 1997;22:441–451. doi: 10.1016/S0306-4565(97)00063-6. [DOI] [Google Scholar]
- Dingemanse NJ, Van der Plas F, Wright J, Reale D, Schrama M, Roff DA, et al. Individual experience and evolutionary history of predation affect expression of heritable variation in fish personality and morphology. Proc R Soc B Biol Sci. 2009;276:1285–1293. doi: 10.1098/rspb.2008.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellers J, Driessen G. Genetic correlation between temperature-induced plasticity of life-history traits in a soil arthropod. Evol Ecol. 2011;25:473–484. doi: 10.1007/s10682-010-9414-1. [DOI] [Google Scholar]
- Etterson JR, Shaw RG. Constraint to adaptive evolution in response to global warming. Science. 2001;294:151–154. doi: 10.1126/science.1063656. [DOI] [PubMed] [Google Scholar]
- Falconer DS, Mackay TFC. Introduction to quantitative genetics. 4th edn. Harlow, UK: Longman Publishing Group; 1996. [Google Scholar]
- Feldman MJ, Paul RE, Banan D, Barrett JF, Sebastian J, Yee MC et al. (2017) Time dependent genetic analysis links field and controlled environment phenotypes in the model C4 grass Setaria. PLOS Genet 13:e1006841 [DOI] [PMC free article] [PubMed]
- Franke K, Dierks A, Fischer K. Directional selection on cold tolerance does not constrain plastic capacity in a butterfly. BMC Evol Biol. 2012;12:235. doi: 10.1186/1471-2148-12-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhardt-Henrich SG, van Noordwijk AJ. Nestling growth in the Great Tit I. Heritability estimates under different environmental conditions. J Evol Biol. 1991;4:341–362. doi: 10.1046/j.1420-9101.1991.4030341.x. [DOI] [Google Scholar]
- Gerken AR, Eller OC, Hahn DA, Morgan TJ. Constraints, independence, and evolution of thermal plasticity: probing genetic architecture of long- and short-term thermal acclimation. Proc Natl Acad Sci USA. 2015;112:4399–4404. doi: 10.1073/pnas.1503456112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goenaga J, Mensch J, Fanara JJ, Hasson E. The effect of mating on starvation resistance in natural populations of Drosophila melanogaster. Evol Ecol. 2012;26:813–823. doi: 10.1007/s10682-011-9540-4. [DOI] [Google Scholar]
- Guerra D, Cavicchi S, Krebs R, Loeschcke V. Resistance to heat and cold stress in Drosophila melanogaster: intra and inter population variation in relation to climate. Genet Sel Evol. 1997;29:497–510. doi: 10.1186/1297-9686-29-4-497. [DOI] [Google Scholar]
- Gunderson AR, Stillman JH (2015) Plasticity in thermal tolerance has limited potential to buffer ectotherms from global warming. Proc R Soc B Biol Sci 282:1–8 [DOI] [PMC free article] [PubMed]
- van Heerwaarden B, Sgrò CM. The effect of developmental temperature on the genetic architecture underlying size and thermal clines in Drosophila melanogaster and D. simulans from the east coast of Australia. Evolution. 2011;65:1048–1067. doi: 10.1111/j.1558-5646.2010.01196.x. [DOI] [PubMed] [Google Scholar]
- Hercus MJ, Hoffmann AA. Maternaland grandmaternal age influence offspring fitness in Drosophila. Proc R Soc B Biol Sci. 2000;267:2105–2110. doi: 10.1098/rspb.2000.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann AA. Physiological climatic limits in Drosophila: patterns and implications. J Exp Biol. 2010;213:870–880. doi: 10.1242/jeb.037630. [DOI] [PubMed] [Google Scholar]
- Hoffmann AA, Merilä J. Heritable variation and evolution under favourable and unfavourable conditions. Trends Ecol Evol. 1999;14:96–101. doi: 10.1016/S0169-5347(99)01595-5. [DOI] [PubMed] [Google Scholar]
- Hoffmann AA, Merilä J, Kristensen TN. Heritability and evolvability of fitness and nonfitness traits: lessons from livestock. Evolution. 2016;70:1770–1779. doi: 10.1111/evo.12992. [DOI] [PubMed] [Google Scholar]
- Hoffmann AA, Parsons PA (1991) Evolutionary genetics and environmental stress. Oxford University Press, Oxford, UK
- Hoffmann AA, Sørensen JG, Loeschcke V. Adaptation of Drosophila to temperature extremes: bringing together quantitative and molecular approaches. J Therm Biol. 2003;28:175–216. doi: 10.1016/S0306-4565(02)00057-8. [DOI] [Google Scholar]
- Houle D. Comparing evolvability and variability of quantitative traits. Genetics. 1992;130:195–204. doi: 10.1093/genetics/130.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Massouras A, Inoue Y, Peiffer J, Ràmia M, Tarone AM, et al. Natural variation in genome architecture among 205 Drosophila melanogaster Genetic Reference Panel lines. Genome Res. 2014;24:1193–1208. doi: 10.1101/gr.171546.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husby A, Visser ME, Kruuk LEB (2011) Speeding up microevolution: the effects of increasing temperature on selection and genetic variance in a wild bird population. PLOS Biol 9:e1000585 [DOI] [PMC free article] [PubMed]
- de Jong G. Quantitative genetics of reaction norms. J Evol Biol. 1990;3:447–468. doi: 10.1046/j.1420-9101.1990.3050447.x. [DOI] [Google Scholar]
- Kadarmideen HN, Von Rohr P, Janss LLG. From genetical genomics to systems genetics: potential applications in quantitative genomics and animal breeding. Mamm Genome. 2006;17:548–564. doi: 10.1007/s00335-005-0169-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellermann V, Overgaard J, Hoffmann AA, Fløjgaard C, Svenning JC, Loeschcke V. Upper thermal limits of Drosophila are linked to species distributions and strongly constrained phylogenetically. Proc Natl Acad Sci USA. 2012;109:16228–33. doi: 10.1073/pnas.1207553109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellett M, Hoffmann AA, Mckechnie SW. Hardening capacity in the Drosophila melanogaster species group is constrained by basal thermotolerance. Funct Ecol. 2005;19:853–858. doi: 10.1111/j.1365-2435.2005.01025.x. [DOI] [Google Scholar]
- Ketola T, Kellermann V, Kristensen TN, Loeschcke V. Constant, cycling, hot and cold thermal environments: strong effects on mean viability but not on genetic estimates. J Evol Biol. 2012;25:1209–1215. doi: 10.1111/j.1420-9101.2012.02513.x. [DOI] [PubMed] [Google Scholar]
- Kimura MT. Cold and heat tolerance of drosophilid flies with reference to their latitudinal distributions. Oecologia. 2004;140:442–449. doi: 10.1007/s00442-004-1605-4. [DOI] [PubMed] [Google Scholar]
- Kristensen TN, Hoffmann AA, Overgaard J, Sørensen JG, Hallas R, Loeschcke V. Costs and benefits of cold acclimation in field-released Drosophila. Proc Natl Acad Sci USA. 2008;105:216–221. doi: 10.1073/pnas.0708074105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen TN, Overgaard J, Lassen J, Hoffmann AA, Sgrò C. Low evolutionary potential for egg-to-adult viability in Drosophila melanogaster at high temperatures. Evolution. 2015;69:803–814. doi: 10.1111/evo.12617. [DOI] [PubMed] [Google Scholar]
- Levine MT, Eckert ML, Begun DJ. Whole-genome expression plasticity across tropical and temperate Drosophila melanogaster populations from eastern Australia. Mol Biol Evol. 2011;28:249–256. doi: 10.1093/molbev/msq197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay TFC, Richards S, Stone EA, Barbadilla A, Ayroles JF, Zhu D, et al. The Drosophila melanogaster Genetic Reference Panel. Nature. 2012;482:173–178. doi: 10.1038/nature10811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen P, Jensen J, Thompson R (1994). Estimation of (co)-variance components by REML in multivariate mixed linear models using average of observed and expected information. In: FifthWorld Congress of Genetics Applied to Livestock Production, Guelph, Ontario, Canada, pp 455–462.
- Manenti T, Sørensen JG, Moghadam NN, Loeschcke V. Few genetic and environmental correlations between life-history and stress resistance traits affect adaptation to fluctuating thermal regimes. Heredity. 2016;117:149–154. doi: 10.1038/hdy.2016.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murren CJ, Auld JR, Callahan H, Ghalambor CK, Handelsman CA, Heskel MA, et al. Constraints on the evolution of phenotypic plasticity: limits and costs of phenotype and plasticity. Heredity. 2015;115:293–301. doi: 10.1038/hdy.2015.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller A, Swaddle K. Asymmetry, developmental stability and evolution. Oxford, UK: Oxford University Press; 1998. [Google Scholar]
- Noh S, Everman ER, Berger CM, Morgan TJ. Seasonal variation in basal and plastic cold tolerance: adaptation is influenced by both long- and short-term phenotypic plasticity. Ecol Evol. 2017;7:5248–5257. doi: 10.1002/ece3.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyamukondiwa C, Terblanche JS, Marshall KE, Sinclair BJ. Basal cold but not heat tolerance constrains plasticity among Drosophila species (Diptera: Drosophilidae) J Evol Biol. 2011;24:1927–1938. doi: 10.1111/j.1420-9101.2011.02324.x. [DOI] [PubMed] [Google Scholar]
- Overgaard J, Kristensen TN, Mitchell KA, Hoffmann AA. Thermal tolerance in widespread and tropical Drosophila species: does phenotypic plasticity increase with latitude? Am Nat. 2011;178:80–96. doi: 10.1086/661780. [DOI] [PubMed] [Google Scholar]
- Overgaard J, Kristensen TN, Sørensen JG. Validity of thermal ramping assays used to assess thermal tolerance in arthropods. PLOS ONE. 2012;7:1–7. doi: 10.1371/journal.pone.0032758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overgaard J, Sørensen JG, Jensen LT, Loeschcke V, Kristensen TN. Field tests reveal genetic variation for performance at low temperatures in Drosophila melanogaster. Funct Ecol. 2010;24:186–195. doi: 10.1111/j.1365-2435.2009.01615.x. [DOI] [Google Scholar]
- R Core Team . R: a language and environment for statistical computing. Vienna, Austria: Version 3.4.0. R Foundation for Statistical Computing; 2017. [Google Scholar]
- Rellstab C, Fischer MC, Zoller S, Graf R, Tedder A, Shimizu KK, et al. Local adaptation (mostly) remains local: reassessing environmental associations of climate-related candidate SNPs in Arabidopsis halleri. Heredity. 2017;118:193–201. doi: 10.1038/hdy.2016.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roff DA. Life history evolution. Sunderland, UK: Sinauer Associates; 2002. [Google Scholar]
- Rohde PD, Gaertner B, Wards K, Sørensen P, Mackay TFC. Genomic analysis of genotype-by-social environment interaction for Drosophila melanogaster aggressive behavior. Genetics. 2017;206:1969–1984. doi: 10.1534/genetics.117.200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schou MF, Kristensen TN, Kellermann V, Schlötterer C, Loeschcke V. A Drosophila laboratory evolution experiment points to low evolutionary potential under increased temperatures likely to be experienced in the future. J Evol Biol. 2014;27:1859–1868. doi: 10.1111/jeb.12436. [DOI] [PubMed] [Google Scholar]
- Schou MF, Mouridsen MB, Sørensen JG, Loeschcke V. Linear reaction norms of thermal limits in Drosophila: predictable plasticity in cold but not in heat tolerance. Funct Ecol. 2017;31:934–945. doi: 10.1111/1365-2435.12782. [DOI] [Google Scholar]
- Sgrò CM, Blows MW. The genetic covariance among clinal environments after adaptation to an environmental gradient in Drosophila serrata. Genetics. 2004;167:1281–1291. doi: 10.1534/genetics.103.026120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgrò CM, Hoffmann AA. Genetic correlations, trade-offs and environmental variation. Heredity. 2004;93:241–248. doi: 10.1038/sj.hdy.6800532. [DOI] [PubMed] [Google Scholar]
- Sheldon BC, Kruuk LEB, Merilä J. Natural selection and inheritance of breeding time and clutch size in the collared flycatcher. Evolution. 2003;57:406–420. doi: 10.1111/j.0014-3820.2003.tb00274.x. [DOI] [PubMed] [Google Scholar]
- Spichtig M, Kawecki TJ. The maintenance (or not) of polygenic variation by soft selection in heterogeneous environments. Am Nat. 2004;164:70–84. doi: 10.1086/421335. [DOI] [PubMed] [Google Scholar]
- Stearns S, de Jong G, Newman B. The effects of phenotypic plasticity on genetic correlations. Trends Ecol Evol. 1991;6:122–126. doi: 10.1016/0169-5347(91)90090-K. [DOI] [PubMed] [Google Scholar]
- Steinger T, Roy BA, Stanton ML. Evolution in stressful environments II: adaptive value and costs of plasticity in response to low light in Sinapis arvensis. J Evol Biol. 2003;16:313–323. doi: 10.1046/j.1420-9101.2003.00518.x. [DOI] [PubMed] [Google Scholar]
- Stillman JH. Acclimation capacity underlies susceptibility to climate change. Science. 2003;301:65. doi: 10.1126/science.1083073. [DOI] [PubMed] [Google Scholar]
- Stinchcombe JR, Izem R, Heschel MS, McGoey BV, Schmitt J. Across environment genetic correlations and the frequency of selective environments shape the evolutionary dynamics of growth rate in impatiens capensis. Evolution. 2010;64:2887–2903. doi: 10.1111/j.1558-5646.2010.01060.x. [DOI] [PubMed] [Google Scholar]
- Strachan LA, Tarnowski-Garner HE, Marshall KE, Sinclair BJ. The evolution of cold tolerance in Drosophila larvae. Physiol Biochem Zool. 2011;84:43–53. doi: 10.1086/657147. [DOI] [PubMed] [Google Scholar]
- Sunday JM, Bates AE, Dulvy NK. Global analysis of thermal tolerance and latitude in ectotherms. Proc R Soc B Biol Sci. 2011;278:1823–1830. doi: 10.1098/rspb.2010.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swindell WR, Bouzat JL. Associations between environmental stress, selection history, and quantitative genetic variation in Drosophila melanogaster. Genetica. 2006;127:311–20. doi: 10.1007/s10709-005-5240-8. [DOI] [PubMed] [Google Scholar]
- Talloen W, Van Dongen S, Van Dyck H, Lens L. Environmental stress and quantitative genetic variation in butterfly wing characteristics. Evol Ecol. 2009;23:473–485. doi: 10.1007/s10682-008-9246-4. [DOI] [Google Scholar]
- Telonis-Scott M, Hallas R, McKechnie SW, Wee CW, Hoffmann AA. Selection for cold resistance alters gene transcript levels in Drosophila melanogaster. J Insect Physiol. 2009;55:549–555. doi: 10.1016/j.jinsphys.2009.01.010. [DOI] [PubMed] [Google Scholar]
- Tonsor SJ, Elnaccash TW, Scheiner SM. Developmental instability is genetically correlated with phenotypi plasticity, constraining heritability, and fitness. Evolution. 2013;67:2923–2935. doi: 10.1111/evo.12175. [DOI] [PubMed] [Google Scholar]
- Valladares F, Sanchez-Gomez D, Zavala MA. Quantitative estimation of phenotypic plasticity: bridging the gap between the evolutionary concept and its ecological applications. J Ecol. 2006;94:1103–1116. doi: 10.1111/j.1365-2745.2006.01176.x. [DOI] [Google Scholar]
- VanRaden PM. Efficient methods to compute genomic predictions. J Dairy Sci. 2008;91:4414–4423. doi: 10.3168/jds.2007-0980. [DOI] [PubMed] [Google Scholar]
- Via S, Lande R. Genotype-environment interaction and the evolution of phenotypic plasticity. Evolution. 1985;39:505–522. doi: 10.1111/j.1558-5646.1985.tb00391.x. [DOI] [PubMed] [Google Scholar]
- Vieira C, Pasyukova EG, Zeng ZB, Hackett JB, Lyman RF, Mackay TF. Genotype-environment interaction for quantitative trait loci affecting life span in Drosophila melanogaster. Genetics. 2000;154:213–227. doi: 10.1093/genetics/154.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher PM, Hill WG, Wray NR. Heritability in the genomics era—concepts and misconceptions. Nat Rev Genet. 2008;9:255–266. doi: 10.1038/nrg2322. [DOI] [PubMed] [Google Scholar]
- Williams CM, Henry HAL, Sinclair BJ. Cold truths: how winter drives responses of terrestrial organisms to climate change. Biol Rev. 2014;90:214–235. doi: 10.1111/brv.12105. [DOI] [PubMed] [Google Scholar]
- Wilson AJ, Pemberton JM, Pilkington JG, Coltman DW, Mifsud DV, Clutton-Brock TH, et al. Environmental coupling of selection and heritability limits evolution. PLoS Biol. 2006;4:1270–1275. doi: 10.1371/journal.pbio.0040216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood CW, Brodie ED. Evolutionary response when selection and genetic variation covary across environments. Ecol Lett. 2016;19:1189–1200. doi: 10.1111/ele.12662. [DOI] [PubMed] [Google Scholar]
- Zhao K, Tung CW, Eizenga GC, Wright MH, Ali ML, Price AH, et al. Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa. Nat Commun. 2011;2:467. doi: 10.1038/ncomms1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Wang X, Wang H, Tian J, Li B, Chen L, et al. Genome-wide identification of QTL for seed yield and yield-related traits and construction of a high-density consensus map for QTL comparison in Brassica napus. Front Plant Sci. 2016;7:17. doi: 10.3389/fpls.2016.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ørsted M, Malmendal A, Muñoz J, Kristensen TN. Metabolic and functional phenotypic profiling of Drosophila melanogaster reveals reduced sex differentiation under stressful environmental conditions. Biol J Linn Soc. 2018;123:155–162. doi: 10.1093/biolinnean/blx120. [DOI] [Google Scholar]
- Ørsted M, Rohde PD, Hoffmann AA, Sørensen P, Kristensen TN. Environmental variation partitioned into separate heritable components. Evolution. 2018;72:136–152. doi: 10.1111/evo.13391. [DOI] [PubMed] [Google Scholar]
- Ørsted M, Schou MF, Kristensen TN. Biotic and abiotic factors investigated in two Drosophila species—evidence of both negative and positive effects of interactions on performance. Sci Rep. 2017;7:40132. doi: 10.1038/srep40132. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.




