Abstract
Wolbachia bacteria are common insect endosymbionts transmitted maternally and capable of spreading through insect populations by cytoplasmic incompatibility (CI) when infected males cause embryo death after mating with uninfected females. Selection in the Wolbachia endosymbiont occurs on female hosts and is expected to favour strong maternal transmission to female offspring, even at the cost of reduced CI. With maternal leakage, nuclear genes are expected to be selected to suppress cytoplasmic incompatibility caused by males while also reducing any deleterious effects associated with the infection. Here we describe a new type of Wolbachia strain from Drosophila pseudotakahashii likely to have arisen from evolutionary processes on host and/or Wolbachia genomes. This strain is often absent from adult male offspring, but always transmitted to females. It leads to males with low or non-detectable Wolbachia that nevertheless show CI. When detected in adult males, the infection has a low density relative to that in females, a phenomenon not previously seen in Wolbachia infections of Drosophila. This Wolbachia strain is common in natural populations, and shows reduced CI when older (infected) males are crossed. These patterns highlight that endosymbionts can have strong sex-specific effects and that high frequency Wolbachia strains persist through effects on female reproduction. Female-limited Wolbachia infections may be of applied interest if the low level of Wolbachia in males reduces deleterious fitness effects on the host.
Subject terms: Genetics, Phylogenetics
Introduction
Wolbachia are common endosymbionts of insects with diverse effects on insect reproduction. Cytoplasmic incompatibility (CI: reduced egg hatch when uninfected females mate with infected males) is the most common phenotype associated with this endosymbiont (Hoffmann and Turelli 1997). There are also Wolbachia strains with other phenotypic effects on host reproduction, including male killing (Hurst and Jiggins 2000) and feminization (Kern et al. 2015). Although Wolbachia are often described as “manipulating” host reproduction to aid their own spread and persistence, Wolbachia in some populations may also have little impact on host reproduction (Hoffmann et al. 1996). Such Wolbachia persist by generating other fitness advantages for their hosts, perhaps involving nutrition or protection from viruses (Teixeira et al. 2008; Brownlie et al. 2009), although the particular advantages are often unknown (Hamm et al. 2014; Kriesner et al. 2013).
Evolutionary changes in the nuclear genome of hosts or the genome of Wolbachia strains will drive shifts in phenotypic effects on hosts, including both fitness effects and reproductive manipulations, as well as the efficiency of maternal transmission across generations (Bull and Turelli 2013; Prout 1994; Turelli 1994). This raises the issue of which types of changes in CI, fitness, and transmission efficiency might be expected to occur across time, and which are due to changes in host and/or Wolbachia genomes. Microinjection experiments and backcrosses can help to sort out which genomes are involved (Boyle et al. 1993; Braig et al. 1994). Theoretical models (Prout 1994; Turelli 1994) suggest that Wolbachia should be selected on the basis of maternal transmission success, given that a high rate of transmission can ensure that Wolbachia persist in populations even in the absence of reproductive effects. For this reason, Wolbachia genes that favour transmission could be selected even under decreasing levels of incompatibility (Tortosa et al. 2010). If transmission is imperfect, selection also acts on host genes that decrease the impact of Wolbachia on host reproduction. This reflects the fact that females that lose Wolbachia will be sterilized by males carrying Wolbachia unless they carry genes that overcome the incompatibility. Selection pressures acting on the sexes are quite different, because inheritance of Wolbachia normally involves female hosts and paternal transmission appears rare (Hoffmann and Turelli 1988; Turelli et al. 2018); any fitness effects of Wolbachia that are male-specific and associated with the bacterium would not be passed on. Selection on Wolbachia, therefore, should proceed via the female (Frank 1997; Turelli 1994). For this reason, feminization caused by Wolbachia is favoured by selection, and male-killing is also favoured if there is an advantage to females developing at a relatively lower density, for example, with reduced sib competition or cannibalism, as well as for the avoidance of inbreeding (Hurst et al. 2002).
Despite the sex-specific nature of selection effects generated by Wolbachia, there is, so far, limited evidence for sex-specific differences in the density of Wolbachia in insects. In some mosquito studies, lower densities have been found in males, particularly in Aedes albopictus (Dutton and Sinkins 2004; Tortosa et al. 2010) and Culex pipiens (Berticat et al. 2002). In other cases, densities are similar between the sexes, as in Aedes aegypti (Ross et al. 2017). Where differences have been detected, as in the examples cited above, they typically involve 2–15 fold differences in density, although in one species of planthopper, a more substantial difference between the sexes has been noted and this has been associated with a low level of CI (Noda et al. 2001). There are still relatively few Wolbachia infections for which phenotypic effects have been investigated in detail, particularly across both sexes and with field-derived lines, raising the issue of whether Wolbachia strains that are strongly sex-limited in density or even occurrence might yet exist on a broader scale.
Here we describe a Wolbachia infection in a Drosophila species, D. pseudotakahashii, that seems to have sex-specific effects associated with differences in Wolbachia density. This strain, wPse, has previously been identified in D. pseudotakahashii (14022-0301.01, Tucson Stock Centre, origin Atherton Tableland, Australia) (Mateos et al. 2006) using wsp primers, but no phenotypic or molecular studies on the strain have been done. Drosophila pseudotakahashii falls in the takahashii species subgroup, which is in the D. melanogaster species group. We show that the wPse strain is detected in females at a high frequency, but that it appears at a much lower frequency in males. Moreover, there is strong CI in crosses involving uninfected females, even when the males from infected mothers have low-density Wolbachia or it is not detected. We show that maternal transmission of the strain to females is high and that there are no obvious host fitness effects associated with this infection.
Methods
D. pseudotakahashii field collections and laboratory lines
Samples of female D. pseudotakahashii (Appendix 1) were obtained from two locations in New South Wales, three locations in south eastern Queensland, and six locations in northern Queensland. Of these samples, all females except some from New South Wales were used to initiate isofemale lines. Species identification was based on male offspring and involved examination of the configuration of the sex combs on tarsomeres I and II of the male foreleg and the dark abdominal banding on the posterior tergites (see Bock. 1976). Lines were screened for Wolbachia infection by PCR and RT-PCR as outlined below using a single female and multiple males where possible.
Several laboratory lines of D. pseudotakahashii were used in crosses and experiments (Appendix 1). One line (designated Moor+ with the + symbol indicating an infected line) was established by pooling flies from three infected isofemale lines collected from Moorland, New South Wales. Four other infected isofemale lines from northern Queensland were used in crosses and experiments and originated from Smithfield and Townsville with designations Smith+, Town1+, Town2+ and Town3+. Females from all lines were infected by Wolbachia based on PCR characterization, however males were not consistently infected (see below).
Because there were no naturally uninfected lines (see below), uninfected lines were generated from three of the isofemale lines (Moor+, Smith+, Town3+) by treatment with 0.03% tetracycline (Sigma, Castle Hill, NSW, Australia) in cornmeal media for one to two generations as outlined in Hoffmann et al. (1986). Derived Wolbachia-negative lines are designated with a “-“ symbol (i.e., Moor− and Town3−). Curing of Wolbachia infection was verified via PCR (see below). Lines were maintained in the laboratory on cornmeal media at 19 °C with a 12:12 L:D cycle and treated lines were allowed to recover in the absence of tetracycline for at least two generations before being used in experiments. Note that the uninfected status of these lines was confirmed when flies were used in experimental crosses.
Wolbachia infection detection
A preliminary screen for Wolbachia infection was conducted for all field isofemale lines and preserved samples. DNA extractions were performed using a 5% Chelex® (Bio-Rad Laboratories, Gladesville, NSW, Australia; Cat. No. 142-1253; w/v in distilled water) based plate method on single individuals as outlined by Richardson et al. (2016), with extractions diluted to 1 in 10 for all screening work. Samples were screened for Wolbachia via PCR with the gatB primers for the Multilocus Sequence Typing System (MLST) for Wolbachia (Baldo et al. 2006) or via RT-PCR using the wsp_validation primers (Kriesner et al. 2013; Lee et al. 2012). PCR conditions and visualisation techniques are outlined in Richardson et al. (2016).
To investigate the Wolbachia infection in more detail, we used the forward and reverse coxA, hcpA, ftsZ, fpbA and gatB MLST primers (Baldo et al. 2006) and wsp_validation primers (Kriesner et al. 2013; Lee et al. 2012). While whole genome sequencing would have been ideal (Bleidorn and Gerth. 2017; Turelli et al. 2018), it is more expensive, and MLST is an established system that also allows links to be made to previous work. The MLST and wsp_validation primers were used to screen additional PCR amplifications for two individuals from the isofemale lines from Moorland and Smithfield as well as Lake Placid (Appendix 1). Conditions were as outlined in Richardson et al. (2016). PCR products were sent to Macrogen (Korea) for purification and Sanger sequencing. Sequencing chromatograms were examined and processed using Finch TV v1.4.0 (Geospiza, Seattle, WA) and MEGA version 6 (Tamura et al. 2013). Allelic profiles were compared to those in the MLST database at http://pubmlst.org/wolbachia/ (Jolley and Maiden 2010).
For routine scoring of Wolbachia in the flies, we used a genotyping assay (Kriesner et al. 2013; Lee et al. 2012) which determines Wolbachia infection from the melting temperature (Tm) of the wsp validation PCR amplicons. High-resolution melt analysis on the Roche LightCycler® 480 system produced a Tm range from approximately 81.5 to 81.9 °C for D. pseudotakahashii. The same system also determines the cycle number at detection threshold (Cp) which can be used for relative quantification; if we assume that the efficiency of the reaction is perfect (i.e., R = 2-ΔΔCp), and identical gDNA template concentration, the difference in Cp values between samples can give an estimate of the relative concentration of Wolbachia DNA when computed as 2(Cp1 –Cp2), assuming the housekeeping gene has similar copy numbers of DNA in both samples. We do not consider Cp value as a tool for definitive quantification, but it provides a general guide such that lower Cp values correlate with stronger Wolbachia infection. To verify that high Cp values indicated true Wolbachia infection, we Sanger sequenced 14 samples with Cp values that ranged from 24.99 to 34.14 with the wsp validation primers as outlined previously. For comparative purposes, we also measured Cp values of other strains of Wolbachia in males and females of D. pandora (wRi-like infection, pl+ line), D. melanogaster (wMel infection, HAW+ line) and D. simulans (wRi, Burnley line and wAu, Coffs line) (for strain descriptions see Kriesner et al. 2013, 2016; Richardson et al. 2016). Note that these are strain rather than population samples.
To create control primers to verify DNA extraction, Drosophila universal primers were designed, targeting the ribosomal protein gene L40 (RpL40). RpL40 coding sequences of 12 Drosophila species were retrieved from Flybase (flybase.org) and aligned using Sequencher 5.4 (Gene Codes Corporation). Based on the nucleotide alignment of these 12 RpL40 orthologues, primers (Dros_RpL40_F: 5′-AACTGCCGCAAGAAGAAGTG-3′; Dros_RpL40_R: 5′-CTACTTCAACTTCTTCTTGGG-3′) were placed at two conserved regions, amplifying the last 64 bases of the RpL40 gene (Figure S1).
To verify the relationship between Cp value and Wolbachia density, selected Drosophila samples were screened for Wolbachia using a QX100™ Droplet Digital™ PCR (ddPCR™) system (Bio-Rad Laboratories Pty., Ltd. Hercules, CA, USA) with a hydrolysis probe assay (PrimePCR™, Bio-Rad Laboratories, Inc. Cat. No. 10031261) to obtain absolute quantification of Wolbachia copies/µL of DNA. Primers and probe were designed from the Wolbachia 16S ribosomal RNA gene (GenBank accession no. AF093510) to amplify a 62 bp product (Rao et al. 2006). The assay was converted to a ddPCR™ assay by labelling the 5′ end of the probe with the fluorophore FAM (6-carboxyfluorescein) and a Black Hole Quencher (BHQ) at the 3′ end. Sequences used were forward primer, 5′-CCAGCAGCCGCGGTAAT-3′; reverse primer, 5′CGCCCTTTACGCCCAAT-3′; probe, 5′-CGGAGAGGGCTAGCGTTATTCGGAATT3′ and were synthesized by Bio-Rad Laboratories, Inc.
Genomic DNA (2 µL) (Chelex®-extracted and not diluted or digested) was added to a PCR master mix containing 12.5 µL 2x ddPCR™ Supermix for probes (no dUTP) (Bio-Rad Laboratories, Inc. Cat. No.186-3024), 1.25 µL PrimePCR™ assay (Wol16S) and 9.25 µL PCR-grade water to a total volume of 25 µL. 20 µL of the master mix were added to a DG8™ cassette (Bio-Rad Laboratories, Inc. Cat. No. 1864008) followed by 70 µL of Droplet Generation Oil for Probes (Bio-Rad Laboratories, Inc. Cat. No. 1863005) and droplets were generated in a QX100™ Droplet Generator (Bio-Rad Laboratories, Inc.) according to the manufacturer’s instructions. After generation, droplets were transferred to a 96-well PCR plate (Eppendorf, Hamburg, Germany) which was heat sealed with pierceable foil (Bio-Rad Laboratories, Inc. Cat. No. 1814040) and then subjected to PCR in a C1000 Touch™ thermal cycler (Bio-Rad Laboratories, Inc. Cat. No. 1851197). The PCR cycling conditions were: 95 °C for 10 min, 40 cycles of 94 °C for 30 s and 60 °C for 1 min, 98 °C for 10 min with a 12 °C hold on completion. Ramp rate throughout the PCR program was 2.0 °C/s. The data were analysed using Quantasoft™ Analysis Pro Software v 1.0.
Wolbachia tree-building
To assess similarity of the D. pseudotakahashii Wolbachia infection to other known Wolbachia strains in Drosophila species, we built a Bayesian species tree. We obtained sequence data from Drosophila-infecting Wolbachia (which are all in Wolbachia supergroup A) and representatives of the other Wolbachia supergroups. Taxa were selected that had sequences available for the five MLST markers (Baldo et al. 2006) but not wsp which is known to show recombination (Baldo et al. 2005). The majority of sequences were obtained from GenBank (http://www.ncbi.nlm.nih.gov/genbank), and several unpublished sequences for Folsomia candida were kindly provided by Laura Baldo from the University of Barcelona and John H. Werren from the University of Rochester. These are summarised in Table S1. Sequence alignment was performed with Geneious v6.1 http://geneious.com (Kearse et al. 2012) and alignments were trimmed to remove primers and ensure consistent coverage across taxa. Nucleotide evolution and codon position partitioning models were selected for each gene region in turn by Bayesian Information Criterion (BIC) value in PartitionFinder v1.1.1 (Lanfear et al. 2012).
We performed an initial round of tree-building for each locus individually in BEAST v2.4.3. 10 M generations were run with all free priors left to vary widely. These runs were used to choose more stringent priors for the proportion of invariant sites, gamma category shape, and other nucleotide evolution model parameters (see Table S2) by examining the trace in Tracer v1.6.0 (Rambaut et al. 2014) and setting bounds to capture roughly 0.95 CI.
These estimated priors were then used in a *BEAST run (Bouckaert et al. 2014; Heled and Drummond 2010) combining all gene regions. This approach estimates a species tree along with individual gene trees for each region included. Molecular clocks and gene trees were unlinked between markers, but linked across partitions within each marker. This allowed for the possibility of horizontal gene transfer or gene conversion and avoided problems that were caused by topology differences in individual genes. Strict molecular clocks were used with gamma priors (shape = 2, scale = 2); ploidy was set to ‘Y or mitochondrial’ for each marker; the species tree population size was set as linear, the population mean size was estimated, and the species tree was modelled with a Yule model with birth rate distribution 1/X. All SubtreeSlide operator sizes were set to 0.02.
Five independent runs were initiated and run for 50 M generations each, sampling every 5000 generations. After confirming all effective sample size (ESS) values were >200 for the combined runs (checked in Tracer), and that independent runs converged on species tree topology (checked in Densitree v2.0), we combined the five runs with LogCombiner (Bouckaert et al. 2014), removing 20% of each run as burn-in. We then calculated the maximum clade credible tree in Treeannotator v2.4.3 (Bouckaert et al. 2014), using median branch lengths. This tree was plotted with the ape package v3.5 (Paradis et al. 2004) in R v3.3.1.
We also generated bootstrapped (1000 replications) maximum-likelihood trees using MEGA version 5 (Tamura et al. 2011) and default settings for individual genes as well as combined genes except for wsp because of reported problems with intra-genic recombination (Baldo and Werren 2007).
Phenotype characterization
The infection phenotypes were characterized in the ‘Moor+’, ‘Smith+’, ‘Town1+’ and ‘Town2+’ infected D. pseudotakahashii lines by conducting a series of experiments investigating CI, fitness and maternal transmission of Wolbachia. Experiments were conducted at 19 °C with a 12:12 L:D cycle.
CI and fecundity
Incompatibility and fecundity associated with the Moor+ line were assessed in an experiment in July 2014 in which the Moor+ and Moor− lines were reciprocally crossed in addition to control crosses within lines (Cross 1. ♀- × ♂ + , 2. ♀+ × ♂-, 3. ♀+ × ♂ + , 4. ♀- × ♂-) to control for nuclear effects. Virgins were collected within 6 h of emergence and holding vials were checked for absence of progeny after 10 days to confirm virgin status. Fifteen replicates of each cross were set up when virgins were five days old; mating was observed and males were removed and stored in ethanol. Females were provided with spoons containing cornmeal media and a brush of yeast paste to encourage egg laying. Spoons were scored for egg number and replaced every 24 h for the following nine days. A duration of nine days was chosen because initial egg production was delayed, and nine days was the point at which most females had laid sufficient eggs, but had not yet stopped producing viable eggs (males had been removed immediately after mating). Spoons were scored for fecundity immediately upon collection. Those collected from days 6 to 9 (when egg laying was more consistent) were assessed for CI after a further 24 h by scoring hatched and unhatched eggs and then transferring the spoons to 6 ml of cornmeal media to provide food for growing larvae. The egg laying rate for this species is low relative to species like D. melanogaster, although not necessarily other endemic species. We therefore used spoons where eight or more eggs over three days had been laid to assess CI. Progeny took approximately 18 days to develop, emerging adults were stored in 100% ethanol and sexed.
CI was also tested by crossing the infected Town1+ and Town2+ lines with a tetracycline cured line (Town3-) with the same crosses as outlined above. Prior screening indicated variable infection levels in males from these lines (see below), so all males involved in the crosses were screened. Ten to 16 replicates were set up for all crosses when males and females were 4–7 days post eclosion. Eggs were collected and scored in 24-h increments until a minimum of ten eggs had been collected or until 13 days after mating. Because of particularly low egg numbers produced in this experiment, we included all replicates with five or more eggs in analyses. Where pairs did not mate, or laid fewer than five eggs, they were excluded.
The effect of male age on CI was assessed following previous protocols for D. simulans (Hoffmann et al. 1994; Hoffmann et al. 1986); we crossed Wolbachia infected virgin males of various ages from the Smith+ line with uninfected females from the Moor− line (×15 replicates). Males were 3, 5, 8, 11 and 14 days old and females were 5 days old. Wolbachia-infected males and females of the same ages (×3 replicates) were crossed as a control. Mating, egg collection, egg scoring and progeny collection were carried out as described previously (Hoffmann et al. 1994). Replicates that did not mate, or that laid fewer than five eggs, were excluded from further analyses.
Incompatibility relationships between lines
Given the different rates of detection of Wolbachia in males from the Smith+ line compared to those from the Moor+ line (see below), compatibility between these lines was explored in an experiment aimed at separating potential Wolbachia strain effects from nuclear host effects where the Wolbachia background came from one line and the nuclear background was 75% from one line and 25% from the other line (i.e., four combinations of background were produced, as given in Table 4). Initially, reciprocal crosses between the two lines were set up at 19 °C with 31–32 replicates per cross type (Cross1 (C1) = ♀Moor+ × ♂Smith+, Cross 2 (C2) = ♀Smith 2+ × ♂Moor+). Crosses consisted of pairwise matings on cornmeal media which were maintained for ten days before males and females were placed into ethanol for later PCR screening. Male progeny from these reciprocal crosses collected as virgins were backcrossed with females from the two parental lines (Backcross: 1 (B1) = ♀Moor+ × ♂C1; 2 (B2) - ♀Moor+ × ♂C2; 3 (B3) - ♀Smith+ × ♂C1; 4 (B4) = ♀Smith+ × ♂C2) to produce males with the four nuclear/Wolbachia background combinations. Where possible, 2–3 replicate backcrosses were set up per reciprocal cross line, however emergence times and progeny numbers meant this was not possible for all lines. Males and females used in backcrosses were retained in ethanol for screening Wolbachia later. To assess the extent to which CI was retained in males after backcrossing had been completed, twenty male progeny per backcross were collected. Where possible, two males were collected for a representative ten lines per backcross, however where this was not possible, single males were collected from as many lines as possible. To score CI, males were crossed 3 days after eclosion with virgin females from the uninfected Moor− line. Eggs were collected, and hatch rates scored for 7 days as above. Males were screened for Wolbachia using the wsp validation primers and LightCycler® assay outlined above.
Table 4.
Average % hatch for CI crosses involving males with different combinations of Smith and Moor genetic backgrounds and different Wolbachia backgrounds from the maternal line
| Wolbachia from maternal line | Male genetic background | N | Average % hatch | Range % hatch | Mean wsp Cp ± SD of infected males | % Males uninfected |
|---|---|---|---|---|---|---|
| Moor | 75%Moor, 25%Smith | 17 | 0 | 0 | 30.70 ± 2.00 | 0 |
| Smith | 75%Moor, 25%Smith | 18 | 0.8 | 0–9.1 | 29.12 ± 1.83 | 5.6 |
| Moor | 25%Moor, 75%Smith | 19 | 1.4 | 0–22.2 | 30.30 ± 1.90 | 0 |
| Smith | 25%Moor, 75%Smith | 18 | 0.7 | 0–5.7 | 31.98 ± 2.18 | 16.7 |
wsp Cp values and infection levels relate to males involved in the CI crosses
N is the number of replicates after excluding those with fewer than eight eggs
Maternal transmission to adults
Wolbachia transmission in the Moor+ and Smith+ lines to adults of the next generation was assessed following a three-step protocol: (1) eggs were collected from an individual female from each line; (2) 19 and 28 sub-lines (Smith+ and Moor+ lines respectively) were created by crossing the resulting female progeny (all of which were infected) with uninfected males from the Moor− line; and (3) 2–3 male progeny from each sub-line were crossed with an equivalent number of uninfected females from the Moor− line. Individuals for steps 2 and 3 of this experiment were collected as virgins and crossed at age 6 days ♂/ 7–8 days ♀ for step 2 and 4-5 days ♂/ 6 days ♀ for step 3. Eggs were collected on spoons over 24 h and scored for hatched and unhatched eggs after a further 24 h. Progeny were collected into 100% ethanol and sexed. For analysis, we only used crosses producing 8 or more eggs. Males involved in the step 3 crosses and their mothers were screened for Wolbachia using the wsp-validation primers and LightCycler® assay outlined previously.
Statistical analysis
To compare infection frequencies among samples, we ran contingency analysis and tested significance with the likelihood ratio (G) statistic unless an Exact test could be used. Exact tests were also used to compare the incidence of infected and uninfected males that produced hatch rates of 0 or >0 in crosses. For the quantitative measures (hatch rate, fecundity) we used each pairwise cross as a replicate rather than individual eggs or offspring. While this meant that we sometimes computed hatch rates based on low replicate numbers, it was considered an appropriately conservative approach particularly given variability in infection status among the males. Data often tended to deviate from normality and we therefore compared hatch rate and fecundity across crosses with Kruskal-Wallis (non-parametric) tests. Analyses were run in IBM® SPSS® Statistics version 25.0.
Results
Wolbachia infection frequency
Females from all lines (N = 178) of D. pseudotakahashii across the regions sampled were infected with Wolbachia and most were of a similar infection density to other Drosophila species (Table 1). In contrast, many D. pseudotakahashii males appeared to be either infected at a lower level (indicated by high wsp Cp values), or uninfected altogether, despite the control gene RpL40 amplifying strongly and consistently in these samples across the sexes in all lines (indicated by low average Cp values and narrow Cp ranges). For the Townsville lines for example, the mean Cp value for RpL40 was 25.36 ± 0.52 in females and 26.16 ± 0.59 in males. At the F1/F2 stage, a sex difference in Wolbachia infection frequency was particularly evident for the lines from northern Queensland where males had a much lower infection frequency compared to the lines from south-eastern Queensland and New South Wales populations (contingency test, G = 34.43, df = 2, P < 0.001). When Wolbachia was detected in the males, the relative strength of infection (i.e., wsp Cp value) was similar between populations and much lower in males than females (Table 1). Assuming perfect amplification efficiency, the difference in wsp Cp values between males and females provides an estimate of the relative density (R = 2(Cp1 –Cp2), see 'methods'). Based on an average wsp Cp of 33.237 for males and 25.097 for females, males were estimated to have on average 0.3% of the concentration of Wolbachia of females.
Table 1.
Percentage of individuals tested that were infected with Wolbachia and mean wsp Cp value ± SD for F1/F2 males and females of D. pseudotakahashii compared to other Drosophila species
| Species/infection | N | % Wolbachia infected | Mean wsp Cp value ± SD | |||
|---|---|---|---|---|---|---|
| Male | Female | Male | Female | Malea | Female | |
| D. pseudotakahashii | ||||||
| wPse | ||||||
| Northern Queensland | 62 | 26 | 35.5 | 100 | 35.25 ± 2.70 | 27.44 ± 2.70 |
| South eastern Queensland | 122 | 128 | 91.8 | 100 | 31.69 ± 3.13 | 23.41 ± 1.60 |
| New South Wales | 83 | 24 | 85.5 | 100 | 32.58 ± 3.19 | 24.44 ± 2.47 |
| D. melanogaster | ||||||
| wMel (HAW lline) | 4 | 4 | 100 | 100 | 24.63 ± 0.42 | 23.88 ± 0.09 |
| D. pandora | ||||||
| wRi | 4 | 4 | 100 | 100 | 23.81 ± 0.56 | 24.18 ± 0.18 |
| D. simulans | ||||||
| wAu (Coffs Harbour line) | 4 | 4 | 100 | 100 | 22.51 ± 0.49 | 23.29 ± 1.53 |
| wRi (Burnley line) | 4 | 4 | 100 | 100 | 21.63 ± 0.15 | 21.55 ± 0.52 |
A single female for each line was tested, while multiple males were used in some cases. Lower wsp Cp values correlate with higher Wolbachia infection density
N is the number of samples tested
aOnly infected males were used for determining Cp values using the wsp validation primers
In this experiment, high Cp values obtained using primers that amplify wsp (Wolbachia surface protein gene) are associated with a low number of copies/µL of Wolbachia amplified with primers that target 16S rRNA. For the nine samples screened using ddPCR™, Cp values ranged from 22 to 40 and the number of copies in the original DNA extraction ranged from 9.5 to 78,033.6 per microlitre with 95% confidence intervals from the Poisson distribution spanning 4.3 to 81,764.8 across all samples. The relationship between Cp value and copies/µL closely fits an exponential function (y = 1E + 10e−0.539x) (R2 = 0.985). Samples which had no Cp value assigned showed either no copies or a very low number per microlitre (2.9–6.2 with 95% confidence intervals spanning 0.7 to 13.5). Therefore, the use of Cp value as an indication of Wolbachia density is justified.
We found that infections were more frequently detected in males after lines had been held for a time in the laboratory and wsp Cp values were also lower (reflecting a higher density) (Table 2). For example, Wolbachia was detected in the Town2+ line from Townsville at a frequency of 75% after eight generations in the laboratory (Table 2), but was not detectable in the four males tested for this line at the F1 stage (comparison of frequency in F1 versus lab population, contingency (Exact) test, P = 0.061). Similarly, males collected from Nowra were initially detected as being 50% (N = 20) infected, but this increased to 86% (N = 70) in the F1 generation. This difference is significant by a contingency test (G = 10.21, df = 1, P = 0.001). Females were always detected as being 100% infected in field and laboratory samples. Regardless of time in the laboratory, the Wolbachia infection strength in male D. pseudotakahashii remained much lower than that of females. For example, after 29 generations in the lab, the mean wsp Cp value of 70 males tested from the three lines from Smithfield was 30.13 (SD = 3.31), compared to a mean wsp Cp value of 22.00 (SD = 1.54) for 97 females tested from the same lines (males again estimated to have a Wolbachia concentration of 0.3% that of females assuming perfect amplification efficiency).
Table 2.
Percent Wolbachia infection and mean wsp Cp value ± SD for F1 and F8 males from Townsville lines of D. pseudotakahashii
| F1 | F8 | |||||
|---|---|---|---|---|---|---|
| Line | N | % Infected | Mean wsp Cp ± SD | N | % Infected | Mean wsp Cp ± SD |
| Town 1+ | 8 | 0 | 8 | 100 | 29.83 ± 3.62 | |
| Town 2+ | 8 | 0 | 8 | 75 | 30.86 ± 2.68 | |
| Town 3+ | 8 | 0 | ||||
| PH34 | 8 | 0 | ||||
| PH29 | 8 | 25 | 36.39 ± 0.72 | |||
| PH41 | 8 | 25 | 34.28 ± 0.01 | |||
| PH36 | 8 | 37.5 | 37.92 ± 0.57 | |||
| KH50 | 8 | 50 | 32.29 ± 0.41 | 9 | 100 | 31.08 ± 4.01 |
| PH26 | 8 | 62.5 | 36.26 ± 1.68 | |||
| PH27 | 8 | 75 | 32.43 ± 2.63 | |||
| PH28 | 8 | 100 | 36.62 ± 1.98 | 8 | 100 | 27.33 ± 1.41 |
N is number of males tested
To characterise the Wolbachia infection further, nucleotide sequences from multiple locations were obtained for the five MLST loci and wsp (Genbank accession numbers MF348256-MF348261). The Wolbachia sequences were identical for all source locations and the wsp sequence was the same as that identified as wPse by Mateos et al. (2006). For this reason, only one D. pseudotakahashii Wolbachia representative was included in the species tree (see below). Wolbachia infection was also verified for samples with Cp values ranging from 24.99 to 34.14 using the wsp_validation primers, by sequencing the PCR products.
Wolbachia relationships
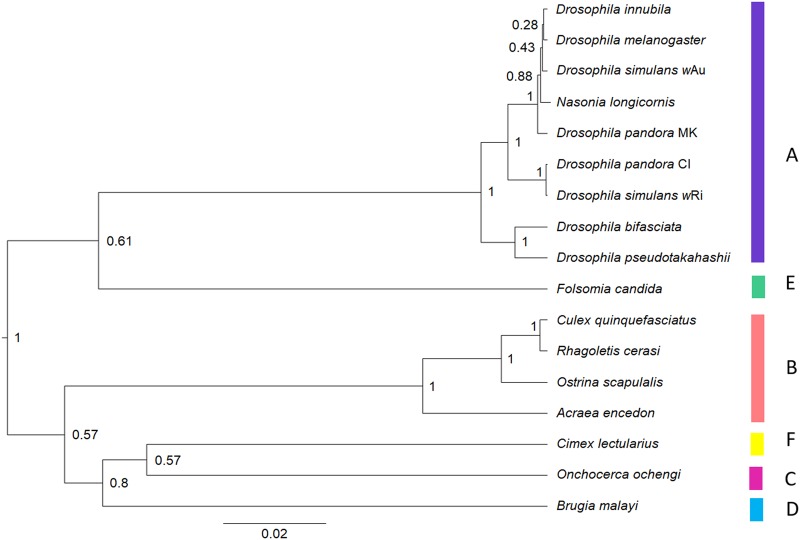
In the Bayesian species tree, the Wolbachia isolate from D. pseudotakahashii fell into Supergroup A (Fig. 1) and D. bifasciata was indicated as being the nearest relative. A similar result was obtained with the maximum likelihood analysis (Appendix 2) which led to the same arrangement across the supergroups. Proximity of D. pseudotakahashii Wolbachia to D. bifasciata was supported by Bayesian analysis of several individual genes including coxA, fbpA, and hcpA as well as wsp (Appendix 2), but this relationship would need to be clarified with additional genes and based ideally on the entire Wolbachia genome sequence. The relationships between strains evident for the individual genes in the Bayesian analyses were similar to those obtained in the maximum likelihood analyses (Appendix 2).
Fig. 1.
Maximum clade credibility tree generated from BEAST v2.4.3 (Bouckaert et al. 2014) on concatenated gene set excluding wsp. Gamma time-reversible nucleotide evolution model was used with 4 gamma categories. All other priors were left as per the default settings. Ten million generations were run, sampling every 1000 generations. The maximum clade credible tree was then calculated in TreeAnnotator v2.4.3 (Bouckaert et al. 2014) after removing the first 20% of the run as burn-in, producing median node heights. The posterior probability value is shown at each node. The known supergroups are labelled (coloured bars and upper-case letters) (colour figure online)
Phenotypic characterization
Cytoplasmic incompatibility
CI of the Moor lines of D. pseudotakahashii was assessed by crossing within and between infected and uninfected lines. Crosses differed significantly in egg hatch rate (Kruskal-Wallis test, X2 = 21.11, df = 3, P < 0.001): only one cross between infected Moor+ males and uninfected Moor− females produced any hatched eggs (11% hatching) in contrast to the expected compatible crosses, indicating high levels of CI but some heterogeneity in hatch rates (Table 3). The successful cross between infected Moor+ males and uninfected Moor− females produced one male and one female offspring. Cross results also indicated high levels of CI for the Town+ lines, despite weaker Wolbachia infections than in the Moor+ strain from NSW (Table 3), and differences between crosses were again significant (Kruskal-Wallis test, X2 = 39.51, df = 3, P < 0.001)
Table 3.
Results of crosses between and within a) Moor+ and Moor− lines and b) Town+ and Town− lines
| Cross | N | Mean % hatch | Range % hatch | Range of egg # scored | % Male Progeny ± SD | Male parent wsp Cp ± SD |
|---|---|---|---|---|---|---|
| (a) Moor line crosses | ||||||
| ♀- × ♂+ | 8 | 3.5 | 0.0–11.1 | 8–19 | 50 (N = 1) | 28.97 ± 2.79 |
| ♀+ × ♂- | 11 | 87.2 | 70.0–100 | 8–32 | 49.0 ± 21.0 | |
| ♀+ × ♂+ | 10 | 95.9 | 70.0–100 | 8–28 | 42.6 ± 17.4 | 28.36 ± 1.84 |
| ♀- × ♂- | 12 | 65.2 | 43.7–100 | 9–25 | 64.6 ± 21.8 | |
| (b) Town line crosses | ||||||
| ♀- × ♂+ | 19 | 8.6 | 0.0–36.6 | 5–42 | 41 (N = 1) | 32.95 ± 2.91 |
| ♀+ × ♂- | 20 | 91.1 | 50.0–100 | 5–44 | 44.9 ± 15.3 | |
| ♀+ × ♂+ | 19 | 86.5 | 9.7–100 | 5–35 | 43.7 ± 18.6 | 34.28 ± 3.11 |
| ♀- × ♂- | 5 | 98.1 | 95.4–100 | 22–31 | 43.0 ± 28.3 | |
N is the number of replicates after excluding those that did not mate
We also crossed Moor+ and Smith+ lines reciprocally and found no obvious CI between these strains (data not shown, hatch rates >80%). To investigate potential differences between these lines in the ability of males to produce CI associated with nuclear or Wolbachia effects that might lead to differences in Cp values and male infection incidence, we created males with different combinations of Moor+ and Smith+ Wolbachia and nuclear backgrounds (Table 4) and tested males for CI. The male progeny continued to show strong CI with uninfected females from the Moor− line regardless of their background (Table 4) and the difference between hatch rates across the four crosses was not significant (Kruskal-Wallis, X2 = 2.9895, df = 3, P = 0.394). Note that four males were negative for the Wolbachia infection (Table 4), and many of the males were not strongly infected (high wsp Cp values). For the four crosses with apparently uninfected males, the hatch rate was zero even though the males were known to have mated. For the same two crosses where males did have detectable Wolbachia (N = 32), we had hatch rates of 0 in all but five of these. This difference in incidence between 0 hatch rates and some eggs hatching is not significantly different between males with and without detectable Wolbachia by an Exact test (P = 0.645). This indicates that CI is expressed in male offspring from infected females regardless of whether the adult males have or lack the infection at the time of crossing.
CI and male age
The effect of male age on CI was tested by crossing Smith+ infected males of 3, 5, 8, 11 and 14 days old with Moor− uninfected females and scoring egg hatch rate. Egg hatch rate increased significantly as males aged indicating that CI was decreasing with time (Kruskal-Wallis test, X2 = 14.99, df = 4, P = 0.005); while no eggs hatched for 3-day-old males, the average hatch rate was 31% when males were 14 days old (Table 5). The wsp Cp value did not differ significantly between the male age groups (Kruskal-Wallis test, X2 = 6.61, df = 4, P = 0.16)
Table 5.
Results of crosses between Smith+ males and Moor− females showing effect of male age on average % egg hatch
| Male age (days) | N | Average % hatch | Range % hatch | Male parent wsp Cp ± SD |
|---|---|---|---|---|
| 3 | 8 | 0 | 0 | 31.80 ± 1.97 |
| 5 | 8 | 14 | 0–50 | 31.93 ± 3.03 |
| 8 | 12 | 10.7 | 0–42.9 | 30.24 ± 4.45 |
| 11 | 6 | 15.1 | 0–50 | 31.39 ± 2.12 |
| 14 | 7 | 30.7 | 4.8–64.7 | 29.47 ± 2.15 |
Mean male parent wsp Cp values are also reported
N is the number of replicates after excluding those which did not mate, or which had fewer than five eggs. Male wsp Cp values were based on all males crossed, regardless of hatch rates or mating
Fecundity
Fecundity assessed for crosses between the Moor+ and Moor− lines was similar for the control and reciprocal crosses with a mean of around 25 eggs over the 9 days although the incompatible cross had a relatively low fecundity (Table 6). Egg production did not differ significantly across the crosses (Kruskal-Wallis test, X2 = 7.289, df = 3, P = 0.063). While there was no large cost for infected females, the low and variable egg production of D. pseudotakahashii mean that there is not a lot of power to detect effects on fecundity. For instance, with a mean of 24 eggs and standard deviation of 10, it is only possible to detect a difference of 11 eggs between two groups with 80% power given a sample size of 13.
Table 6.
Fecundity of D. pseudotakahashii crosses between ‘Moor+’ and ‘Moor−’ lines scored over nine days
| Cross | N | Mean Fecundity ± SD |
|---|---|---|
| ♀- × ♂+ | 11 | 15.64 ± 12.19 |
| ♀+ × ♂- | 15 | 25.87 ± 10.20 |
| ♀+ × ♂+ | 10 | 24.50 ± 8.11 |
| ♀- × ♂- | 15 | 24.93 ± 11.84 |
N is the number of replicates after excluding those which did not mate
Maternal transmission
We assessed maternal transmission of Wolbachia in D. pseudotakahashii from mothers to adult daughters by testing the infection status of daughters in the 19 Smith+ and 28 Moor+ lines set up in this experiment. All 42 of the 47 daughters able to be tested were infected with Wolbachia (mean wsp Cp value = 23.58 ± 0.56), suggesting 100% maternal transmission (binomial confidence intervals, 92.4, 100%).
We assessed maternal transmission of Wolbachia to adult sons phenotypically by generating 1–3 sublines from an individual female, crossing their male progeny with uninfected females and assessing the hatch rates of their eggs. We also assessed maternal transmission at a molecular level by screening adult males from crosses using the wsp_validation primers. Uninfected males and high hatch rates were expected where adult transmission was imperfect, and infected males and low hatch rates were expected in most crosses when maternal transmission was high, consistent with the high CI that occurs when an infected male mates with an uninfected female.
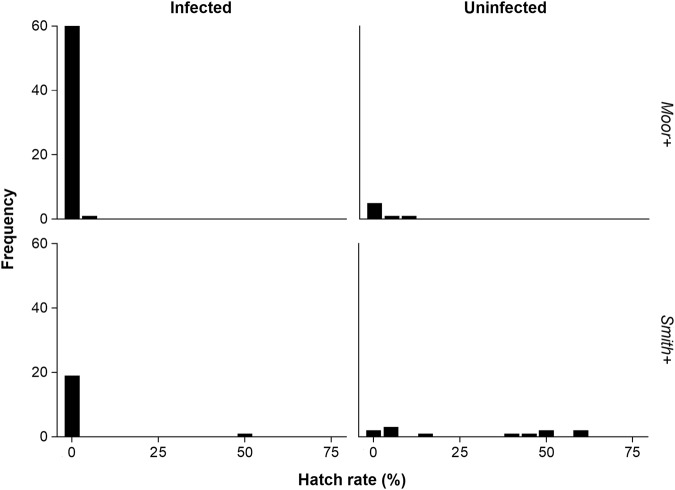
Phenotypic assessment suggested that maternal transmission was high and near 100% for the Moor+ and Smith+ lines. Of 70 crosses involving the Moor+ line (26 sub-lines × 2–3 replicates), only five replicates had any hatched eggs (3.7,10% hatch), to produce an overall hatch rate of 0.25% (95% confidence interval 0.05, 0.73%). Of 37 crosses involving the Smith+ line (17 sub-lines × 1–3 replicates), 12 replicates had hatched eggs (4.5, 63.6% hatch) with an overall hatch rate of 8.3% (95% confidence intervals, 6.0, 10.4%). These results are summarized in Fig. 2 which highlights the marked difference in distribution of hatch rates in crosses involving the two strains.
Fig. 2.
Maternal transmission of Wolbachia effects. Histograms showing number of males from two strains (Moor+, Smith+) producing different hatch rates when crossed with uninfected females. All males were taken from lines where females were infected, however some adult males screened as uninfected and these are presented separately
Molecular screening suggested a more moderate maternal transmission of Wolbachia to adult males of 88% (95% confidence intervals 81.1, 95.6%) for the Moor+ males and 59% (confidence intervals 39.1, 75.5%) for the Smith+ males, despite 100% maternal infection. Most crosses with hatched eggs were associated with uninfected males (Fig. 2) suggesting that imperfect maternal transmission to adults was involved (we found only three infected males of the 16 involved in crosses in which any progeny emerged, and those were infected at a low level with wsp Cp values ranging from 33.45–40). Males with moderate to weak infections (suggested by Cp values) appeared to produce mixed results with complete CI observed for some individuals, but incomplete CI for others. Despite being uninfected, six males were associated with 0% hatch rates, consistent with our earlier observations. Four of these were from the Moor+ line, with three apparently uninfected males from this line that had hatch rates >0; this differed (Exact test, P = 0.006) from the 59 infected males producing 0 hatch rates compared to two infected males with hatched eggs. The equivalent numbers for Smith+ were two uninfected males producing no hatched eggs (versus 10 with hatched eggs) and comparable figures of 18 (2) for infected males (Exact test, P < 0.001). Note that control crosses with Moor− females run around this time (and throughout the experimental period) always led to vials with hatched eggs. The RpL40 housekeeping gene indicated that the extractions were sound, so we attribute these results to two possible causes: either Wolbachia infection is present but below the detection limit of our current methods; or CI is induced early in male development, prior to mating, and the Wolbachia infection is lost from adult males.
Discussion
The infection we describe here is the first example of a Wolbachia strain in Drosophila that has become partly sex-limited. We found that the infection appears to be at a high frequency in field females. However, when flies were tested after being cultured for one or two generations under laboratory conditions, the infection was often not detectable in adult males based on PCR and RT-PCR assays. Where it was detected, the densities of Wolbachia in adult males were low. We also showed that transmission appears near-perfect to female offspring, whereas transmission to adult male offspring is variable. These effects depend to some extent on the strain of Drosophila used in crosses. The results indicate that either transmission to male embryos is low or male embryo and/or immature development leads to a loss of Wolbachia.
The mechanism behind this difference in Wolbachia density and transmission between the sexes is unclear. Wolbachia density is thought to be controlled partly by the Wolbachia genome and partly by the host nuclear genome (Boyle et al. 1993; Mouton et al. 2003; Kondo et al. 2005), and density (as well as tissue tropism) is thought to, in turn, control some phenotypes affected by Wolbachia. These phenotypes include virus blocking (Teixeira et al. 2008; Martinez et al. 2014) and the magnitude of deleterious fitness effects (Hoffmann et al. 2015). For strains that cause CI, there is often an association between density of Wolbachia in a strain and the level of CI (Bourtzis et al. 1996). Consistent with this pattern, we found that CI was lower on average when males lacked detectable Wolbachia. However, in the case of D. pseudotakahashii and unlike in previous work, adult male offspring lacking detectable Wolbachia are still capable of inducing strong CI when mated with uninfected females. When exactly in their lifetime male hosts lose detectable infection levels needs further investigation but may provide insights into mechanisms underlying CI in this system.
Under assumptions of no fitness costs in females and 100% maternal transmission to daughters, with strong CI despite the loss of infection in males, we anticipate that this Wolbachia infection should spread to fixation in natural populations of D. pseudotakahashii. This hypothesis is consistent with the limited data we have collected so far, but more information is needed, particularly under different environmental conditions known to influence Wolbachia fitness effects on hosts including heat stress (Ross et al. 2017). The decrease in CI in older males and increased detection of infected males in established laboratory stocks also seen in other Drosophila-Wolbachia systems (Hoffmann et al. 1986) suggests that this infection and its phenotypic effects are likely influenced by a range of internal and external factors. Drosophila pseudotakahashii is a tropical/subtropical species which is attracted to fruit bait, but its ecology is not well understood (Bock 1976) and it will be critical to expand information about this species within its natural environment to understand Wolbachia strain dynamics.
The Wolbachia strain we have isolated falls within a clade that includes D. bifasciata, an infection that is associated with stable and non-reversible male killing (Longdon et al. 2012) whereas for the D. pseudotakahashii infection, there is no evidence of an effect on sex ratio. Related infections in this clade (and others in the takahashii group) have been detected with molecular probes (Mateos et al. 2006) but have yet to be investigated; these infections could provide an opportunity for using comparative genomics to investigate the molecular basis of Wolbachia-induced phenotypes. Such comparisons have recently shown that loci associated with CI (Bonneau et al. 2018; LePage et al. 2017) are likely to be under strong selection in wRi Wolbachia infections that have spread rapidly across species and resulted in divergent (CI and non-CI) phenotypes (Turelli et al. 2018).
The presence of CI in males with low Wolbachia load raises issues around the mechanisms responsible for CI. Recent data from Drosophila and mosquitoes point to CI being associated with a toxin-antitoxin mechanism encoded by prophage genes of Wolbachia (Beckmann et al. 2017; Bonneau et al. 2018; LePage et al. 2017) with two genes inducing CI and one of these in the maternal lineage rescuing CI (Shropshire et al. 2018). Supporting evidence includes the fact that expression of these genes changes with male age in a way that is correlated with changes in CI with male age (LePage et al. 2017). In D. pseudotakahashii data we also see a decrease in CI with male age, but Wolbachia are absent or at a very low density in adults. If prophage genes are involved in CI in D. pseudotakahashii, perhaps the prophage persist independently of Wolbachia in males, and/or there is maternal transmission of CI effectors. On the other hand, CI in this system could also be associated with a completely different mechanism given that multiple factors can influence CI in species (Ju et al. 2017).
Although the wPse infection is currently at a high frequency in populations, partial loss from males may affect the dynamics of CI in populations and even equilibrium frequencies of Wolbachia. If male fitness is compromised by Wolbachia, there may be an ongoing shift towards loss of the infection in male offspring and an associated reduction in CI. This would make it harder for such Wolbachia to invade new uninfected populations. Wolbachia-based selection normally acts on females rather than males (Frank 1997) and reproductive effects mediated through males might be incidental to other effects of Wolbachia and female transmission fidelity which is expected to be under strong selection (Turelli 1994). Nevertheless, infections such as wPse may be of interest from an applied perspective. Females maintain a high density of the infection and may block virus transmission after transfer to novel hosts. While any loss of CI due to low densities of the infection in males may decrease the rate at which the infection invades populations (Hoffmann et al. 1990), invasion is still likely given that substantial CI seems to persist even when the infection is at a low density. Moreover, male fitness costs may be low for wPse, increasing their competitiveness with uninfected males from natural populations. Of course, it is unclear whether the sex-specific densities will be maintained when wPse is transferred to other species, which will depend on whether the phenotype described here has evolved as a consequence of genomic changes in Wolbachia or in the host genome.
Data Archiving
Sequences have been deposited in GenBank; accession numbers MF348256 - MF348261. Experimental data are available via Figshare at 10.26188/5b5a630cce4ba
Electronic supplementary material
Acknowledgements
This research was supported by a grant from the National Institute of Health (1R01GM104325-01) and a Fellowship from the National Health and Medical Research Council. The authors thank Vanessa Tsui, Valerie Elizabeth, Xinyue Gu and Sally Richardson for assistance, and Vanessa White for running the maximum likelihood analysis. The authors also thank L. Baldo from the University of Barcelona and J. H. Werren from the University of Rochester for providing some of the sequences used in this publication, and John Beckmann as well as Michael Turelli for comments.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
The online version of this article (10.1038/s41437-018-0133-7) contains supplementary material, which is available to authorized users.
References
- Baldo L, Lo N, Werren J. Mosaic nature of the Wolbachia surface protein. J Bacteriol. 2005;15-16:5406. doi: 10.1128/JB.187.15.5406-5418.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldo L, Hotopp JCD, Jolley KA, Bordenstein SR, Biber SA, Choudhury RR, et al. Multilocus sequence typing system for the endosymbiont Wolbachia pipientis. Appl Environ Microbiol. 2006;72:7098–7110. doi: 10.1128/AEM.00731-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldo L, Werren JH. Revisiting Wolbachia supergroup typing based on WSP: Spurious lineages and discordance with MLST. Curr Microbiol. 2007;55:81. doi: 10.1007/s00284-007-0055-8. [DOI] [PubMed] [Google Scholar]
- Beckmann JF, Ronau JA, Hochstrasser M. A Wolbachia deubiquitylating enzyme induces cytoplasmic incompatibility. Nat Microbiol. 2017;2:17007. doi: 10.1038/nmicrobiol.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berticat C, Rousset F, Raymond M, Berthomieu A, Weill M. High Wolbachia density in insecticide-resistant moquitoes. Proc R Soc Lond B. 2002;269:1413–1416. doi: 10.1098/rspb.2002.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleidorn C, Gerth M. A critical re-evaluation of mulilocus sequence typing (MLST) efforts in Wolbachia. FEMS Microbiol Immunol. 2017;94:fix163.. doi: 10.1093/femsec/fix163. [DOI] [PubMed] [Google Scholar]
- Bock I. Drosophilidae of Australia. I. Drosophila (Insecta: Diptera) Aust J Zool Supp Ser. 1976;24:1–105. doi: 10.1071/AJZS040. [DOI] [Google Scholar]
- Bonneau M, Atyame C, Beji M, Justy F, Cohen-Gonsaud M, Sicard M, et al. Culex pipiens crossing type diversity is governed by an amplified and polymorphic operon of Wolbachia. Nat Commun. 2018;9:10. doi: 10.1038/s41467-017-02449-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu CH, Xie D, et al. BEAST 2: A software platform for Bayesian evolutionary analysis. PLoS Comput Biol. 2014;10:e1003537. doi: 10.1371/journal.pcbi.1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourtzis K, Nirgianaki A, Markakis G, Savakis C. Wolbachia infection and cytoplasmic incompatibility in Drosophila species. Genetics. 1996;144:1063–1073. doi: 10.1093/genetics/144.3.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle L, O’Neill SL, Robertson HM, Karr TL. Interspecific and intraspecific horizontal transfer of Wolbachia in Drosophila. Science. 1993;260:1796–1799. doi: 10.1126/science.8511587. [DOI] [PubMed] [Google Scholar]
- Braig HR, Guzman H, Tesh RB, O’Neill SL. Replacement of the natural Wolbachia symbiont of Drosophila simulans with a mosquito counterpart. Nature. 1994;367:453–455. doi: 10.1038/367453a0. [DOI] [PubMed] [Google Scholar]
- Brownlie JC, Cass BN, Riegler M, Witsenburg JJ, Iturbe-Ormaetxe I, McGraw EA, et al. Evidence for metabolic provisioning by a common invertebrate endosymbiont, Wolbachia pipientis, during periods of nutritional stress. Plos Pathog. 2009;5:e1000368. doi: 10.1371/journal.ppat.1000368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull JJ, Turelli M. Wolbachia versus dengue: evolutionary forecasts. Evol Med Public Health. 2013;2013:197–207. doi: 10.1093/emph/eot018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutton TJ, Sinkins SP. Strain-specific quantifications of Wolbachia density in Aedes albopictus and effect of larval rearing conditions. Insect Mol Biol. 2004;13:317–322. doi: 10.1111/j.0962-1075.2004.00490.x. [DOI] [PubMed] [Google Scholar]
- Frank SA. Cytoplasmic incompatibility and population structure. J Theor Biol. 1997;184:327–330. doi: 10.1006/jtbi.1996.0276. [DOI] [PubMed] [Google Scholar]
- Hamm CA, Begun DJ, Vo A, Smith CC, Saelao P, Shaver AO, et al. Wolbachia do not live by reproductive manipulation alone: infection polymorphism in Drosophila suzukii and D. subpulchrella. Mol Ecol. 2014;23:4871–4885. doi: 10.1111/mec.12901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heled J, Drummond AJ. Bayesian inference of species trees from multilocus data. Mol Biol Evol. 2010;27:570–580. doi: 10.1093/molbev/msp274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann AA, Turelli Unidirectional cytoplasmic incompatibility in Drosophila simulans- inheritance, geographic variation and fitness effects. Genetics. 1988;119:435–444. doi: 10.1093/genetics/119.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann AA, Turelli M (1997). Cytoplasmic incompatibility in insects. In: Influential Passenger: Inherited Microorganisms and Arthropod Reproduction, Oxford University Press, pp. 42–80.
- Hoffmann AA, Clancy D, Duncan J (1996) Naturally-occurring Wolbachia infection in Drosophila simulans that does not cause cytoplasmic incompatibility. Heredity 76:1-8 [DOI] [PubMed]
- Hoffmann AA, Clancy DJ, Merton E. Cytoplasmic incompatibility in Australian populations of Drosophila melanogaster. Genetics. 1994;136:993–999. doi: 10.1093/genetics/136.3.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann AA, Ross PA, Rasic G. Wolbachia strains for disease control: ecological and evolutionary considerations. Evol Appl. 2015;8:751–768. doi: 10.1111/eva.12286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann AA, Turelli M, Harshman LG. Factors affecting the distribution of cytoplasmic incompatibility in Drosophila simulans. Genetics. 1990;126:933–948. doi: 10.1093/genetics/126.4.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann AA, Turelli M, Simmons GM. Unidirectional incompatibility between populations of Drosophila simulans. Evolution. 1986;40:692–701. doi: 10.1111/j.1558-5646.1986.tb00531.x. [DOI] [PubMed] [Google Scholar]
- Hurst G, Jiggins FM. Male-killing bacteria in insects: mechanisms, incidence, and implications. Emerg Infect Dis. 2000;6:329. doi: 10.3201/eid0604.000402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst GDD, Jiggins FM, Pomiankowski A. Which way to manipulate host reproduction? Wolbachia that cause cytoplasmic incompatibility are easily invaded by sex ratio-distorting mutants. Am Nat. 2002;160:360–373. doi: 10.1086/341524. [DOI] [PubMed] [Google Scholar]
- Jolley KA, Maiden MCJ. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinforma. 2010;11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju JF, Hoffmann AA, Zhang YK, Duan XZ, Guo Y, Gong JT, et al. Wolbachia-induced loss of male fertility is likely related to branch chain amino acid biosynthesis and iLvE in Laodelphax striatellus. Insect Biochem Mol Biol. 2017;85:11–20. doi: 10.1016/j.ibmb.2017.04.002. [DOI] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern P, Cook JM, Kageyama D, Riegler M. Double trouble: combined action of meiotic drive and Wolbachia feminization in Eurema butterflies. Biol Lett. 2015;11:20150095. doi: 10.1098/rsbl.2015.0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo N, Shimada M, Fukatsu T. Infection density of Wolbachia endosymbiont affected by co-infection and host genotype. Biol Lett. 2005;1:488–491. doi: 10.1098/rsbl.2005.0340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriesner P, Conner WR, Weeks AR, Turelli M, Hoffmann AA. Persistence of a Wolbachia infection frequency cline in Drosophila melanogaster and the possible role of reproductive dormancy. Evolution. 2016;70:979–997. doi: 10.1111/evo.12923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriesner P, Hoffmann AA, Lee SF, Turelli M, Weeks AR. Rapid sequential spread of two Wolbachia variants in Drosophila simulans. PLoS Pathog. 2013;9:e1003607. doi: 10.1371/journal.ppat.1003607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfear R, Calcott B, Ho S, Guindon S. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 2012;29:1695–1701. doi: 10.1093/molbev/mss020. [DOI] [PubMed] [Google Scholar]
- Lee SF, White VL, Weeks AR, Hoffmann AA, Endersby NM. High-throughput PCR assays to monitor Wolbachia infection in the dengue mosquito (Aedes aegypti) and Drosophila simulans. Appl Environ Microbiol. 2012;78:4740–4743. doi: 10.1128/AEM.00069-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LePage DP, Metcalf JA, Bordenstein SR, On J, Perlmutter JI, Shropshire JD, et al. Prophage WO genes recapitulate and enhance Wolbachia-induced cytoplasmic incompatibility. Nature. 2017;543:243–247. doi: 10.1038/nature21391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longdon B, Fabian DK, Hurst GDD, Jiggins FM. Male-killing Wolbachia do not protect Drosophila bifasciata against viral infection. BMC Microbiol. 2012;12(Suppl 1):S8–S8. doi: 10.1186/1471-2180-12-S1-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J, Longdon B, Bauer S, Chan YS, Miller WJ, Bourtzis K, et al. Symbionts commonly provide broad spectrum resistance to viruses in insects: a comparative analysis of Wolbachia strains. PLoS Pathog. 2014;10:e1004369. doi: 10.1371/journal.ppat.1004369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateos M, Castrezana SJ, Nankivell BJ, Estes AM, Markow TA, Moran NA. Heritable endosymbionts of Drosophila. Genetics. 2006;174:363–376. doi: 10.1534/genetics.106.058818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouton L, Henri H, Bouletreau M, Vavre F. Strain-specific regulation of intracellular Wolbachia density in multiply infected insects. Mol Ecol. 2003;12:3459–3645. doi: 10.1046/j.1365-294X.2003.02015.x. [DOI] [PubMed] [Google Scholar]
- Noda H, Koizumi Y, Zhang Q, Deng K. Infection density of Wolbachia and incompatibility level in two planthopper species, Laodelphax striatellus and Sogatella furcifera. Insect Biochem Mol Biol. 2001;31:727–737. doi: 10.1016/S0965-1748(00)00180-6. [DOI] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- Prout T. Some evolutionary possibilities for a microbe that causes incompatibility in its host. Evolution. 1994;48:909–911. doi: 10.1111/j.1558-5646.1994.tb01371.x. [DOI] [PubMed] [Google Scholar]
- Rambaut A, Suchard M, Xie D, Drummond A. (2014). Tracer v1.6. http://beast.bio.ed.ac.uk/Tracer.
- Rao RU, Atkinson LJ, Ramzy RMR, Helmy H, Farid HA, Bockarie MJ, et al. A real-time PCR-based assay for detection of Wuchereria bancrofti DNA in blood and mosquitoes. Am Soc Trop Med Hyg. 2006;74:826–832. doi: 10.4269/ajtmh.2006.74.826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson KM, Schiffer M, Griffin PC, Lee SF, Hoffmann AA. Tropical Drosophila pandora carry Wolbachia infections causing cytoplasmic incompatibility or male killing. Evolution. 2016;70:1791–1802. doi: 10.1111/evo.12981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross PA, Wiwatanaratanabutr I, Axford JK, White VL, Endersby-Harshman NM, Hoffmann AA. Wolbachia infections in Aedes aegypti differ markedly in their response to cyclical heat stress. PLoS Pathog. 2017;13:e1006006. doi: 10.1371/journal.ppat.1006006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shropshire JD, On J, Layton EM, Zhou H, Bordenstein SR (2018). One prophage WO gene rescues cytoplasmic incompatibility in Drosophila melanogaster. Proc Natl Acad Sci USA. 10.1073/pnas.1800650115 [DOI] [PMC free article] [PubMed]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony method. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira L, Ferreira A, Ashburner M. The bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster. Plos Biol. 2008;6:2753–2763. doi: 10.1371/journal.pbio.1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortosa P, Charlat S, Labbé P, Dehecq J, Barré H, Weill M. Wolbachia age-sex-specific density in Aedes albopictus: a host evolutionary response to cytoplasmic incompatibility? PLoS ONE. 2010;5:e9700. doi: 10.1371/journal.pone.0009700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli M. Evolution of incompatibility‐inducing microbes and their hosts. Evolution. 1994;48:1500–1513. doi: 10.1111/j.1558-5646.1994.tb02192.x. [DOI] [PubMed] [Google Scholar]
- Turelli M, Cooper BS, Richardson KM, Ginsberg PS, Peckenpaugh B, Antelope CX, et al. Rapid global spread of wRi-like Wolbachia across multiple Drosophila. Curr Biol. 2018;28:963–971. doi: 10.1016/j.cub.2018.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequences have been deposited in GenBank; accession numbers MF348256 - MF348261. Experimental data are available via Figshare at 10.26188/5b5a630cce4ba